Abstract
Introduction
Chronic obstructive pulmonary disease (COPD) is the fourth leading cause of death in the United States, with high associated costs. Most of the cost burden results from acute exacerbations of COPD (AE-COPD), events associated with heightened symptoms and mortality. Cellular mechanisms underlying AE-COPD are poorly understood, likely because they arise from dysregulation of complex immune networks across multiple tissue compartments.
Methods
To gain systems-level insight into cellular environments relevant to exacerbation, we applied data-driven modeling approaches to measurements of immune factors (cytokines and flow cytometry) measured previously in two different human tissue environments (sputum and peripheral blood) during the stable and exacerbated state.
Results
Using partial least squares discriminant analysis, we identified a unique signature of cytokines in serum that differentiated stable and AE-COPD better than individual measurements. Furthermore, we found that models integrating data across tissue compartments (serum and sputum) trended towards being more accurate. The resulting paracrine signature defining AE-COPD events combined elevations of proteins associated with cell adhesion (sVCAM-1, sICAM-1) and increased levels of neutrophils and dendritic cells in blood with elevated chemoattractants (IP-10 and MCP-2) in sputum.
Conclusions
Our results supported a new hypothesis that AE-COPD is driven by immune cell trafficking into the lung, which requires expression of cell adhesion molecules and raised levels of innate immune cells in blood, with parallel upregulated expression of specific chemokines in pulmonary tissue. Overall, this work serves as a proof-of-concept for using data-driven modeling approaches to generate new insights into cellular processes involved in complex pulmonary diseases.
Electronic supplementary material
The online version of this article (10.1007/s12195-019-00567-2) contains supplementary material, which is available to authorized users.
Keywords: Systems biology, Inflammation, Immune system, Data-driven models, Pulmonary disease
Introduction
Chronic obstructive pulmonary disease (COPD) is a progressive and heterogeneous lung disease that is the fourth leading cause of death in the United States,31 with yearly U.S. medical costs expected to increase to nearly $50 billion in 2020.19 A large portion of these costs is attributed to acute exacerbations of COPD (AE-COPD), characterized by increased symptoms (dyspnea, coughing, sputum production, and fatigue) beyond day-to-day variation that requires treatment with antibiotics or corticosteroids.49 Severe exacerbations (that require hospitalization) have an in-hospital all-cause mortality rate of 5–7%,29,48 and account for most of the financial burden of COPD.53 Accordingly, the prediction and treatment of AE-COPD events are top priorities.
Nonetheless, pathogenic cellular mechanisms underpinning AE-COPD are largely undefined. Local tissue and systemic inflammatory pathways are hallmarks of COPD, and are further increased during AE-COPD. Most AE-COPD are also associated with evidence of viral or bacterial infections or both,7,46,51 with upregulation of IL-8, TNF-α and reactive oxygen species in cells and tissue environments.51 Some AE-COPD are also highly eosinophilic.7 COPD patients with persistent systemic inflammation have higher mortality and exacerbation rates compared to non-inflamed patients.3 AE-COPD frequency is reduced by several types of therapies, including inhaled corticosteroids (ICS), long-acting muscarinic antagonists, scheduled azithromycin, and roflumilast.4,25,37,55 The success of these treatments, which share immunomodulatory effects, support acutely increased inflammation as contributing to AE-COPD, though fundamental mechanisms driving AE-COPD remain elusive.
Despite identification of individual cell types and cytokines that are differentially expressed between stable and exacerbated COPD,9,52,54 no single factor entirely accounts for AE-COPD, and therapies based on single targets have been unsuccessful. In the past 25 years, only one new class of medicine has been accepted for COPD treatment.45 Plasma fibrinogen was recently qualified by the Food and Drug Administration as a prognostic biomarker, but only for subject enrichment in clinical trials of exacerbation and mortality.40 Both serum C-reactive protein (CRP)11,28 and IL-611,32 are upregulated in the secreted systemic environment during AE-COPD, but CRP alone is insufficiently sensitive as an AE-COPD biomarker,42 and IL-6 elevations are inconsistently associated with exacerbations.30 New approaches to understanding cellular mechanisms underpinning AE-COPD pathogenesis are clearly required.
As COPD is a complex condition exhibiting evidence of immunological involvement,13,14 it is plausible that AE-COPD events result from disrupted networks of immune cells and cytokine communication, rather than from individual mediators. Data-driven modeling approaches offer the opportunity to infer these systems-level relationships by identifying small signatures of proteins or other cellular immune factors that co-vary with each other and are associated with disease state. These signatures can then be linked to mechanisms or cell types involved in phenotypes or pathogenic states, providing insight into specific disease biology and potential targets for follow-up experiments and therapeutic intervention. Partial least squares discriminant analysis (PLSDA) is a useful tool for highlighting covariance among variables that best classify groups of interest, which could lead to the identification of potential proteomic and cellular networks associated with AE-COPD. We have previously illustrated that PLSDA is able to identify and aid in visualizing biologically relevant proteomic and cellular signatures that may give insight into inflammatory pathways. We have used it to evaluate inflammatory signatures in the female reproductive tract mucosa6 and the blood of interstitial pulmonary fibrosis (IPF) patients,44 in both cases identifying new biomarkers and generating novel insight into key cellular mechanisms.
In this study we apply data-driven modeling approaches to gain insight into the proteomic networks and cellular mechanisms in blood and lung environments that underpin AE-COPD using a prospective cohort study,22 which collected paired sputum and peripheral blood samples from COPD subjects when clinically stable and again before treatment for an AE-COPD. We show that data-driven modeling approaches are able to (1) identify cytokine networks that may be better for classifying AE-COPD than individual cytokines, (2) determine key relationships between cytokines in different tissue compartments, and (3) integrate information measured in different assays to provide a more complete picture of pathogenic processes involved in AE-COPD.
Methods
Study Design, Ethics and Subject Populations
All samples and data in this analysis derived from a published prospective observational trial (ClinicalTrials.gov NCT00281216),22 which followed subjects at increased risk of AE-COPD for up to three years. Patients were recruited at the VA Ann Arbor Healthcare System (VAAAHS) and the University of Michigan Health System (UMHS). All parts of the study adhered to the Declaration of Helsinki and obtained approval of each site’s Institutional Review Board, with all subjects giving written consent to the study before any procedures occurred. At enrollment and quarterly, participants underwent spirometry, pulmonologist clinical evaluations, collection of peripheral blood and spontaneously expectorated sputum, and a post-visit questionnaire. An exacerbation of COPD was said to occur if the subject reported an increase in dyspnea, cough or sputum production, and if the study physician ordered antibiotics or oral steroid for the patient after a physical examination and chest radiographs to rule out pneumonia. Only if a diagnosis of AE-COPD was made were sputum and peripheral blood samples collected at these unscheduled visits. After all data and sample collection occurred, then each subject began treatment for AE-COPD.
Sample Collection, Processing, and Measurements
Peripheral blood was used for both leukocyte immunophenotyping and to measure 40 analytes in serum; after collection, it was stored at − 80 °C until analysis. Spontaneously expectorated sputum was immediately processed in a 9:1 mixture of distilled water to Sputolysin® (EMD Millipore, Billercia, MA) as described,22 and the resulting supernatant was stored at − 80 °C until used to measure 36 analytes. Serum and sputum samples were unfrozen and protein concentrations were measured simultaneously either using a Luminex 200 System® (Luminex Corporation, Austin TX) or ELISA (GDF-15, IL-18, IL-23p19 and IFN-β).22
Whole blood was stained with directly conjugated monoclonal antibodies on the day of the visit as described in the text and supplemental information of Freeman et al.22 Cells were analyzed using a LSR II flow cytometer (BD Bioscience, San Jose, CA) as reported in McCubbrey et al.,39 using FACSDiva software (BD Biosciences) data with automatic compensation and FlowJo software (Tree Star, Ashland, OR).
Data Processing and Systems Analysis
Samples with multiple missing measurements were removed from analysis if missing values were recorded for more than 25% of the proteins that were measured in each assay (serum protein, sputum protein or blood cell marker); proteins were then removed if more than two measurements were missing for any one protein. We identified and illustrated individual proteins that were differentially expressed in stable and exacerbated states using a volcano plot. First, a non-parametric, two-sided, paired Wilcoxon signed rank test was used to determine significance in the non-normalized proteomic or cell marker expression during the stable and exacerbation states, with significance being defined as p < 0.05. Then, the relative fold change in protein or cell marker level was calculated by dividing the average concentration during exacerbation by the average concentration during stability. Each protein or cell marker was then plotted in one figure, with fold change on the x-axis and the p value on the y-axis. Minor differences between these results and the previously published univariate results (Freeman et al.) can be attributed to variation in which subset of patients were included in each analysis.22
PLSDA, which was performed using the Eigenvector PLS Toolbox in MATLAB, was used to identify signatures of multivariate cytokine and cellular markers that differentiated stable and AE-COPD.34 Taking a supervised approach, PLSDA assigns a loading to each variable and selects a linear combination of all variables (a latent variable) that best separates pre-defined groups. A higher value of a protein loading on a latent variable indicates the protein is of more importance in differentiating the groups of interest. Each sample is then scored based on its protein expression and are visualized in the scores plot. The loadings can then be used for hypothesis generation based on how the subsets of the protein signature are associated with each of the groups in the scores plot. Each PLSDA model was cross-validated as a measure of model accuracy. Cross-validation was performed by iteratively excluding ~ 10% of the data for all models based on serum proteins only, ~ 17% of the data from the serum and sputum protein PLSDA model, and ~ 20% from the serum and sputum protein and blood cell marker model, which in each case resulted in 3–4 samples being excluded. The excluded data was then used to test the trained model. Care was taken when designing the training and test sets to ensure that no test set had more than one measurement from a unique patient. All missing data points included in the PLSDA models were filled in by the Eigenvector software’s “best guess.” All models were orthogonalized to enable clear visualization of the results, and all data were mean centered and variance scaled before being used to create the model. Variable importance in projection (VIP) scores were used to reduce model dimensionality by determining the importance of each variable in differentiating the groups of interest.57 Proteins with a VIP score < 1 were removed from the model, and a new PLSDA model was built then on the remaining proteins or cellular factors.
In order to facilitate a more quantitative comparison across PLSDA analyses, we calculated the cross-validation accuracy associated with each training and test set that was created during cross-validation. We then statistically compared cross-validation accuracies across the models based on different folds by using a one-way ANOVA with Tukey’s post hoc test. A p value of less than 0.05 was considered significant after application of Tukey’s test.
We visualized the distinct proteomes associated with stable and AE-COPD events through unsupervised average linkage hierarchical clustering; Spearman’s correlation coefficient was used as the distance metric. Correlation heat maps were constructed based on the Spearman rank correlation calculated between the difference in cell marker and protein concentration from the stable to the exacerbated state, where correlation coefficients that had a p value of greater than 0.05 were set to be zero for the figure. When creating hierarchical clusters or correlation heat maps, all missing data points were imputed using the MATLAB function knnimpute, with the pairwise distances between patients calculated based on the Spearman rank correlation.
All PLSDA models, VIP scores, Wilcoxon signed rank tests, hierarchical clusters, heat maps, and Spearman correlation testing were created or calculated using MATLAB (MATLAB, Natick, MA); PLSDA models and VIP scores were specifically generated using the PLS toolbox in MATLAB (Eigenvector, Manson, WA). ANOVA and Tukey’s tests were performed using Prism version 7.00 (GraphPad Software, San Diego, CA).
Results
Patient Enrollment and Demographics
We analyzed data from 13 COPD subjects who completed both the baseline visit and at least one AE-COPD visit. They were a predominantly middle-aged (mean age 67.9 years), male (9 of 11) group with advanced COPD (mean FEV1 33.4% predicted) comprised of both current and former smokers. Specifics of their demographics, clinical characteristics and in which data-driven models their data were used is shown in Table 1. In summation, this study captured 18 total paired stable and AE-COPD events among the 13 subjects, with some subjects experiencing more than one AE-COPD during the course of the study.
Table 1.
Summary of demographic, smoking, spirometry and model inclusion information.
| Age (yrs) | Sex | FEV1 (% predicted) | FEV1/FVC | Pack-years | Smoking status | # AE-COPD during study | ICS use (Y/N) | Use in modelsa |
|---|---|---|---|---|---|---|---|---|
| 74 | Female | 51 | 0.5 | 50 | Former | 3 | Yes | Allb |
| 77 | Male | 28 | 0.5 | 50 | Former | 1 | Yes | All |
| 69 | Male | 14 | 0.34 | 98 | Former | 3 | Yes | Allc |
| 59 | Male | 47 | 0.63 | 18 | Former | 1 | Yes | All |
| 72 | Male | 36 | 0.55 | 39 | Former | 2 | Yes | All |
| 58 | Male | 26 | 0.44 | 25 | Former | 1 | Yes | Serum |
| 67 | Male | 52 | 0.61 | 108 | Current | 1 | Yes | All |
| 66 | Male | 29 | 0.43 | 40 | Current | 1 | No | All |
| 67 | Male | 20 | 0.46 | 84 | Current | 1 | Yes | Serum |
| 72 | Male | 31 | 0.25 | 120 | Current | 1 | Yes | Serum |
| 66 | Female | 33 | 0.35 | 104 | Current | 1 | Yes | Serum |
| 67.9d | 9/2 | 33.4 | 0.5 | 66.9 | 6/5 | 1.5 | 10/1 |
aExcept where indicated, shows if any paired stable and exacerbation measurement from that patient was used in a data-driven model. “All” indicates at least one stable or AE-COPD measurement from that patient was used in all three data-driven models, and “Serum” means at least one paired stable and AE-COPD measurement from that patient was used only in the serum model
bOnly an exacerbation measurement was used from this patient in the data-driven model based on serum, sputum and flow data
cOnly a stable measurement was used from this patient in the data-driven model based on serum, sputum and flow data
dData are presented as averages, except in the cases of gender (Male/Female), Smoking status (Former/Current) and ICS use (Yes/No)
Evaluation of Individual Immune Factors Associated with AE-COPD
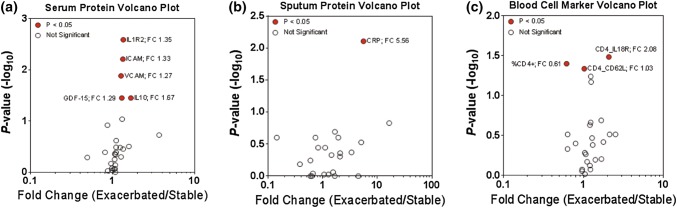
We first identified individual cellular immune factors and receptors that differed significantly between stable and AE-COPD, similar to our previously published work.22 Out of 35 serum proteins (see Materials and Methods), five were found to be significantly different (p < 0.05): interleukin 1 receptor 2 (IL-1R2; fold change 1.35), soluble intercellular adhesive molecule 1 (sICAM-1; fold change 1.33), soluble vascular cellular adhesion molecule 1 (sVCAM-1; fold change 1.27), growth differentiation factor (GDF-15; fold change 1.29) and interleukin 10 (IL-10; fold change 1.66) (Fig. 1a). From 30 proteins measured in sputum, only CRP was significantly different between stable and AE-COPD (fold change 5.56) (Fig. 1b). Three of 26 cellular markers measured by flow cytometry were differentially expressed: percent of CD4+ cells (%CD4+; fold change 0.61), CD4+ CD62L cells (CD4_CD62L, fold change 1.03), and CD4+ IL-18R cells (CD4_IL18; fold change 2.08) (Fig. 1c). The expression of both CD62L and IL-18R indicate activation of CD4+ T cells. While the significance levels indicated in the volcano plots are based on average concentration data, the grouped scatter plots in Supplemental Figures S1, S2 and S3 track individual changes across the two COPD states in specific patients. All immune factors were significantly elevated during exacerbation with the exception of %CD4+ cells. Overall, these results reflect observations made in the original study,22 in which only a small number of proteins and individual blood cell types and activation markers were significantly different between stable and exacerbation. None of the proteins or cell markers in the three volcano plots were found to be significant after application of the Bonferroni correction, and many of the fold changes measured were small (close to 1).
Figure 1.
Individual proteins and cell populations measured in stable and exacerbation states. (a) Volcano plot illustrates serum proteins that are both differentially expressed (x axis) and significantly different (y axis) between the stable and exacerbated state. Significance was determined using non-normalized data (Supplemental Figures S1, S2 and S3), and points in red indicate significantly different expression between the stable and exacerbated state via paired Wilcoxon signed rank test, with significance being defined as p < 0.05. (b) Volcano plot highlighting significantly different sputum proteins across the stable and exacerbated state. Significance was determined as described above (p < 0.05). (c) Volcano plot illustrating blood cell marker measurements that were significantly different between stable and AE-COPD. Significance was determined as described above (p < 0.05).
In our data there were three patients who experienced more than one exacerbation event. We explored the effects of this by additionally analyzing the data after averaging multiple stable and multiple exacerbation measurements within the same patient. Overall, we found that our results were similar, both in individual significant proteins identified and in fold change in the exacerbated state (Supplemental Figure S4).
PLSDA Identified a Signature of Serum Proteins that Differentiated Stable and Exacerbated COPD
To obtain new insight into key systems-level relationships between networks of immune factors in sputum and blood that associated with AE-COPD, we next employed data-driven modeling approaches to integrate matched stable and exacerbation data in both blood and pulmonary immune environments from the same COPD patients. We first examined serum protein measurements alone with PLSDA.34 PLSDA is a useful tool due to its ability to highlight covariance among variables that best classify groups of interest, which could lead to the identification of potential proteomic networks associated with AE-COPD. Calibration accuracy and k-fold cross-validation were used to assess model accuracy (see Materials and Methods). To focus on the cytokines that were best at differentiating stable and AE-COPD, we used variable importance in projection (VIP) scores57 as a feature selection technique. The value of using PLSDA with VIP feature selection is the identification of small protein “signatures” that differentiate groups of interest and are potentially biologically meaningful, which helps with generating new mechanistic hypotheses.
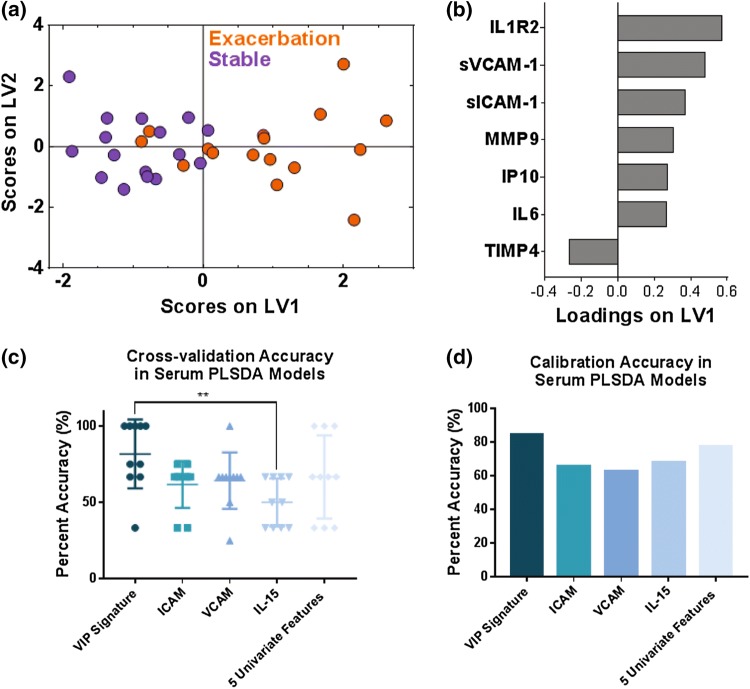
We found that a two-latent variable PLSDA model based on the serum VIP-selected protein signature best classified stable and exacerbation points with 81.25% cross-validation accuracy and an 84.38% calibration accuracy (Fig. 2a). Latent variable 1 (LV1) differentiated most stable visits (negative scores on LV1) from AE-COPD (positive scores on LV1; Fig. 2b). Six of the seven proteins were loaded positively on LV1, indicating positive association with AE-COPD, while only tissue inhibitor of metalloproteinases (TIMP-4) was loaded negatively on LV1, indicating negative association with AE-COPD. The six positively associated proteins were IL-1R2, sVCAM-1, sICAM-1, matrix metalloproteinase 9 (MMP-9), interferon gamma-induced protein 10 (IP-10, the chemokine also known as CXCL10), and IL-6.
Figure 2.
VIP scores and PLSDA identified a signature of 7 serum proteins that differentiated stable from exacerbation measurements in 16 paired stable and AE-COPD events experienced by 11 unique patients. (a) VIP scores identified a 7-protein serum signature that differentiated stable (purple) and exacerbation (orange) events with 81.25% cross-validation accuracy and 84.38% calibration accuracy. Latent variable 1 (LV1) accounted for 25.00% of the variance in the data, and latent variable 2 (LV2) accounted for 16.75% of the variance in the data. (b) The loadings plot shows how much each protein contributes to the signature, with positive loadings associated with exacerbation events, and negative loadings comparatively reduced in exacerbation. (c) Comparison of the differentiation between stable and exacerbated states based on individual factors vs. multivariate signatures. The VIP signature identified by the PLSDA models trended towards higher cross-validation accuracy than individual factors that were most significantly different. A one-way ANOVA determined that this signature was significantly better than IL-15 alone, with ** indicating a p value less than 0.01 after Tukey’s test for multiple comparisons. (d) Comparison of the calibration accuracies for individual factors vs. the VIP signature identified by the PLSDA model.
We next compared the classification ability of this signature to the classification ability of the top individual factors identified in univariate analysis of these data.22 The univariate model indicated that IL-10, IL-15, GDF-15, sICAM-1, and sVCAM-1 were individual factors that were significantly increased during exacerbation.22 For the purpose of comparing multivariate with univariate results, we took each of the top significant individual mediators from the previous analysis (sICAM-1, sVCAM-1, and IL-15) and assessed their individual ability to classify stable and AE-COPD. We then made a PLSDA model where we combined all five significant proteins previously identified through univariate analysis. We compared the performance of these four analyses to our VIP-selected PLSDA model described above, using the cross-validation accuracy and the calibration accuracy as comparison metrics. The cross-validation accuracy of the VIP-selected PLSDA model trended towards being higher than all analyses based on single significant proteins, but was only significantly better than the cross-validation based on IL-15 alone (p < 0.01, one-way ANOVA with Tukey’s HSD) (Fig. 2c). The VIP-selected PLSDA model did have the highest calibration accuracy out of all five accuracies that were compared (Fig. 2d). Overall, these figures serve to highlight the use of co-varying features, or “signatures,” in differentiating exacerbation events.
Insight into Cross-Tissue Compartment Cellular Interactions Associated with AE-COPD
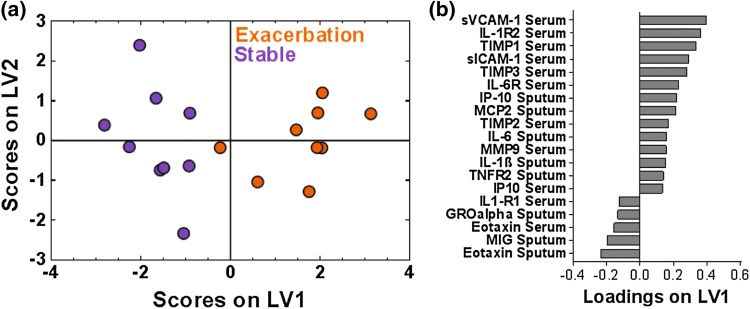
To gain deeper insight into relationships between immune factors in lung and serum tissue compartments involved in AE-COPD, we used PLSDA to integrate data from serum and sputum measurements in stable and exacerbated states. We first evaluated proteins for which both paired sputum and serum results were available (n = 9 matched stable and AE measurements), creating a PLSDA model based on 60 total analytes and employing VIP feature selection to eliminate those not contributing to differentiation. A one-latent variable PLSDA model separated exacerbation and stable measurements with a cross-validation and calibration accuracy of 88.89%, though a two-latent variable PLSDA model scores plot is presented to facilitate interpretation of group clustering (Fig. 3a). LV1 largely differentiated the stable state (negative scores on LV1) from AE-COPD (Fig. 3b). Fourteen of the 19 proteins were loaded positively on LV1, indicating positive association with AE-COPD, whereas five proteins were associated with stable COPD. Of the fourteen proteins that were positively associated with exacerbation, many of the serum proteins have been established as adhesion or chemoattraction factors (sICAM-1,56 sVCAM-1,2 IP-10,36 MCP-259), while most of the sputum proteins were known inflammatory factors (IL-6,27 IL-1β,16 TNFR-217). Similar to the serum-only model, this signature suggests migration and activation of innate immune cells in the serum during exacerbation, yet the addition of sputum data to the model demonstrates the corresponding importance of lung inflammation and chemokine secretion. As classification accuracy of the combined serum-sputum model was better than either separately, these results highlight the importance of the parallel relationship between chemokine secretion in lung and innate immune cell activation in serum.
Figure 3.
A one latent variable PLSDA model of VIP-selected proteins from the combined serum and sputum samples combined resulted in clear differentiation between stable and exacerbation measurements across 9 paired stable and AE-COPD events experienced by 7 unique patients. (a) PLSDA and VIP scores identified a signature of 19 proteins that differentiated the stable (purple) from exacerbation (orange) states with 88.89% cross-validation and calibration accuracy. Latent variable 1 accounted for 21.73% of the variance in the data. The scores plot shown is based on a two latent variable model to enable better visualization of group separation. (b) The loadings plot illustrates the protein contributions to the VIP-selected signature, with positive loadings positively associated with the exacerbation measurements, and negative loadings comparatively reduced during exacerbation.
Integration of Data Across Experimental Assays Gives Additional Insight into the Cellular and Proteomic Mechanisms Associated with AE-COPD
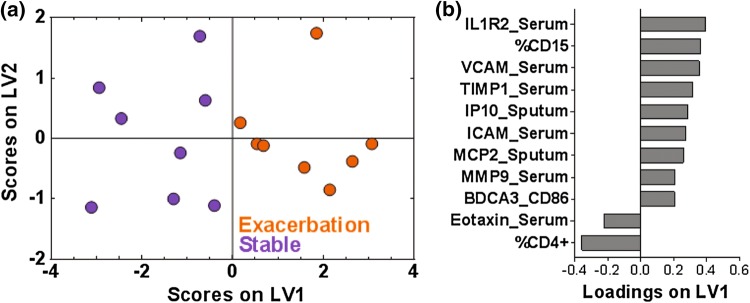
We also used our systems approach to integrate data across experimental assays by adding flow cytometry measurements, which were performed only on whole blood samples. We specifically explored whether PLSDA might help us integrate measurements made in different experimental assays. PLSDA and two rounds of VIP selection identified a one-latent variable model and a signature of 11 cell markers and proteins that differentiated stable COPD from AE-COPD with a cross-validation accuracy and a calibration accuracy of 87.5%. Differentiation between states (Fig. 4a) was driven by the loadings on LV1, which separated most individuals by exacerbation status (Fig. 4b). Nine of the cytokines and cell markers were loaded positively on LV1, indicating positive association with exacerbation, and two were loaded negatively on LV1, indicating negative association with exacerbation. Cellular factors associated with exacerbation in the integrated PLSDA model included CD86 expression by BDCA-3+ dendritic cells (DC) and the percentage of CD15+ granulocytes (reported in the original study to be neutrophils).22 In contrast, the percent of CD4+ T-cells was found to be associated with the stable measurements in this model.
Figure 4.
A one latent variable PLSDA model based on two rounds of VIP selection from serum and sputum proteins and blood flow markers shows clear differentiation between stable and exacerbation events across 8 pairs of patient samples, which included 7 paired stable and AE-COPD events experienced by 6 unique patients and one stable and one exacerbation measurement that were not patient matched. (a) PLSDA and two rounds of VIP analysis identified a signature of eleven factors that differentiated the stable (purple) from the exacerbation (orange) events, with 87.5% calibration and cross-validation accuracy. Latent variable 1 (LV1) accounted for 41.51% of the variance in the data. The scores plot shown is based on a two latent variable model to enable better visualization of group separation. (b) The loadings plot highlights factor contributions to the VIP-selected signature, with positive loadings positively associated with AE-COPD, and negative loadings comparatively reduced during an exacerbation event.
We next compared the cross-validation accuracies across all three of the VIP-selected models that consisted of varying amounts of tissue compartment and assay data. Although none of these three models were significantly different from each other according to Tukey’s post hoc test (one-way ANOVA), inclusion of data from more tissues and assays in the model trended toward a tighter and higher range of cross-validation accuracies (Supplemental Figure S5).
To visualize the unbiased classification ability of this signature, we also employed hierarchical clustering and created a heat map (Supplemental Figure S6). We found this clustering algorithm based on distance metrics was not as useful for classification, with three stable and four exacerbation samples misclassified out of sixteen total samples (56.25% classification accuracy). As our data contained measurements from three individuals with more than one exacerbation event, we also examined our scores plot after labeling the points with the patient’s exacerbation status and visit number. The resulting scores plot (Supplemental Figure S7) indicates no clear intra-patient clustering, though this study was not powered for a thorough statistical analysis in this direction.
We further explored potential relationships between cell number and protein concentration across the stable and exacerbated states in our identified signature using Spearman rank correlation coefficients and a heat map. Overall we found that MMP-9 in the serum was positively correlated with CD4+ cells expressing the IL-18 receptor, and TIMP-1 in the serum was positively correlated with CD4+ cells expressing the CD122 activation marker. The BDCA3+ CD86+ and the %CD15 neutrophils were not correlated with the other proteins in the signature, but were correlated with other measured proteins (Supplemental Figure S8). Overall, this suggests that changes in cell number from the stable to the exacerbated state may be related to simultaneous increases in concentration of some inflammatory proteins across the two states.
Discussion
Using systems analysis of paired data points from cellular factors measured in blood and sputum in exacerbated and stable COPD states, we identified a signature that differentiated stable and AE-COPD with > 87% cross-validation accuracy. This signature trended towards being better than any previously identified individual cellular factors for differentiating stable and exacerbated COPD states, though more measurements would be needed to determine statistical significance. Biologically, the signature indicated that parallel increases in inflammatory cytokines in sputum environments, adhesion/chemoattractive cytokines in serum environments, and greater numbers of BDCA-3+ DC and an increased percent of CD15+ neutrophils in the blood were all associated with AE-COPD. These results highlight the value of computational approaches when integrating measurements across tissue compartments and from different experimental assays, and motivate use of these approaches to gain new perspective into cellular systems involved in this prevalent, lethal, but understudied disorder.
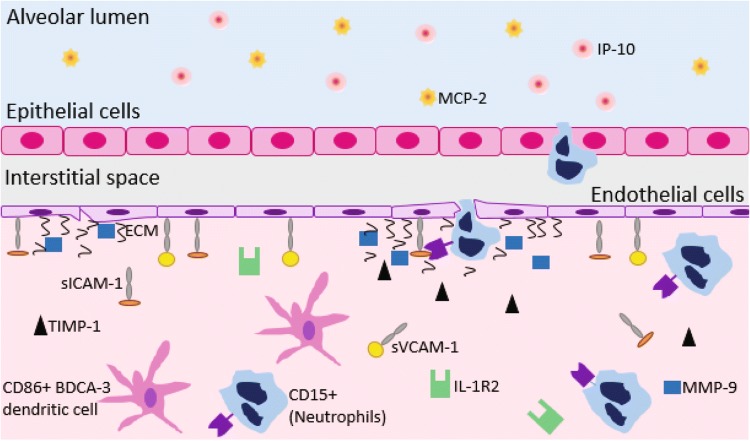
One important strength of our approach is the ability to define parsimonious cellular signatures by selecting the most significant co-varying cellular immune factors. This approach may be valuable as a means of defining key cellular systems involved in disease progression, and using these to efficiently choose end-points in clinical trials and guide future experimental endeavors. This approach is especially useful for integrating cellular measurements made in multiple tissue compartments, which is important given the central role of sputum production in AE-COPD. Based on these findings, we propose a model of key networks in AE-COPD (Fig. 5) involving specific immune cell types, metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs), and chemokines. We discuss our findings in that framework.
Figure 5.
A hypothesis of cross-tissue mechanisms of action in the lungs and blood of patients experiencing an AE-COPD. Adhesion molecules aid in moving immune cells from the blood to the lung, which is further promoted by the presence of the chemokine interferon gamma-induced protein 10 (IP-10) and monocyte chemoattractive protein 2 (MCP-2) in the sputum. sICAM: soluble intercellular adhesion molecule. sVCAM: vascular cell adhesion molecule. IP: interferon gamma-induced protein. MCP: monocyte chemoattractive protein. TIMP: tissue inhibitor of metalloproteinases. MMP: matrix metalloproteinase. R2: receptor 2. ECM: extracellular matrix. CD: cluster of differentiation. BDCA: blood dendritic cell antigen.
In terms of peripheral blood leukocyte participation in AE-COPD, we extend the observation from univariate analysis of these data22 that CD4+ T cells decreased in blood during exacerbation, which is compatible with trafficking to lung or regional lymph nodes (or both), by showing the importance of the simultaneous increase in blood of BDCA-3+ DC. We have previously demonstrated the physical interaction of this DC subset with CD4+ T cells in lung tissue from COPD patients.21 BDCA-3+ DC were previously termed mDC2, but are now designated as cDC1;20 they are the counterpart of murine CD103+ DC, which are essential for cross-presentation of viral antigens to CD8+ T cells. Our model suggests recruitment to the lungs of cDC1, likely from the bone marrow, as a crucial step driving lung inflammation during AE-COPD. The other type of leukocyte in our signature, neutrophils, has been shown by other studies to be linked to AE-COPD,5 one of which related their numbers to exacerbation severity.46
Key soluble factors in our signature agree with and extend previous individual associations of inflammatory mediators with AE-COPD. These not only include the anticipated agreement with previous univariate analysis of these data,22 but also several serum proteins involved in adhesion and chemoattraction of inflammatory cells. Chief among these is the neutrophil chemoattractant IP-10/CXCL10, also found to be elevated in AE-COPD in two studies.7,28 Our signature also included IL-6, a pro-inflammatory cytokine27 that has been vigorously investigated as a possible biomarker for AE-COPD. Increased IL-6 in serum and sputum during AE-COPD was reported by several large studies using longitudinal design;7,8 this association was questioned in a systematic review which, however, included many studies of cross-sectional design.12 Our results illustrate the superior power of comparing paired results from the same subjects across stable and exacerbated states. We also identified elevations in levels of sICAM-1 and sVCAM-1, truncated forms of transmembrane adhesion molecules that interact with leukocyte integrins. sVCAM is chemotactic for murine neutrophils in vitro.41 sICAM-1 is expressed both by leukocytes and by activated endothelial cells, and levels of sICAM-1 correlate to endothelial cell ICAM expression in vitro.35 Each of these proteins are elevated in stable COPD,18,26 though to our knowledge, no study (other than our original data) has linked it to AE-COPD in longitudinal data. sICAM has been reported to be elevated in subjects admitted for AE-COPD compared with healthy control subjects.23 Higher plasma sICAM-1 levels were also independently associated with emphysema progression in the MESA Lung cohort, a general population sample.1
Our signature identified elevated serum MMP-9 as a crucial feature of AE-COPD, in agreement with a previous study.33 Also known as gelatinase B, MMP-9 is released by activated neutrophils.10 It has an unique ability to induce self-perpetuating lung inflammation by degrading extracellular matrix, thus liberating the neutrophil chemoattractant tripeptide N-acetyl Proline-Glycine-Proline.58 Along with IL-6, MMP-9 was one of 34 serum analytes found to be highly reproducible over a 6 week period of clinical stability in COPD patients,50 further supporting our findings. Our MMP-9 finding is interesting in light of the disparity between the association with exacerbation of TIMP-1, TIMP-2, and TIMP-3, which stoichiometrically inhibit MMP activity,38,43 and TIMP-4, which associated with the stable state in the VIP signature. Unlike the other three TIMP family members, which act as soluble inhibitors, TIMP-3 is typically bound to matrix sulfated glycosaminoglycans,38 suggesting that its presence in the serum during AE-COPD might reflect matrix degradation.
All of our models identified IL-1R2 as a crucial serum factor increased during AE-COPD, in agreement with two studies from the group in Maastricht of patients admitted for AE-COPD.15,24 IL-1R2 (Gene ID: 7850) is an early response gene47 whose product is a decoy receptor that inhibits activity of its three ligands: IL-1α, IL-1β, and the type I IL-1 receptor. Together with associations for TIMP1-3, our results highlight the importance of counter-regulatory factors during AE-COPD. Although all the subjects in the original dataset were successfully treated as outpatients with resolution, not all patients regain lung function following AE-COPD; an intriguing possibility is that those who do not recover entirely might exhibit relatively deficient up-regulation of IL-1R2 and TIMPs during AE-COPD.
There are several limitations to this analysis. Although our original study22 recruited a larger group of subjects, many sought treatment for AE-COPD locally, rather than returning when acutely ill. Some measurements had to be excluded from this analysis due to missing data. Collectively, these factors reduced our sample size, making it all the more noteworthy that our approach identified AE-COPD cellular signatures that could be used to gain biological insight. However, the small sample size did limit our ability to find signatures that could be used in diagnostic contexts. Even though our identified signature trended towards being better than individual factors, it was only statistically significant in one case. Furthermore, additional unknown test data in different patient cohorts would be needed to truly assess signature classification ability for diagnostic purposes. A second limitation is the necessary dependence on proteins measured in the original study, which used a “candidate gene” approach based in part on prior knowledge, and not an unbiased screen of the entire proteome. Because our original study involved flow cytometric analysis of peripheral blood leukocytes collected in part during AE-COPD, there is, to our knowledge, no current exacerbation cohort available for validation testing. However, to prevent model overfitting as much as possible, we did employ internal cross-validation.
Results of this work support exciting future research in several directions. First, if similar data from other cohorts of paired stable and exacerbation measurements were to become available, generated models could be tested and validated. Data-driven approaches such as these could be applied as a classification tool to identify differences in exacerbation endotypes or in AE-COPD events resulting from different upstream causes (including viruses, bacteria, etc.), thus providing insight into systems-level mechanisms of action that could result in personalized treatment options. Unbiased data-driven models applied to multiplex COPD data from across tissue compartments may also prove useful to characterize COPD endotypes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors would like to thank Lisa McCloskey, RRT, Christi Getty, RRT, and Candace Flaherty, RRT for interactions with subjects in the original study.
Funding
This work was supported by NIH R01 HL144849-01 (to K.B.A.). K.C.N. was supported by a Department of Education Graduate Assistance in Areas of National Need (GAANN) Fellowship awarded to the biomedical engineering department at the University of Michigan (PR Award Number: P200A150170). C.M.F. was supported by Merit Review Awards I01 CX001553 from the Department of Veterans Affairs and by MedImmune, Ltd. M.K.H. reports a grant from the National Heart, Lung and Blood Institute. F.J.M. has received grants from the National Institute of Health. J.L.C. was supported by Merit Review Awards I01 CX000911 from the Department of Veterans Affairs and by MedImmune, Ltd.
Conflict of interest
K.C.N., C.M.F., N.S.B, J.L.C. and K.B.A. reported no conflicts of interest. M.K.H. reports consultant arrangements with GlaxoSmithKline, Boehringer Ingelheim, Novartis, Sunovion, and AstraZeneca. F.J.M. has received personal fees from Forest, Janssen, GlaxoSmithKline, Nycomed/Takeda, Amgen, AstraZeneca, Boehringer Ingelheim, Ikaria/Bellerophon, Genentech, Novartis, Pearl, Pfizer, Roche, Sunovion, Theravance, Axon, CME Incite, California Society for Allergy and Immunology, Annenberg, Integritas, InThough, Miller Medical, National Association for Continuing Education, Paradigm, Peer Voice, UpToDate, Haymarket Communications, Western Society of Allergy and Immunology, Informa, Bioscale, Unity Biotechnology, ConCert, Lucid, Methodist Hospital, Prime, WebMD, Bayer, Ikaria, Kadmon, Vercyte, American Thoracic Society, Academic CME, Falco, Axon Communication, Johnson & Johnson, Clarion, Continuing Education, Potomac, Afferent, and Adept; and has collected nonfinancial support from Boehringer Ingelheim, Centocor, Gilead, and Biogen/Stromedix; and declares other interests with Mereo, Boehringering Ingelheim, and Centocor.
Ethical Standards
All human subjects research was carried out in accordance with the Declaration of Helsinki and were approved by the Institutional Review Boards of the Veterans’ Affairs Ann Arbor Healthcare System (VAAHS) and of the University of Michigan Health System (UMHS).
Research Involved in Human or animal rights
No animal studies were carried out by the authors for this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Katy C. Norman and Christine M. Freeman are co-first authors. Jeffrey L. Curtis and Kelly B. Arnold are co-senior authors.
Contributor Information
Jeffrey L. Curtis, Email: jlcurtis@umich.edu
Kelly B. Arnold, Phone: 734-763-5230, Email: kbarnold@umich.edu
References
- 1.Aaron CP, Schwartz JE, Bielinski SJ, Hoffman EA, Austin JHM, Oelsner EC, Donohue KM, Kalhan R, Berardi C, Kaufman JD, Jacobs DR, Tracy RP, Barr RG. Intercellular adhesion molecule 1 and progression of percent emphysema: The MESA Lung Study. Resp. Med. 2015;109:255–264. doi: 10.1016/j.rmed.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agouridakis P, Kyriakou D, Alexandrakis MG, Prekates A, Perisinakis K, Karkavitsas N, Bouros D. The predictive role of serum and bronchoalveolar lavage cytokines and adhesion molecules for acute respiratory distress syndrome development and outcome. Resp. Res. 2002;3:25–25. doi: 10.1186/rr190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agustí A, Edwards LD, Rennard SI, MacNee W, Tal-Singer R, Miller BE, Vestbo J, Lomas DA, Calverley PMA, Wouters E, Crim C, Yates JC, Silverman EK, Coxson HO, Bakke P, Mayer RJ, Celli B, ECLIPSE Investigators Persistent systemic inflammation is associated with poor clinical outcomes in COPD: a novel phenotype. PLoS ONE. 2012;7:e37483–e37483. doi: 10.1371/journal.pone.0037483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert RK, Connett J, Bailey WC, Casaburi R, Cooper JAD, Criner GJ, Curtis JL, Dransfield MT, Han MK, Lazarus SC, Make B, Marchetti N, Martinez FJ, Madinger NE, McEvoy C, Niewoehner DE, Porsasz J, Price CS, Reilly J, Scanlon PD, Sciurba FC, Scharf SM, Washko GR, Woodruff PG, Anthonisen NR, Network CCR. Azithromycin for prevention of exacerbations of COPD. New Engl. J. Med. 2011;365:689–698. doi: 10.1056/NEJMoa1104623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andelid K, Andersson A, Yoshihara S, Âhrén C, Jirholt P, Ekberg-Jansson A, Lindén A. Systemic signs of neutrophil mobilization during clinically stable periods and during exacerbations in smokers with obstructive pulmonary disease. Int. J. Chronic Obstr. 2015;10:1253–1263. doi: 10.2147/COPD.S77274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnold KB, Burgener A, Birse K, Romas L, Dunphy LJ, Shahabi K, Abou M, Westmacott GR, McCorrister S, Kwatampora J, Nyanga B, Kimani J, Masson L, Liebenberg LJ, Abdool Karim SS, Passmore J-AS, Lauffenburger DA, Kaul R, McKinnon LR. Increased levels of inflammatory cytokines in the female reproductive tract are associated with altered expression of proteases, mucosal barrier proteins, and an influx of HIV-susceptible target cells. Mucosal Immunol. 2016;9:194–205. doi: 10.1038/mi.2015.51. [DOI] [PubMed] [Google Scholar]
- 7.Bafadhel M, McKenna S, Terry S, Mistry V, Reid C, Haldar P, McCormick M, Haldar K, Kebadze T, Duvoix A, Lindblad K, Patel H, Rugman P, Dodson P, Jenkins M, Saunders M, Newbold P, Green RH, Venge P, Lomas DA, Barer MR, Johnston SL, Pavord ID, Brightling CE. Acute exacerbations of chronic obstructive pulmonary disease: identification of biologic clusters and their biomarkers. Am. J. Resp. Crit. Care. 2011;184:662–671. doi: 10.1164/rccm.201104-0597OC. [DOI] [PubMed] [Google Scholar]
- 8.Barnes PJ. The cytokine network in chronic obstructive pulmonary disease. Am. J. Resp. Cell Mol. 2009;41:631–638. doi: 10.1165/rcmb.2009-0220TR. [DOI] [PubMed] [Google Scholar]
- 9.Cane JL, Mallia-Millanes B, Forrester DL, Knox AJ, Bolton CE, Johnson SR. Matrix metalloproteinases -8 and -9 in the airways, blood and urine during exacerbations of COPD. COPD. 2016;13:26–34. doi: 10.3109/15412555.2015.1043522. [DOI] [PubMed] [Google Scholar]
- 10.Chakrabarti S, Patel KD. Regulation of matrix metalloproteinase-9 release from IL-8-stimulated human neutrophils. J. Leukocyte Biol. 2005;78:279–288. doi: 10.1189/jlb.1004612. [DOI] [PubMed] [Google Scholar]
- 11.Chang C, Guo Z, Shen N, He B, Yao W, Zhu H, Zhao J. Dynamics of inflammation resolution and symptom recovery during AECOPD treatment. Sci. Rep. 2014;4:5516–5516. doi: 10.1038/srep05516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y-WR, Leung JM, Sin DD. A systematic review of diagnostic biomarkers of COPD exacerbation. PLoS ONE. 2016;11:e0158843–e0158843. doi: 10.1371/journal.pone.0158843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosio MG, Saetta M, Agustí A. Immunologic aspects of chronic obstructive pulmonary disease. New Engl. J. Med. 2009;360:2445–2454. doi: 10.1056/NEJMra0804752. [DOI] [PubMed] [Google Scholar]
- 14.Curtis JL, Freeman CM, Hogg JC. The immunopathogenesis of chronic obstructive pulmonary disease: Insights from recent research. Proc. Am. Thorac. Soc. 2007;4:512–521. doi: 10.1513/pats.200701-002FM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dentener MA, Creutzberg EC, Schols AM, Mantovani A, van’t Veer C, Buurman WA, Wouters EF. Systemic anti-inflammatory mediators in COPD: increase in soluble interleukin 1 receptor II during treatment of exacerbations. Thorax. 2001;56:721–726. doi: 10.1136/thorax.56.9.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720–3732. doi: 10.1182/blood-2010-07-273417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douni E, Kollias G. A critical role of the p75 tumor necrosis factor receptor (p75TNF-R) in organ inflammation independent of TNF, lymphotoxin alpha, or the p55TNF-R. J. Exp. Med. 1998;188:1343–1352. doi: 10.1084/jem.188.7.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Deek SE, Makhlouf HA, Saleem TH, Mandour MA, Mohamed NA. Surfactant protein D, soluble intercellular adhesion molecule-1 and high-sensitivity C-reactive protein as biomarkers of chronic obstructive pulmonary disease. Med. Prin. Pract. 2013;22:469–474. doi: 10.1159/000349934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford ES, Murphy LB, Khavjou O, Giles WH, Holt JB, Croft JB. Total and state-specific medical and absenteeism costs of COPD among adults aged ≥ 18 years in the United States for 2010 and projections through 2020. Chest. 2015;147:31–45. doi: 10.1378/chest.14-0972. [DOI] [PubMed] [Google Scholar]
- 20.Freeman CM, Curtis JL. Lung dendritic cells: shaping immune responses throughout COPD progression. Am. J. Resp. Cell Mol. 2017;56:152–159. doi: 10.1165/rcmb.2016-0272TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman CM, Martinez FJ, Han MK, Ames TM, Chensue SW, Todt JC, Arenberg DA, Meldrum CA, Getty C, McCloskey L, Curtis JL. Lung dendritic cell expression of maturation molecules increases with worsening chronic obstructive pulmonary disease. Am. J. Resp. Crit. Care. 2009;180:1179–1188. doi: 10.1164/rccm.200904-0552OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman CM, Martinez CH, Todt JC, Martinez FJ, Han MK, Thompson DL, McCloskey L, Curtis JL. Acute exacerbations of chronic obstructive pulmonary disease are associated with decreased CD4+ & CD8+ T cells and increased growth & differentiation factor-15 (GDF-15) in peripheral blood. Resp. Res. 2015;16:94–94. doi: 10.1186/s12931-015-0251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerritsen WBM, Asin J, Zanen P, van den Bosch JMM, Haas FJLM. Markers of inflammation and oxidative stress in exacerbated chronic obstructive pulmonary disease patients. Resp. Med. 2005;99:84–90. doi: 10.1016/j.rmed.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 24.Groenewegen KH, Dentener MA, Wouters EFM. Longitudinal follow-up of systemic inflammation after acute exacerbations of COPD. Resp. Med. 2007;101:2409–2415. doi: 10.1016/j.rmed.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 25.Halpin DMG, Miravitlles M, Metzdorf N, Celli B. Impact and prevention of severe exacerbations of COPD: a review of the evidence. Int. J. Chronic. Obstr. 2017;2891–2908:2017. doi: 10.2147/COPD.S139470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollander C, Sitkauskiene B, Sakalauskas R, Westin U, Janciauskiene SM. Serum and bronchial lavage fluid concentrations of IL-8, SLPI, sCD14 and sICAM-1 in patients with COPD and asthma. Resp. Med. 2007;101:1947–1953. doi: 10.1016/j.rmed.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 27.Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 28.Hurst JR, Donaldson GC, Perera WR, Wilkinson TMA, Bilello JA, Hagan GW, Vessey RS, Wedzicha JA. Use of plasma biomarkers at exacerbation of chronic obstructive pulmonary disease. Am. J. Resp. Crit. Care. 2006;174:867–874. doi: 10.1164/rccm.200604-506OC. [DOI] [PubMed] [Google Scholar]
- 29.Johannesdottir SA, Christiansen CF, Johansen MB, Olsen M, Xu X, Parker JM, Molfino NA, Lash TL, Fryzek JP. Hospitalization with acute exacerbation of chronic obstructive pulmonary disease and associated health resource utilization: a population-based Danish cohort study. J. Med. Econ. 2013;16:897–906. doi: 10.3111/13696998.2013.800525. [DOI] [PubMed] [Google Scholar]
- 30.Karadag F, Karul AB, Cildag O, Yilmaz M, Ozcan H. Biomarkers of systemic inflammation in stable and exacerbation phases of COPD. Lung. 2008;186:403–409. doi: 10.1007/s00408-008-9106-6. [DOI] [PubMed] [Google Scholar]
- 31.Kochanek, K. D., S. Murphy, J. Xu, and E. Arias. Mortality in the United States, 2016. NCHS Data Brief 1–8, 2017. [PubMed]
- 32.Krommidas G, Kostikas K, Papatheodorou G, Koutsokera A, Gourgoulianis KI, Roussos C, Koulouris NG, Loukides S. Plasma leptin and adiponectin in COPD exacerbations: associations with inflammatory biomarkers. Resp. Med. 2010;104:40–46. doi: 10.1016/j.rmed.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 33.Kwiatkowska S, Noweta K, Zieba M, Nowak D, Bialasiewicz P. Enhanced exhalation of matrix metalloproteinase-9 and tissue inhibitor of metalloproteinase-1 in patients with COPD exacerbation: a prospective study. Respiration. 2012;84:231–241. doi: 10.1159/000339417. [DOI] [PubMed] [Google Scholar]
- 34.Lau KS, Juchheim AM, Cavaliere KR, Philips SR, Lauffenburger DA, Haigis KM. In vivo systems analysis identifies spatial and temporal aspects of the modulation of TNF-α-induced apoptosis and proliferation by MAPKs. Sci. Signal. 2011;4:ra16. doi: 10.1126/scisignal.2001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leeuwenberg JF, Smeets EF, Neefjes JJ, Shaffer MA, Cinek T, Jeunhomme TM, Ahern TJ, Buurman WA. E-selectin and intercellular adhesion molecule-1 are released by activated human endothelial cells in vitro. Immunology. 1992;77:543–549. [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M, Guo S, Hibbert JM, Jain V, Singh N, Wilson NO, Stiles JK. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev. 2011;22:121–130. doi: 10.1016/j.cytogfr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez FJ, Calverley PMA, Goehring U-M, Brose M, Fabbri LM, Rabe KF. Effect of roflumilast on exacerbations in patients with severe chronic obstructive pulmonary disease uncontrolled by combination therapy (REACT): a multicentre randomised controlled trial. Lancet. 2015;385:857–866. doi: 10.1016/S0140-6736(14)62410-7. [DOI] [PubMed] [Google Scholar]
- 38.Masciantonio MG, Lee CKS, Arpino V, Mehta S, Gill SE. The balance between metalloproteinases and TIMPs. Prog. Mol. Biol. Transl. Sci. 2017;147:101–131. doi: 10.1016/bs.pmbts.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 39.McCubbrey AL, Sonstein J, Ames TM, Freeman CM, Curtis JL. Glucocorticoids relieve collectin-driven suppression of apoptotic cell uptake in murine alveolar macrophages through downregulation of SIRPα. J. Immunol. 2012;189:112–119. doi: 10.4049/jimmunol.1200984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller BE, Tal-Singer R, Rennard SI, Furtwaengler A, Leidy N, Lowings M, Martin UJ, Martin TR, Merrill DD, Snyder J, Walsh J, Mannino DM. Plasma fibrinogen qualification as a drug development tool in chronic obstructive pulmonary disease. Perspective of the chronic obstructive pulmonary disease biomarker qualification consortium. Am. J. Resp. Crit. Care. 2016;193:607–613. doi: 10.1164/rccm.201509-1722PP. [DOI] [PubMed] [Google Scholar]
- 41.Mishra A, Guo Y, Zhang L, More S, Weng T, Chintagari NR, Huang C, Liang Y, Pushparaj S, Gou D, Breshears M, Liu L. A critical role for P2X7 receptor-induced VCAM-1 shedding and neutrophil infiltration during acute lung injury. J. Immunol. 2016;197:2828–2837. doi: 10.4049/jimmunol.1501041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller B, Tamm M. Biomarkers in acute exacerbation of chronic obstructive pulmonary disease: among the blind, the one-eyed is king. Am. J. Resp. Crit. Care. 2006;174:848–849. doi: 10.1164/rccm.200607-922ED. [DOI] [PubMed] [Google Scholar]
- 43.Navratilova Z, Kolek V, Petrek M. Matrix metalloproteinases and their inhibitors in chronic obstructive pulmonary disease. Arch. Immunol. Ther. Exerc. 2016;64:177–193. doi: 10.1007/s00005-015-0375-5. [DOI] [PubMed] [Google Scholar]
- 44.O’Dwyer DN, Norman KC, Xia M, Huang Y, Gurczynski SJ, Ashley SL, White ES, Flaherty KR, Martinez FJ, Murray S, Noth I, Arnold KB, Moore BB. The peripheral blood proteome signature of idiopathic pulmonary fibrosis is distinct from normal and is associated with novel immunological processes. Sci. Rep. 2017;7:46560–46560. doi: 10.1038/srep46560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oba Y, Lone NA. Efficacy and safety of roflumilast in patients with chronic obstructive pulmonary disease: a systematic review and meta-analysis. Ther. Adv. Respir. Dis. 2013;7:13–24. doi: 10.1177/1753465812466167. [DOI] [PubMed] [Google Scholar]
- 46.Papi A, Bellettato CM, Braccioni F, Romagnoli M, Casolari P, Caramori G, Fabbri LM, Johnston SL. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am. J. Resp. Crit. Care. 2006;173:1114–1121. doi: 10.1164/rccm.200506-859OC. [DOI] [PubMed] [Google Scholar]
- 47.Peters VA, Joesting JJ, Freund GG. IL-1 receptor 2 (IL-1R2) and its role in immune regulation. Brain Behav. Immun. 2013;32:1–8. doi: 10.1016/j.bbi.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roche N, Zureik M, Soussan D, Neukirch F, Perrotin D, B. S. C. Urgence and Investigators Predictors of outcomes in COPD exacerbation cases presenting to the emergency department. Eur. Respir. J. 2008;32:953–961. doi: 10.1183/09031936.00129507. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez-Roisin R. Toward a consensus definition for COPD exacerbations. Chest. 2000;117:398S–401S. doi: 10.1378/chest.117.5_suppl_2.398S. [DOI] [PubMed] [Google Scholar]
- 50.Röpcke S, Holz O, Lauer G, Müller M, Rittinghausen S, Ernst P, Lahu G, Elmlinger M, Krug N, Hohlfeld JM. Repeatability of and relationship between potential COPD biomarkers in bronchoalveolar lavage, bronchial biopsies, serum, and induced sputum. PLoS ONE. 2012;7:e46207–e46207. doi: 10.1371/journal.pone.0046207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santos S, Marín A, Serra-Batlles J, de la Rosa D, Solanes I, Pomares X, López-Sánchez M, Muñoz-Esquerre M, Miravitlles M. Treatment of patients with COPD and recurrent exacerbations: the role of infection and inflammation. Int. J. Chronic Obstr. 2016;11:515–525. doi: 10.2147/COPD.S98333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomsen M, Ingebrigtsen TS, Marott JL, Dahl M, Lange P, Vestbo J, Nordestgaard BG. Inflammatory biomarkers and exacerbations in chronic obstructive pulmonary disease. JAMA. 2013;309:2353–2361. doi: 10.1001/jama.2013.5732. [DOI] [PubMed] [Google Scholar]
- 53.Toy EL, Gallagher KF, Stanley EL, Swensen AR, Duh MS. The economic impact of exacerbations of chronic obstructive pulmonary disease and exacerbation definition: a review. COPD. 2010;7:214–228. doi: 10.3109/15412555.2010.481697. [DOI] [PubMed] [Google Scholar]
- 54.Vaitkus M, Lavinskiene S, Barkauskiene D, Bieksiene K, Jeroch J, Sakalauskas R. Reactive oxygen species in peripheral blood and sputum neutrophils during bacterial and nonbacterial acute exacerbation of chronic obstructive pulmonary disease. Inflammation. 2013;36:1485–1493. doi: 10.1007/s10753-013-9690-3. [DOI] [PubMed] [Google Scholar]
- 55.Wedzicha JA, Banerji D, Chapman KR, Vestbo J, Roche N, Ayers RT, Thach C, Fogel R, Patalano F, Vogelmeier CF, Investigators F. Indacaterol-glycopyrronium versus salmeterol-fluticasone for COPD. New Engl. J. Med. 2016;374:2222–2234. doi: 10.1056/NEJMoa1516385. [DOI] [PubMed] [Google Scholar]
- 56.Witkowska AM, Borawska MH. Soluble intercellular adhesion molecule-1 (sICAM-1): an overview. Eur. Cytokine Netw. 2004;15:91–98. [PubMed] [Google Scholar]
- 57.Wold S, Johansson E, Cocchi M. PLS-partial least squares projections to latent structures. In: Kubinyi H, editor. 3D QSAR in Drug Design: Theory Methods and Applications. Dordrecht: Escom; 1993. pp. 523–550. [Google Scholar]
- 58.Xu X, Jackson PL, Tanner S, Hardison MT, Roda MA, Blalock JE, Gaggar A. A self-propagating matrix metalloprotease-9 (MMP-9) dependent cycle of chronic neutrophilic inflammation. PLoS ONE. 2011;6:e15781. doi: 10.1371/journal.pone.0015781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yung S. C. and J. M. Farber. Chemokines. edited by A. J. KastinElsevier, pp. 656–663, 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.