Abstract
Introduction
Triple negative breast cancer (TNBC) is a highly aggressive type of breast cancer with high resistance to current standard therapies. We demonstrate that phenotypically stratified carbon nanoparticle is highly effective in delivering a novel combinatorial triple drug formulation for synergistic regression of TNBC in vitro and in vivo.
Method
The combinatorial formulation is comprised of repurposed inhibitors of STAT3 (nifuroxazide), topoisomerase-II-activation-pathway (amonafide) and NFκb (pentoxifylline). Synergistic effect of drug combination was established in a panel of TNBC-lines comprising mesenchymal-stem-like, mesenchymal and basal-like cells along with non-TNBC-cells. The delivery of combinatorial drug formulation was achieved using a phenotypically screened carbon nanoparticles for TNBC cell lines.
Results
Results indicated a remarkable five-fold improvement (IC50-6.75 µM) from the parent drugs with a combinatorial index <1 in majority of the TNBC cells. Multi-compartmental carbon nanoparticles were then parametrically assessed based on size, charge (positive/negative/neutral) and chemistry (functionalities) to study their likelihood of crossing endocytic barriers from phenotypical standpoint in TNBC lines. Interestingly, a combination of clathrin mediated, energy and dynamin dependent pathways were predominant for sulfonated nanoparticles, whereas pristine and phospholipid particles followed all the investigated endocytic pathways.
Conclusions
An exactitude ‘omics’ approach helps to predict that phospholipid encapsulated-particles will predominantly accumulate in TNBC comprising the drug-‘cocktail’. We investigated the protein expression effects inducing synergistic effect and simultaneously suppressing drug resistance through distinct mechanisms of action.
Electronic supplementary material
The online version of this article (doi:10.1007/s12195-017-0490-y) contains supplementary material, which is available to authorized users.
Introduction
Although myriad advancement has been made to develop a systemic treatment for human breast carcinomas, the lack of responsive-ness to hormonal therapies due to the absence of molecular targets such as estrogen receptor (ER), progesterone receptor (PR), or C-erbB-2 (HER2), makes it challenging.32,44 For triple-negative breast cancer (TNBC) cells with negative phenotypes, including ER (−), PR (−), and HER2 (−),5 due to shortage of a recognized molecular target, relatively poor prognosis of late-stage patients with TNBC, and frequent chemo-resistance makes it extremely challenging to advance the treatment regimen. Resistance to current standard therapies for previously treated patients with metastatic TNBC pushed the researchers to explore combinatorial therapy regime. It has been reported that eribulin, platinum-based regimens, bevacizumab or anti-EGFR treatment with cetuximab, have shown some aids in combination therapy. STAT3-inhibitors also acted synergistically with metformin, in reducing cell growth and inducing apoptosis in TNBCs.57 Aggressive TNBCs is known to associate closely with BRCA1 mutation or dysregulation.12 Agents targeting DNA repair represents a new approach to TNBC therapy.7 Advancement in treatment may also be facilitated by a combination of drugs, which can target multiple protein cascades. Furthermore, these benefits can be appreciated even more by using currently approved agents and/or in combination with newer targeted drugs.
However, the combinatorial drug delivery is not trivial and can only successfully be achieved through a well-defined nanoparticle platform.13,30,34,39 Design of an ideal nanostructure would allow us to encapsulate the therapeutic agents with varying polarity profiles in a highly stable manner to avoid systemic dis-integration. Due to the inability of current techniques to envisage which patients will be most responsive to these treatments,6,16,17,59 it is critical to identify tumors likely to respond to drugs delivered via nanoparticles providing a better match based on patients’ pathophysiology. We hypothesize that assuming a ‘precision’ nanomedicine approach can envisage a patient- specific response to a drug based on physiological alterations to nanotherapeutics.
Internalization of nanoparticles to a cell is typically guided by penetration, adhesion, hemifusion and endocytosis.8,51,53 Endocytosis is one of the most well-studied cellular barrier, involved in several specialized functions.11,14,20,24,50 Independent studies indicate that endocytotic variations in cancer cells may affect cell surface expression of crucial molecules and significantly impact the cancer-relevant phenotypes, with potential implications for interventions to control cancer by modulating nanoparticle surface chemistry and other parametric function. Various signaling pathways do persist in the endocytic route and emerging evidence ties endocytosis as a whole, or individual proteins to mitosis, apoptosis and cell fate determination. Internalization of nanoparticles in cells are often dictated by their size,27,52,60 functionality,23,25,46 and surface charge.3,10 However, the mechanistic understanding of nanoparticle internalization dependent on the stepwise progression of cancer leading to tumor formation and subsequent metastasis, is still complex and not well understood.
With this unmet need, our approach follows the field of genetics to predict an individual’s response to a drug, based on the histopathological information obtained from biopsy. A diagnostic cohort will be generated for stratification of ‘right’ delivery platform candidate relying on cancer phenotypes for potentially improved efficacy and reduced toxicity. We parametrically assessed carbon nanoparticles based on their charge and surface chemistry through endocytic stratification from pathophysiological differences. Carbon nanodots were chosen as a base platform due to its biodegradable nature, ease of large scale production, and facile surface characteristics.26,43 We anticipate that uniting biopsy results with a nanomedicine of superior probabilities to cross the endocytic “barrier” to deliver combinatorial drug can be proposed.1,2,4,9,18,21,22,28,31,36,38,40,42,45,47–49,54–56,58
Towards this aim, a series of carbon dots were prepared with cationic, anionic or neutral surface functionalities. Carbon nanoparticles have shown great potential as drug delivery vehicles due to excellent biocompatibility, ease of synthesis and surface modification properties. These particles can easily penetrate cytoplasmic and nuclear membranes, allowing for enhanced delivery of the intact drug to tumor sites for more effective treatment compared to free drug.19,29,33 Anticancer drugs have been tested in a carbon nanoparticle system both in vitro and in vivo with promising results. Our laboratory and others have demonstrated that these particles can also be utilized as a contrast probe for imaging various biological processes.15,35,37,41 Our results for the first time demonstrated that carbon nanoparticles, as a function of their surface charge, behave uniquely in presence of TNBC cells in comparison to non-TNBC cells. Although the understanding of pathophysiological contributions of endocytic alterations is not robust, we studied the major endocytic routes, i.e. clathrin, lipid-raft mediated, dynamin, and energy-dependent pathways. Selective endocytic pathway depending on the TNBC nature of breast cancer can be selected and this understanding of surface characteristics and cancer stage-driven internalization of particles guided us to select the best delivery platforms with optimized cytotoxicity and maximum selectivity.
Recently, we have invented a combinatorial drug formulation comprising repurposed inhibitors of STAT3 (nifuroxazide), topoisomerase-II-activation-pathway (amonafide) and NFκb (pentoxifylline) for optimized effect. The drug screening and details of molecular biology-based studies will be published elsewhere. In the previous report, our formulation included niclosamide (NSA, i.e. 5-chloro-N-(2-chloro-4-nitrophenyl)-2-hydroxybenzamide), a known STAT3 inhibitor and amonafide (AMF, 5-amino-2-[2-(dimethylamino)ethyl]-1H-benzo[de]isoquinoline-1,3(2H)-dione), an agent known to exert its effect through topoisomerase-II activation pathways.2,4,38 NSA, an FDA approved anthelmintic drug, has been established as a highly potent small-molecule inhibitor of the STAT3 signaling pathway acting against stem-like breast cancer cells.2,45,54 Learning from these previous studies, the purpose of this study was tuned to determine the ability of combinatorial drug formulation including repurposed inhibitors of STAT3 (nifuroxazide), topoisomerase-II-activation-pathway (amonafide) and NFκb (pentoxifylline) to selectively target the TNBC subtype of breast cancer cells. We also propose a novel solution for their controlled delivery using a phenotypically screened multi-compartmental carbon dot system. We anticipate that a controlled delivery of this combinatorial drug formulation would stimulate synergism against TNBCs and suppress drug resistance through distinct mechanisms of action.
Materials and Methods
Materials
Sucrose, Poly (ethylene glycol) (PEG, average M n = 400, M n = 10,000 and M n = 20,000), Polyethylenimine, branched (PEI, average M w ≈ 25,000), Sodium azide (NaN3, ≥99.5%), Nystatin, 2-Deoxy-d-Glucose (DOG, ≥98%), Pentoxifylline were purchased from Sigma-Aldrich (MO, USA) and used without further purification. Polyethylene glycol phosphate (PO4−, M w = 5000) was purchased from Chemicell (Berlin, Germany). Methoxy PEG Thiol (SH, ≥97%, M w = 20,000) was purchased from JenKem Tech. (TX, USA). Polyethylene glycol monomethyl ether mesylate (SO3H, M w = 5000), Chlorpromazine (CPM, 97%), Amonafide were purchased from Santa Cruz Biotechnology (TX, USA). Nifuroxazide was purchased from A.K.Scientific (Union City, CA, USA). Lecithin was purchased from Coatsome, NOF America Corporation, CA, USA. Dynasore (>99%) was purchased from Fisher Scientific (PA, USA). The 0.2 and 0.45 μM filters (Millex, Merck Millipore Ltd., Tullagreen, Carrigtwohill, County Cork, Ireland) were used wherever applicable.
Preparation of Pristine Carbon Nanoparticles
Pristine CNPs were prepared by dissolving 250 mg of agave nectar in 2 ml of nanopure water (0.2 μM, 18 MΩ cm). Then the aqueous solution was heated on a hot plate at 270 °C for roughly 30 min and re-suspended in 4 ml of nanopure water. Followed by 20 min probe sonication (Q700™, Qsonica Sonicators, CT, USA) (Amp:1, On: 2 sec, Off: 1 sec), the solution was passed through a syringe filter with a 0.2 μm pore size (Millex®, Merck Millipore Ltd., County Cork, Ireland) prior to use.
Preparation of Carbon Nanoparticles by Pre-passivation
A mixture of agave nectar and polymer (PEGylated CNPs, CNP PO4, CNP SO3H, CNP SH) was taken in glass sample vial of 20 mL capacity. To this, nanopure water was added and the solution was mixed well to form a homogenous mixture. The concentration ratio of agave nectar to polymeric passivating agent was maintained at 1:1 (w/w). For the nucleation process, a hot plate surface was maintained at 270 °C while glass vials were kept un-capped to allow the slow evaporation of water. The temperature was maintained for around 30 mins, which led to a generation of brownish to black mass (varies with passivating agent). The reaction mixture changed color from light yellow to dark brown to black along the course of the heating process. The as-synthesized particle mass was suspended in 4 mL of nanopure water followed by 20 min probe-sonication (Q700™, Qsonica Sonicators, CT, USA) (Amp: 1, on: 2 sec, off: 1 sec). Sonicated suspension was centrifuged at 12,000×g for 20 min, followed by collecting the supernatant by filtering through a 0.22 μm syringe filter.
Preparation of Carbon Nanoparticles by Post-passivation
To make CNP-PEI, 10 μL of branched PEI solution (1 mg/10 μL) were added to 1 mg of CNPs. After vortexed and left at room temperature for 30 min, the solution was centrifuged for 30 min at 75,000 rpm at 4 °C (Optima™ MAX-XP Ultracentrifuge, Beckman-Coulter, CA, USA) to remove the suspension. The collected pellet was then redispersed in nanopure water and probe-sonicated for 2 min (Amp: 1, On: 2 sec, Off: 1 sec). All CNP formulations were made same concentration (5 mg/ml) before using for the study. Similar techniques were adopted for the preparation of CNP PolyLys.
Physico-chemical Characterization
Hydrodynamic diameters and zeta potential of CNPs were determined by a Malvern Zetasizer Nano ZS90 (Malvern Instruments Ltd., Worcestershire, UK) in hydrated state. The averaged hydrodynamic diameter was obtained from the peak values of the intensity-weighted distribution with n ≥ 3 and results were reported as mean ± standard deviation. Micrographs of transmission electron microscopy (TEM) was collected by JEOL 2010 LaB6 (JEOL Ltd., Tokyo, Japan) at an acceleration voltage of 200 kV. Samples were deposited on 200-mesh Quantifoil holey carbon grids (Structure Probe, Inc., PA, USA) to visualize their anhydrous sizes and morphology. For FT-IR studies, an aqueous suspension of the CNP-Tri Comb was dried onto a MirrIR IR-reflective glass slides (Kelvey Technologies, Chesterland, Ohio, USA) for Fourier Transform Infrared (FT-IR) measurements using a Nicolet Nexus 670 FT-IR (Fredrick Seitz Material Research Laboratories (FSMRL), Urbana, Illinois, USA). For each measurement 100 × 100 μm2 images were collected at 1 cm-1 spectra resolution with 64 scans per pixel and a 25 × 25 μm2 pixel size and individual spectra were corrected for atmospheric contributions.
Inhibitor Studies
HCC 1806 cells (105) were plated in 96 well plates. Cells were grown for 24 h before being incubated with different endocytic inhibitors. Inhibitor formulations were made with reconstituted medium having sodium azide, DOG (deoxyglucose), CPM (chlorpromazine), Nystatin and Dynasore at a concentration of 10 μM, 50 μM, 28 μM, 180 nM and 80 μM, respectively, which were used as described in some previous reports.32,44 Cells were incubated with inhibitors for 1 h at ambient condition. Inhibitors were then replaced with CNPs suspensions in reconstituted medium at concentrations of 5% (v/v). All the treatments were performed in triplicate. Cells that had only inhibitor treatments were considered as negative controls whereas cells treated with only CNP formulations without any pre-inhibitor treatment were used as positive controls. The cytotoxic effect of CNPs were investigated with a 3-(4,5-dime thylthiazole-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Sigma-Aldrich, MO, USA). Cells were further grown for 44 h and at the end of incubation, a 20 μL (5 mg/mL) of MTT solution was added to each well and cells were further incubated for another 4.5 h. After incubation, media were aspirated and 200 μL of dimethyl sulfoxide (DMSO, ≥99%, MP Biomedicals, USA) was added to dissolve formazan crystals. Absorption of the samples was determined by Synergy HT (BioTek, USA) with a reference wavelength of 592 nm.
Endocytic Blockers to Decrease Cell Internalization of CNPs
Response to cellular entry of CNPs against endocytic blockers were studied by fold decrease in cell death or increase in cell viability. It measures how the cell viability values for different CNP formulations, changed with/without the presence of inhibitors. The values were calculated by the following formula:
Fold increase in Cell viability = ((Cell viability of CNP in presence of inhibitor − Cell viability of CNP in absence of inhibitors)/Cell viability of CNP in absence of inhibitors).
Statistical Analysis
Statistical analysis on various biological results was carried out by using one-way ANOVA method. CNP Pristine was treated as control and compared to other CNP formulations. Fold increase for the nanoparticle formulations was plotted as mean values ± standard deviation, n = 3. Statistical analysis results were represented as * for p < 0.05, ** for p < 0.01, *** for p < 0.001 and **** for p < 0.0001.
Preparation of Drug-CNP Formulations
We used three commercial drugs, Amonafide (AMF), Pentoxifylline (PTX) and Nifuroxazide (NIFU) wherein they were used to prepare CNP-drug formulations (CNP-AMF, CNP-PTX, and CNP-NIFU) along with one formulation having CNP and all the drugs used (CNP-Triple).
For CNP-AMF Formulation
Firstly, an empty 20 ml vial was taken and its weight was noted (w1). We then took 212 mg of sucrose and 0.2 mg of AMF in the 20 ml vial and recorded the weight again (w2). After that, 950 μL of water and 50 μL of DMSO was added to the 20 ml vial, following which microwave treatment was done. For microwave treatment, P2 cycle (200 W) was applied for 5 min, 5 cycles until the solution evaporated. After this, P10 cycle (1200 W) was applied for 2 min, 2 cycles resulting in deposition of char in the vial, and after cooling it at room temperature, its weight was recorded (w3). This char was broken into small pieces using a spatula, and 4 ml of autoclaved water was added, followed by 20 min probe-sonication (Q700™, Qsonica Sonicators, CT, USA) (Amp: 1, on: 2 sec, off: 1 sec). Sonicated suspension was centrifuged at 3000 rpm for 30 min, followed by collecting the supernatant by filtering through a 0.22 μm syringe filter. The pellets were re-suspended in water and added to the charred vial, and let to dry in heat oven overnight, and weight was obtained (w4).
For CNP-PTX Formulation
Similar procedure was adopted for CNP-PTX as CNP-AMF, however, the starting amount of sucrose and PTX was 695 mg and 0.7 mg respectively.
For CNP-Nif Formulation
Similar procedure was adopted for CNP-NIFU as CNP-AMF, however, the starting amount of sucrose and PTX was 695 and 0.7 mg respectively.
For CNP-Tri Comb Formulation
Similar procedure was adopted for CNP-Triple as CNP-AMF, however, the starting amount of sucrose, AMF, PTX and NIFU was 1594, 0.2, 0.7 and 0.7 mg respectively.
Furthermore, just bare CNPs were prepared using a similar procedure as CNP-AMF, however no drug was used and starting amount of sucrose was 1594 mg.
HCC1806 Cell Line
HCC1806 is a human breast cancer cell line characterized by lack of estrogen and progesterone receptor and no her2/neu expression. The tumor the cell line was isolated from was classified as TNM Stage IIB.
In Vitro Cell Culture Method
HCC1806 cells were obtained from American Type Culture Collection (ATCC) and cultured according to the standard mammalian tissue culture protocols and sterile technique. Cells were cultured in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin.
MTT Assay Protocol
HCC1806 cells were seeded at 10,000 cells/well in RPMI-1640 medium (200 μL/well) in a 96-well plate and incubated at 37 °C and 5% CO2 for 24 h. Following formulations were used for treatments:
CNP = Carbon nanoparticle, CNP-Am = CNP loaded with Amonafide, CNP-Nif = CNP loaded with Nifuroxazide, CNP-PTX = CNP loaded with Pentoxifylline, CNP-Tri = CNP loaded with Am + Nif + PTX, Am = Amonafide in water, Am-D = Amonafide in DMSO, Nif = Nifuroxazide in water, Nif-D = Nifuroxazide in DMSO, PTX = Pentoxifylline in water, PTX-D = Pentoxifylline in DMSO.
Each treatment of the cells had concentrations ranging from 1.25 to 20 μM for Am, 4–70 μM for Nif and PTX in water, DMSO or as loaded in CNP particles. After treatment, cells were incubated for 44 h. After 44 h, 20 μL MTT solution (5 mg/mL Thiazolyl Blue Tetrazolium Bromide) was added to each well and the plate was incubated for an additional 4 h. Media was then aspirated from each well and 200 μL DMSO was added. The plate absorbance was then read at 560 nm wavelength using Gen5 Microplate Reader and Imager Software.
Protein Extraction and Western Blotting
Western blotting was used to examine the expression of pSTAT3 and a housekeeping protein β-actin. 100 mm2 dishes were plated at 1 × 106 cells per dish and treated with the previously mentioned drug formulations at concentration of 10 μM of CNP-Am and 35 of CNP-Nif and CNP-PTX and same concentration incorporated in CNP-Tri Comb after 24 h of cell growth. After treatment, cells were incubated for 48 h. Total protein content in the cells was extracted using Minute™ Total Protein Extraction Kit (Invent Biotechnologies, USA) as recommended in manufacturer’s protocol. The concentration of the protein samples was determined by a colorimetric assay using Pierce™ BCA Protein Assay Kit (ThermoFisher Scientific, USA). A 15 μg of protein extracts from various cell treatments were separated by gradient (4–20%) sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred to a nitrocellulose membrane (Bio-Rad, USA) using a wet transfer unit (company name). Protein laden membranes were blocked with 2.5% BSA in TBS-Tween buffer (0.12 M Tris-base, 1.5 M NaCl, 0.1% Tween 20) for an hour at room temperature, and incubated overnight at 4 °C with the appropriate primary antibodies; β-actin (1:3000) and pSTAT3 (1:3000). This was followed by incubation with an HRP-conjugated goat anti-mouse secondary antibody (Thermo Scientific, USA) for an hour and subsequent washing in TBST. The blots were developed using Chemiluminescent kit (Thermo Scientific) to detect the target protein as per manufacturer’s protocol.
Animal Studies
Advanced 3D cell culture techniques are known to mimic some of the aspects of the in vivo tumor environment but still, lack in the complexity found in vivo. Thus, to make a logical transition, after optimizing treatment strategies in vitro, it had to be established in animal models. The experimental protocol was approved by the Institutional Animal Care and Use Committee (IACUC), University of Illinois, Urbana–Champaign, and satisfied all University and National Institutes of Health (NIH) rules for the humane use of laboratory animals. All the animal experiments were carried out in accordance with the approved guidelines.
To evaluate the efficacy of triple drug combination loaded on CNPs in TNBCs, tumor regression experiments were performed in xenograft nude animal models. Experiment was designed to minimize the use of animals. In order to detect at least 20% difference in tumor size, we decided to generate four tumors per animal as three mice per group. Athymic mice were bought from Charles River Laboratories International, Inc. USA. Upon arrival, athymic mice were allowed one week for acclimation in animal facility. Animals were single-cage housed and had free access to food and water. Animals were housed in Carl R. Woese Institute for Genomic Biology, university of Illinois at Urbana-Champaign.
Injection of HCC-1806 in Flanks of Athymic Mice
Animals were anesthetized with isoflurane before injecting the HCC 1806 cells. Cells were suspended in matrigel (33%, v/v) and injected using a 1 mL insulin syringe tipped with a 26 gauge 1/2′′ long needle. 5 × 106 HCC 1806 cells suspended in 100 μL of matrigel:buffer mixture were injected into four sites on the flank of each mouse. Mice were monitored during recovery from the anesthesia and placed in recommended facility for allowing the tumor growth. Tumors were grown on the on the cell injection sites with 100% success rate. In the time frame of completing the experiment, grown tumors showed no significant discomfort to the mice. Mice were followed daily for signs of discomfort and any significant behavior change. Mice body weight was measured every 2nd day. Any change in physiological function or abnormal behavior including shortness of breath, unsteady gait, abnormal eating behavior, physical abnormalities, rough hair coat due to lack of grooming, or lethargy were not reported during experiment. Criteria for interventions were set up as animal body weight drop by 20% or tumor increase to 17 × 17 mm2, which was never reached during experiment. Tumor size was determined by measuring the length and width of the tumor using caliper and then calculating the tumor volume via formulae.
Treatment of Tumors
Animals were followed till tumors grew to a minimum of 5 mm × 5 mm before starting the treatment protocol. CNP-Tri Comb was injected to tumors intratumorally (n = 12 tumor) of animals (three animals) post anesthesia with isoflurane-oxygen mixture with 3–4% isoflurane gas from a vaporizer. A second tube was used to remove carbon dioxide and excess of anesthetic. All the personnel involved wore protective lab coats, face masks, sterile gloves during experimental procedures. A total of 60 μL formulation was injected to each tumor after every 48 h including 0, 2, 4 and 6th day after first injection and followed for tumor growth regression.
Results and Discussion
Physico-chemical Characterization
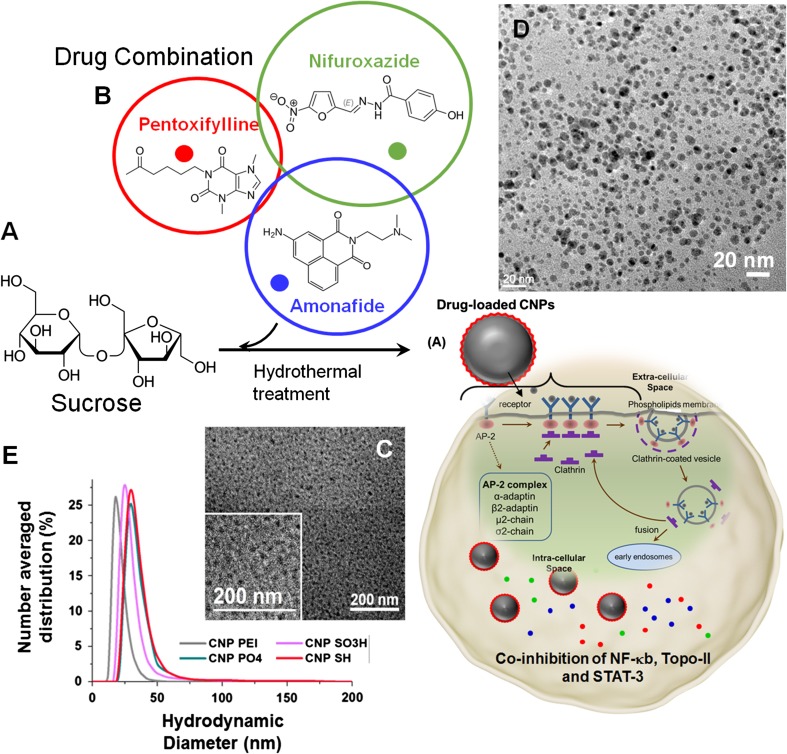
Figure 1d shows the overlaid hydrodynamic diameters of as-synthesized nanoparticles (<50 nm) while the size did not change after drug encapsulation. Anionic CNPs exhibited an electrophoretic potential of −30 ± 5 to −20 ± 2 mv after being passivated with the anionic and neutral molecules, whereas decrease in surface charge potential (+10 ± 2 mv) for cationic-molecule passivated CNPs confirm their successful surface chemistry. Anhydrous particles were studied by transmission electron microscopy (TEM). A representative TEM of combinatorically encapsulated CNPs showed a distribution of sizes to be 20 ± 5 nm (Fig. 2c), slightly smaller than its hydrodynamic size. The loss of hydration layer around surface coating while preparing TEM samples likely accounts for the slight decrease in size. The particle sizes were calculated from multiple TEM images of the same sample and an average of 100 particle count. The FT-IR spectrum of CNP-Tri Comb displayed spectra of both the carbon core as well as the drug molecules which were passivated onto it. It exhibited broad peaks for –O–H and –N–H centering on 3300 and 2900 cm−1, along with asymmetric stretching due to the presence of methyl functionalities (νCH3) at 2840 cm−1 and methylene stretching band (νCH2) at 2760 cm−1. It also exhibited strong peaks at around 2200 cm−1 (νCNO), 2090 cm−1 (νC–N) and 1710 cm−1 (νC=N) which could be due to the passivated drug molecule, PTX. We also observed peaks at 1780 cm−1 (νC = O), 1450 cm−1 (νNO2) and also around 1140 cm−1 (νC–O–C), which is presumably due to the presence of the furan ring present in the drug molecules, nifuroxazide. Presence of a tertiary amine group (νCN) in amonafide is observed in the FT-IR spectra of the triple drug formulation at around 1200 cm−1.
Figure 1.
Schematic representation of CNPs, their plausible mechanism of cellular entry, and conceptual cartoon behind co-inhibition of NF-κb, Topo-II and STAT-3. (a) Synthetic pathway to combinatorically loaded CNPs and schematic representation of CNPs entering TNBC through endocytosis pathways. (b) Chemical structures of three repurposed drug molecules, for inhibition of NF-κb, Topo-II and STAT-3. (c, d) Transmission electron microscope (TEM) images of a representative phospholipid-coated carbon nanoparticles before and after loading, respectively, (e) Overlaid hydrated state diameter of various anionic, neutral and cationic carbon nanoparticle (dynamic light scattering).
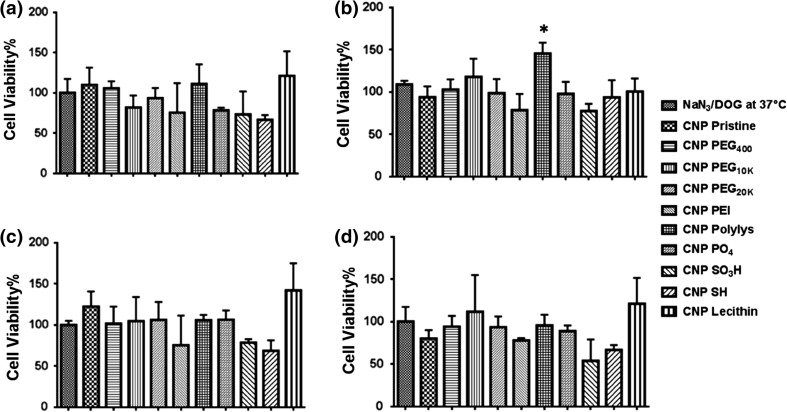
Figure 2.
MTT Assay evaluation of inhibitor efficiency of (a) NaN3/DOG at 37 °C, (b) chlorpromazine, (c) dynasore and (d) nystatin for different CNP formulations (5% v/v) for fold-increase calculations. A non-synchronized population of HCC-1806 cells (10,000) was plated in 96 well plates for 24 h before performing the treatments. Cells were first incubated with inhibitors for 1 h followed by CNP formulation addition. Experiments were performed in triplicates. Fold Increase greater than or equal to 1.30 was considered significant.
Enzymatic Degradation of CNP by Human Myeloperoxidase (hMPO)
Enzymatic degradation of CNPs were confirmed by exposing the samples in presence of human myeloperoxidase (hMPO) enzyme. Raman spectra of the CNP as treated with hMPO monitored by the change in the Raman intensity over a week. The sample with no enzyme treatment was taken as control. The enzymatic degradation can be inferred from the loss of the intensity in the D and G bands. (Figure S2) Four freeze dried CNP samples were dispersed in DPBS with the concentration of 10 mg ml−1 containing 200 mM of hydrogen peroxide. Then the samples were treated with 100 μg of hMPO with activity >50 units mg−1 at 37 °C except for one sample which was taken as control. The activity of the enzyme was impeded at 4 °C after specified time points. These samples were dried and further characterized by Raman spectroscopy using the parameters indicated above.
Inhibitor Studies
To inspect different endocytic mechanisms utilized by pristine CNP and passivated CNPs, pharmacological inhibitors were used. These small molecule inhibitors were selected in such a way that they did not exert any significant toxic effect in a short period of incubations. Furthermore, they were also not supposed to affect the actin cytoskeleton after the treatment.18,42 The essence of using these inhibitors to probe into the uptake mechanisms is that each inhibitor would only block a specific endocytic pathway of interest and consequently CNPs internalizing the cells via that pathway. This would in turn decrease the cellular internalization and subsequently increase the cell viability. A mixture of sodium azide (NaN3) and 2-deoxyglucose (DOG), were used to inhibit glycogenosis and cellular respiration respectively, via energy dependent uptake.1,21,55 Chlorpromazine (CPM), was used to probe into the clathrin dependant entry, as it is known to reduce the formation of clathrin-coated pits due to reversible translocation of clathrin and its adapter proteins from plasma membrane to intracellular vesicles.31,49,56 Dynasore has been used to understand the inhibition of dynamin (GTPase) dependency in endocytosis of cells.22,28 Finally, nystatin, a sterol-binding agent, which dismantles caveolae and cholesterol in the membrane, has been employed to study clathrin-independent inhibition of endocytosis.9,47 Fold-increase calculations were performed relative to the cell-viability measurements obtained for different CNP formulations in the presence and absence of inhibitors. The following equation was used to calculate the fold-increase values from the cell-viability measurements.
Fold-increase in cell viability = ((Cell viability of CNP in presence of inhibitor − Cell viability of CNP in absence of inhibitor)/Cell viability of CNP in absence of inhibitor.
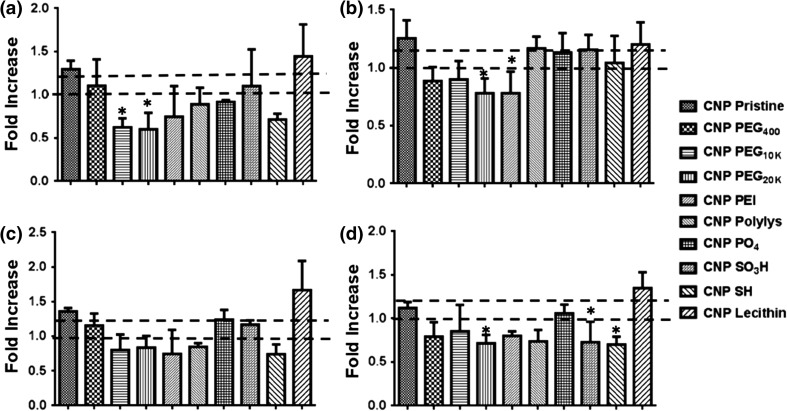
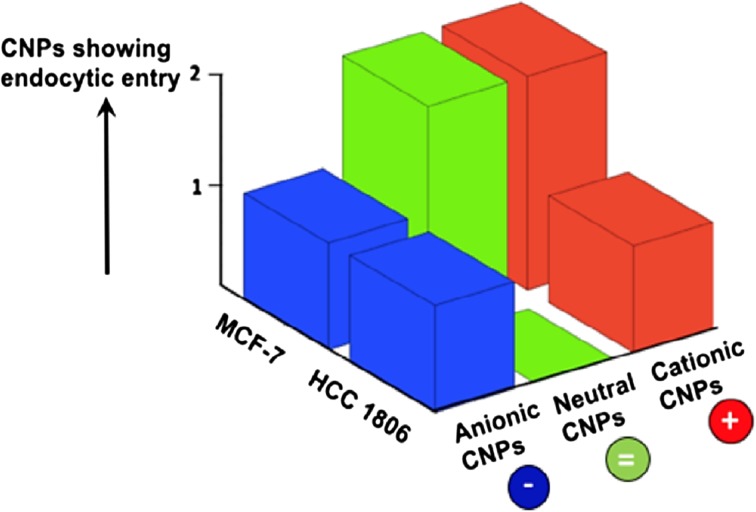
To investigate the mechanisms of entry,36,48 HCC-1806 cells were grown 24 h (10 × 103 cells/well, 96 well plate), were incubated with different inhibitors for 1 h (Table 1) followed by treatment with different formulations of CNPs.19,33,40 All the results obtained from inhibitor studies in terms of cell viability (Fig. 2) were converted to fold increase values (Fig. 3). A mean fold-increase value greater than or equal to 1.30 was considered to have significant uptake inhibition and suggested it to be the predominant internalization pathway as these fold-increase value would consequently lie on a higher confidence interval (~95%), as calculated by confidence interval calculator by Mccallum-Layton, thereby making subsequent comparison amongst rest of data sets significant. It was interesting to observe that CNP Pristine internalized into the cells using a combination of energy dependent, clathrin dependent and lipid-raft pathway whereas CNP passivated with lecithin internalized the cells through all the four inspected endocytic pathways. However, since the variability in the calculation of fold-increase values for pristine CNP is lesser as compared to CNP Lecithin, it would have a higher probability of crossing the endocytic barrier. Remaining CNPs did not show any uptake for the investigated pathways. Parametric evaluation of surface charge and change from non-Triple negative breast cancer (TNBC), MCF-7 to TNBC, HCC 1806 effects on internalization of CNPs can also be appreciated in a 3D map (Fig. 4). Results indicated that with the progression of non-TNBC to TNBC, less number of neutral and cationic CNPs show tendency for internalization via endocytic routes. For a TNBC or non-TNBC cell-line, an initial screening of CNPs having different surface functionalities were done using the inhibitor assay. A combination of cell type (non-TNBC and TNBC) and CNP surface functionality type were assessed using the inhibitor assay to reach at a conclusion that as the cancer progresses from non-TNBC to TNBC, less number of neutral (CNP PEG 400, CNP PEG10K, CNP PEG20K and CNP Lec.) and cationic CNP (CNP PEI) show tendency for internalization via endocytic routes.
Table 1.
Inhibitors and their treatment conditions for achieving endocytic inhibition by defined working mechanism.
| Endocytic inhibitors | Working mechanism | Working concentration | Incubation time (hours) |
|---|---|---|---|
| NaN3/DOG (37°C) | ATP depletion | 10 mM/50 mM | 1 |
| NaN3/DOG (4°C) | ATP depletion + slows down metabolism | 10 mM/50 mM | 1 |
| CPM | Inhibits clathrin mediated endocytosis | 28 nM | 1 |
| Nystatin | Inhibits lipid-raft mediated endocytosis | 180 nM | 1 |
| Dynasore | Inhibits dynamin-dependent endocytosis | 80 μM | 1 |
Figure 3.
Fold-increase values for different CNP formulations (5% v/v) in presence of (a) NaN3/DOG at 37°C, (b) chlorpromazine, (c) dynasore and (d) was obtained via the cell-viability measurements. From the Mccallum-Layton calculator, a cut-off fold increase value of 1.30 was set, as it would lie on a higher confidence interval (~95%), thereby making subsequent comparisons amongst rest of data sets significant. Fold Increase greater than or equal to 1.30 was considered significant.
Figure 4.
Parametric evaluation of surface charge and change from non-triple negative breast cancer (TNBC), MCF-7 to TNBC, HCC 1806 effects on internalization of CNPs. As the cancer progresses non-TNBC to TNBC, less number of neutral and cationic CNPs show tendency for internalization via endocytic routes.
Effect of Trident Drug Combination Selectively on Triple Negative Cell Population
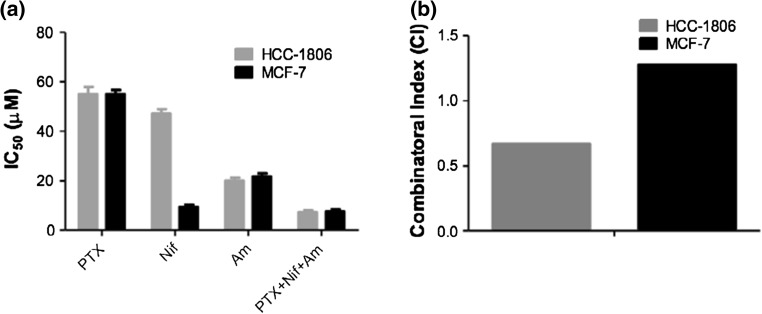
To verify the selective effect of drug cocktail on TNBC cells i.e. HCC 1806, MTT assay performed on TNBC HCC-1806 and results were compared with non-TNBC ER (+) MCF-7 cells. Cytotoxicity was used as a measure for cellular entry due to its functional nature. It was considered as a better option as probability of getting false positive results could be minimized and can be neglected. Cells were treated with 25 μM concentration of NfF-kB modulator PTX, STAT3 modulator Nif and Topo-II modulator Am as individual drugs or in triple combination of PTX + Nif + Am for 48 h before performing the MTT assay. IC50 values of CNP-drug, free drug, CNP-Tri and free Tri combination were calculated by plotting concentration vs. cell viability, with concentrations ranging from 12.5 to 50 μM. From this data, the concentration at which cell viability reached 50% was extrapolated, indicating the IC50 value. A comparison of IC50 for HCC-1806 and MCF-7 cells showed no particular selectivity for PTX, Am and PTX + Nif + Am mixture treatments though Nif introduced a significant change in IC50 values as ~10 μM in MCF-7 compared to ~50 μM in HCC1806 (Fig. 5a). A combinatorial index (CI) calculation on triple drug mixture (PTX + Nif + Am) compared to individual drugs was found to be at least two times more responsive for TNBC HCC-1806 cells (~0.6) compared to non-TNBC MCF-7 cells (~1.2) (Fig. 5b). A lower than one CI value (<1) represented the synergistic effect of drug mixture while higher than one (<1) representative of non-communicating drug molecules and probably their action mechanism pathways in that particular cell lines. This was clear from the results that this drug mixture (PTX + Nif + Am) was synergistic in TNBC HCC1806 cells. Although three representative cell lines were used for the study, a significantly high number of CNPs with different surface functionality were scanned to reach a conclusion. Therefore, a combination of cell type and parametrically screened CNP surface functionality makes it substantial to reach at a conclusion.
Figure 5.
MTT assay performed on TNBC HCC-1806 and non-TNBC ER (+) MCF-7 cells. Cells were incubated for 48 h before performing the MTT assay. (a) Comparison of IC50 for HCC-1806 and MCF-7 cells when being treated with NfF-kB modulator PTX, STAT3 modulator Nif and Topo-II modulator Am as individual drugs or in triple combination of PTX + Nif + Am solubilized in DMSO. (b) Combinatorial index (CI) calculation on triple drug mixture compared to individual drugs as found to be at least two times more responsive for TNBC HCC-1806 cells compared to non-TNBC MCF-7 cells.
Effect of Trident Drug Combination Incorporation in CNPs on Cell Growth Density, Morphology and Viability
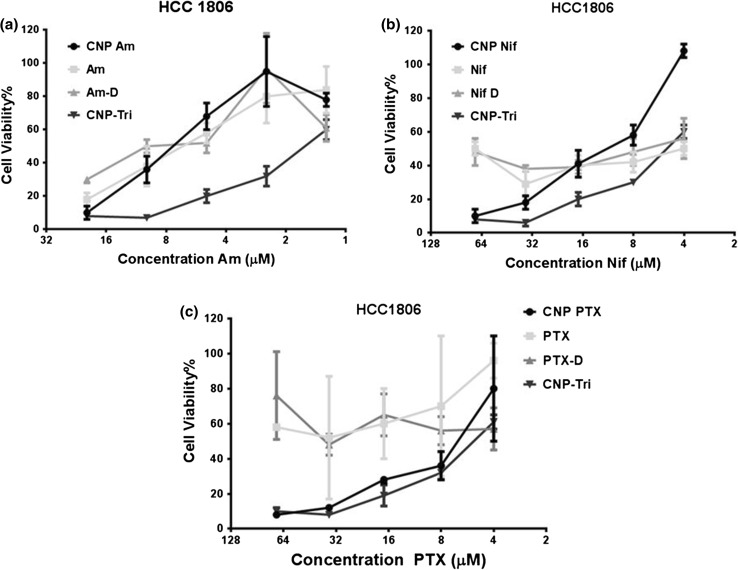
MTT assay performed in TNBC cells, HCC 1806 after incubation with various drug formulations for 48 h were analyzed in terms of cell viability efficiency and their respective effects on cell growth density and morphology. Experiments were performed with 4 to 70 μM concentration of PTX and Nif while Am from 1.25 to 20 μM as optimized for effective dose range in water, DMSO or loaded to CNPs in individual or combination form. Different solution forms (water and DMSO solvents) were used to evaluate effect of solubility of drugs on cell viability (Fig. 6). It was found the CNP-Tri comb was the best formulation with maximum cell death. Additionally, all CNP loaded drugs were better than free drugs in killing the cells at same concentration. DMSO dissolved drug molecules were found be more effective than water solubilized one probably due to improved solubility in DMSO. Am was found better than Nif and PTX in same order all across the formulation in different solvent or loaded to CNP.
Figure 6.
MTT assay performed in TNBC cells, HCC 1806 after incubation with various drug formulations for 48 h. PTX and Nif were used at a concentration range of 4–70 μM while Am from 1.25 to 20 μM as optimized for effective dose range. Concentration of drugs were same in water, DMSO or loaded to CNPs in individual or combination form.
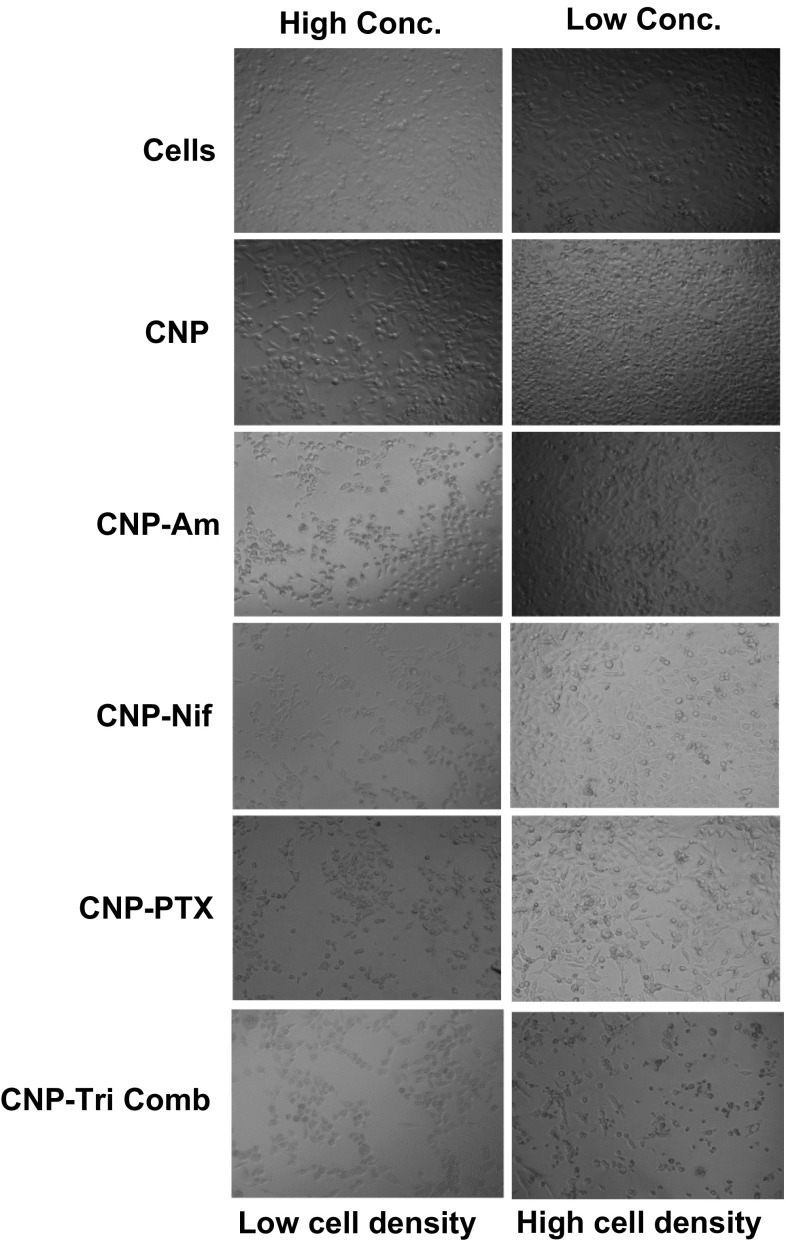
Effects were further observed by studying cell morphology and growth density changes in TNBC cells, HCC 1806 after incubation with various drug formulations for 48 h. PTX and Nif were used at a concentration range of 4 μM (low conc.) 70 μM (high conc) while Am from 1.25 (low conc.) and 20 μM (high conc) (Fig. 7). A clear loss in cell density was reported for high concentrations of cells treated with CNP-Tri Comb with minimum effect in CNP-PTX treatments. Even at lower concentration CNP-Tri Comb was best formulation causing maximum loss in cell density and morphology.
Figure 7.
Cell morphology and growth density study performed in TNBC cells, HCC 1806 after incubation with various drug formulations for 48 h. PTX and Nif were used at a concentration range of 4 μM (low conc.) 70 μM (high conc.) while Am from 1.25 (low conc.) and 20 μM (high conc.).
Protein Expression as Indication of Effective Drug Combination
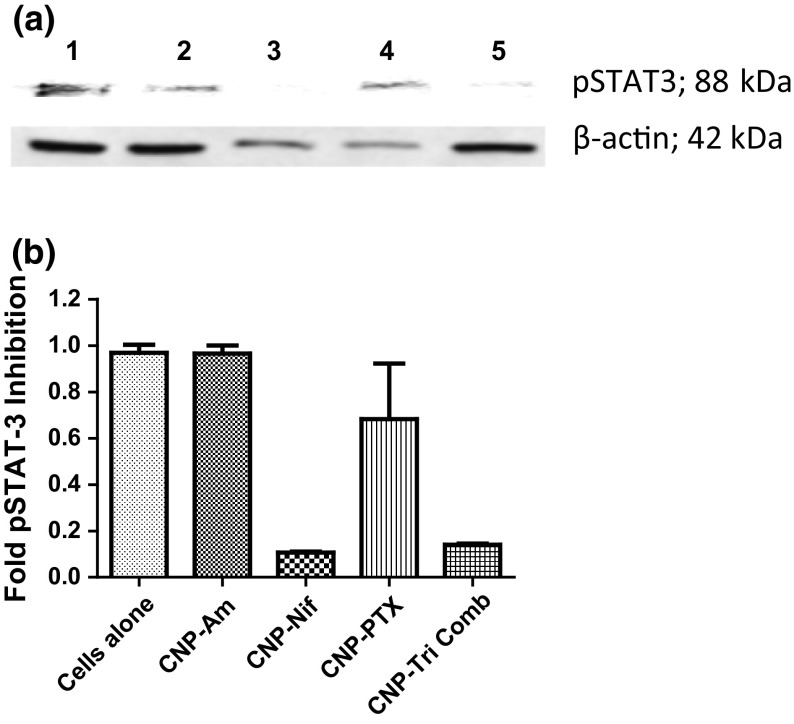
Nifuroxazide inhibits STAT3 by blocking its dimerization and in turn phosphorylation which is an active form of STAT3 involved in the survival of cancer cells including TNBCs. To validate this, western blot analysis was performed to determine the effect of CNP-Tri Comb on the expression of phosphorylated STAT3. Western blot showing expression of the pSTAT3 protein and β-actin extracted from cells treatment with 1: No treatment, 2: CNP-Am, 3: CNP-Nif, 4: CNP-PTX and 5: CNP-Tri Comb (Fig. 8a). It showed that CNP-Tri Comb treatment induced down regulation of phosphorylated STAT3 along with similar effects from CNP-Nif treatments at the same concentration of Nif in CNP-Nif or form of triple drug mixture loaded in CNPs. On the other end, minimum to no difference in pSTAT3 protein expression was noticed between control and treated samples with CNP-PTX and CNP-Am. Gel quantification of expressed pSTAT3 protein respect to unaffected home protein β-actin was performed to reveal an approx. downregulation of 90% pSTAT3 for CNP-Nif and CNP-Tri Comb compare to just 30% for CNP-PTX and almost no effect by CNP-Am treatments (Fig. 8b). Thus, it signifies the partial involvement of PTX, a NF-κB modulator, in STAT3 signaling but none by TOPO-II modulator (Am) to start with but CI index calculation supported the synergism achieved by triple combination.
Figure 8.
Effect of triple drug mixture treatment on HCC 1806 cells protein expression. (a) Western blot showing expression of pSTAT3 protein and β-actin extracted from cells treatment with 1: No treatment, 2: CNP-Am, 3: CNP-Nif, 4: CNP-PTX and 5: CNP-Tri Comb. (b) Quantification of expressed pSTAT3 protein respect to unaffected housekeeping protein β-actin.
Effect of CNP-Tri Comb on TNBC Xenograft Tumors
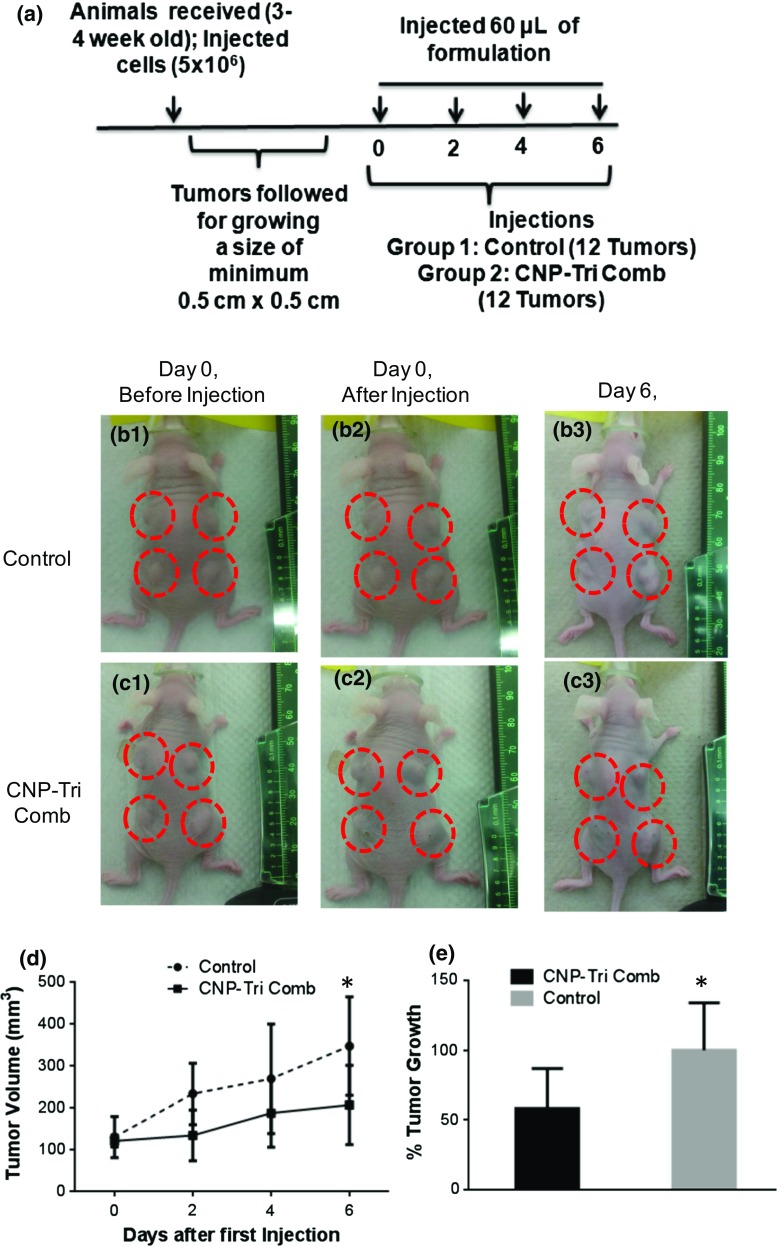
To evaluate the efficacy of triple drug combination loaded on CNPs in TNBCs, tumor regression experiments were performed in xenograft nude animal models. The drug cocktail showed maximum effect when encapsulated to CNPs in vitro. The effect was considerably better than drug combination alone and CNPs alone with no significant effect on cell death in all in vitro studies. As identified during in vitro studies, CNP alone were not significant toxic and did not contribute to overall effects of drug combination. The results from the in vitro studies were considered and in vivo experiments were designed to minimize the use of vertebrate animals. In order to detect at least 20% difference in tumor size, we decided to generate 4 tumors per animal as 3 mice per group and followed by tumor volume all along the experiment (Fig. 9a). Tumors grown to a minimum size of 5 × 5 mm2 (Figs. 9b1 and 9c1) were intratumorally injected with phosphate buffer DPBS (Figs. 9b1 and 9b3) and CNP-Tri Comb (Figs. 9c1 and 9c3) in 60 μL volume on day 0, 2, 4 and 6 days. The growth was followed by caliper and volume was calculated to compare the regression effect. The regression of tumor growth was found to be biologically significant for CNP-Tri Comb (Fig. 9c3) compared to tumors treated with buffer (Fig. 9b3) and could be summarized as tumor growth curves with time (Fig. 9d). A percentage changes in tumor size after treatment with buffer (control) and CNP-Tri Comb was significantly visible at around 100% more growth in buffer control tumors compared CNP-Tri Comb treated tumors with in six days of first treatment (Fig. 9e). Here * represent the p < 0.05 and a bio-statistically significant difference in tumor size treated with CNP-Tri Comb compared to buffer control as obtained after performed t test. We recognize the biological barriers and complexity of systemic circulation presented by intravenous administration route. Thus, intratumoral administration route was chosen for simplification and to reach logical conclusions. An alternative administration route is therefore out of the scope of this immediate study but will be chosen as part of a future extension of this project.
Figure 9.
In vivo results on xenograft mouse model. (a) Timeline of experiment; (b, c) representative animals with tumors before and after treatment with (b) DPBS; (c) CNP-Tri Comb. B1 and C1 represent animals with tumors before injections and B2 and C2 after injections while smaller size of tumors were seen in C3 compared to B3 at 6th day of measurement. (d) Tumor growth curves with time and (e) fold changes after treatment with buffer (control) and CNP-Tri Comb. * represent the p < 0.05 and a bio-statistically significant difference in tumor size treated with CNP-Tri Comb compared to buffer control as obtained after performed t test.
Conclusions
In this work, we demonstrated a combinatorial drug formulation comprising DNA damage repair agents, along with inhibitors for STAT-3 exhibit synergistic effect for triple negative breast cancer. We demonstrate that phenotypically stratified multi-compartmental nanoparticle is highly effective in delivering a novel combinatorial triple drug formulation for synergistic regression of TNBC in vitro and in vivo. Multicompartmental carbon nanoparticles were then parametrically assessed based on size, charge (positive/negative/neutral) and chemistry (functionalities) to study their likelihood of crossing endocytic barriers from phenotypical standpoint in various TNBC lines. Interestingly, a combination of clathrin mediated, energy and dynamin dependent pathways were predominant for sulfonated nanoparticles, whereas phospholipid particles followed all the investigated endocytic pathways. An exactitude ‘omics’ approach helps to predict that phospholipid encapsulated-particles will predominantly accumulate in TNBC comprising the drug-‘cocktail’. After an exhaustive screening process for different CNPs based on their surface functionalities, best “candidate” was chosen for in vivo studies. However, by testing only one particle formulation, we could only draw conclusions on the therapeutic effect of this particular formulation. The added shortcoming of this approach was inability to conclude is the effect of free drug cocktail was considerably different than CNP-Tri comb in vivo. We investigated the gene and protein expression effects inducing synergistic effect and simultaneously suppressing drug resistance through distinct mechanisms of action. Overall, this work is aimed at identifying specific surface chemistries on carbon nanoparticles to facilitate its entry in TNBC and extrapolate such information in a real-world situation of cancer tumors with mixed population of TNBC and non-TNBC. To the best of our knowledge, this is one of the earliest attempt to stratifying nanotherapeutics based on cancer phenotypes. We recognize the complexity of the disease and realize that optimization can only be achieved by conducting multiple studies beyond this preliminary communication. Successful outcome of these studies may have significant translational impact for combinatorial nanotherapeutics.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Materials characterizations were done at Frederick Seitz Materials Research Laboratory, UIUC. We would like to thank Fatemeh Ostadhossein and Enrique Daza for help with the biodegradability studies.
Funding
Funding from UIUC, National Science Foundation, Michael Reese Foundation and Children’s Discovery Institute are acknowledged.
Conflict of Interest
Prof. Pan has received research grants from NIH, NSF, American Heart Association, Children’s Discovery Institute, Michael Reese Foundation and other agencies. Prof. Pan is the founder or co-founder of three start-up companies. None of these entities supported this research. Taylor Kampert, Indrajit Srivastava and Santosh Misra declare that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. This article does not contain any studies with human participants performed by any of the authors.
Footnotes
Prof. Dipanjan Pan, MS, PhD, is a recognized expert in nanomedicine, molecular imaging and drug delivery. He is an Associate Professor in Bioengineering and Materials Science and Engineering and Institute of Sustainability in Energy and Environment in University of Illinois, Urbana-Champaign. He holds a full faculty position with Beckman Institute for Advanced Science and Technology, University of Illinois Cancer Center and Carle Foundation Hospital. Administratively he directs the Professional Masters in Engineering Program in Bioengineering in the College of Engineering. He is also a course director for the newly founded engineering inspired medical school at the University of Illinois. Prior to coming to Illinois, he was an Assistant Professor in Medicine, Research in Washington University School of Medicine, St Louis. His research is highly collaborative and interdisciplinary centering on the development of novel materials for biomedical applications and targeted therapies for stem-like cancer cell and phenotypically screened nanomedicine platforms. Over the years, this research has resulted in more than 100 high impact peer reviewed publications in scientific journals, numerous conference abstracts and has been supported by external funding from NIH, NSF, DoD, American Heart Association and other sources. Prof. Pan edited and co-written two books published from Taylor and Francis (Nanomedicine: A Soft Matter Perspective, ISBN-13: 978-1466572829) and Springer (Personalized Medicine with a Nanochemistry Twist: Nanomedicine (Topics in Medicinal Chemistry, ISBN-13: 978-3319335445). He holds multiple patents, several ongoing clinical trials and is the founder of three University based early start-ups. He is the CEO/President for a biotechnology start-up Vitruvian Biotech dedicated to develop novel image guided therapies. He also co-founded InnSight Technologies dedicated to nanotechnology based application for ocular diseases. His other company Kalocyte, which he cofounded with his clinical collaborators, develops oxygen therapeutics. Technology developed in his laboratories has been licensed for commercial development multiple times. He serves frequently as study section review board member for NIH, CDMRP (DoD) and multiple review committee member for American Heart Association. In 2016 he received Nano-Micro Letter (NML) Researcher award. He is an elected fellow of Royal Society of Chemistry, and a Fellow of American Heart Association. Professor Pan is an editorial board member of Scientific Reports (Nature Publishing) and an editorial advisory board member of Molecular Pharmaceutics (ACS) .
.
This article is part of the 2017 CMBE Young Innovators special issue.
References
- 1.Allen C, Yu Y, Eisenberg A, Maysinger D. Cellular internalization of PCL20-b-PEO44 block copolymer micelles. Biochim. Biophys. Acta. 1999;1421:32. doi: 10.1016/S0005-2736(99)00108-X. [DOI] [PubMed] [Google Scholar]
- 2.Andersson BS, Beran M, Bakic M, Silberman LE, Newman RA, Zwelling LA. In vitro toxicity and DNA cleaving capacity of benzisoquinonlinedione (nafimide; NSC 308847) in human leukemia. Cancer Res. 1040;1987:47. [PubMed] [Google Scholar]
- 3.Arvizo RR, Rana S, Miranda OR, Bhattacharya R, Rotello VM, Mukherjee P. Mechanism of anti-angiogenic property of gold nanoparticles: role of nanoparticle size and surface charge. Nanomedicine. 2011;7:580. doi: 10.1016/j.nano.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brana MF, Sanz AM. Synthesis and cytostatic activity of benz[de]isoqinolin-1,3-diones. Structure-activity relationships. Eur. J. Med. Chem. 1981;16:207. [Google Scholar]
- 5.Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J. Clin. Oncol. 2005;23:7350. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- 6.Brigger I, Dubernet C, Couvreur P. Nanoparticles in cancer therapy and diagnosis. Adv. Drug Deliv. Rev. 2012;64:24. doi: 10.1016/j.addr.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Che-Ming JH, Aryal S, Zhang L. Nanoparticle-assisted combination therapies for effective cancer treatment. Ther. Deliv. 2010;1:323. doi: 10.4155/tde.10.13. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Tian F, Zhang X, Wang W. Internalization pathways of nanoparticles and their interaction with a vesicle. Soft Matter. 2013;9:7592. doi: 10.1039/c3sm50931a. [DOI] [Google Scholar]
- 9.Chen Y, Wang S, Lu X, Zhang H, Fu Y, Luo Y. Cholesterol sequestration by nystatin enhances the uptake and activity of endostatin in endothelium via regulating distinct endocytic pathways. Blood. 2011;117:6392. doi: 10.1182/blood-2010-12-322867. [DOI] [PubMed] [Google Scholar]
- 10.Cho EC, Xie JW, Wurm PA, Xia YN. Understanding the role of surface charges in cellular adsorption versus internalization by selectively removing gold nanoparticles on the cell surface with I2/KI etchant. Nano Lett. 1080;2009:9. doi: 10.1021/nl803487r. [DOI] [PubMed] [Google Scholar]
- 11.Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 12.Crown J, O’Shaughnessy J, Gullo G. Emerging targeted therapies in triple-negative breast cancer. Ann. Oncol. 2012;23(vi5):6. doi: 10.1093/annonc/mds196. [DOI] [PubMed] [Google Scholar]
- 13.Davis ME, Chen ZG, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat. Rev. Drug Discov. 2008;7:771. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 14.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Ann. Rev. Biochem. 2009;78:857. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 15.Elhissi AMA, Ahmed W, Hassan IU, Dhanak VR, D’Emanuele A. Carbon nanotubes in cancer therapy and drug delivery. J. Drug Deliv. 2012;2012:837327. doi: 10.1155/2012/837327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farokhzad OC, Langer R. Impact of nanotechnology on drug delivery. ACS Nano. 2009;3(1):16. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari M. Actin reorganization contributes to loss of cell adhesion in pemphigus vulgaris. Nat. Rev. Cancer. 2005;5:161. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 18.Gliem M, Heupel W-M, Spindler V, Harms GS, Waschke J. Actin reorganization contributes to loss of cell adhesion in pemphigus vulgaris. Am. J. Physiol. 2010;299(3):606. doi: 10.1152/ajpcell.00075.2010. [DOI] [PubMed] [Google Scholar]
- 19.He, H., L. A. Pham-Huy, P. Dramou, D. Xiao, P. Zuo, and C. Pham-Huy. Carbon nanotubes: applications in pharmacy and medicine. BioMed Res. Int., 2013 [DOI] [PMC free article] [PubMed]
- 20.Ivanov AI. Pharmacological inhibition of endocytic pathways: is it specific enough to be useful? Methods Mol. Biol. 2008;440:15. doi: 10.1007/978-1-59745-178-9_2. [DOI] [PubMed] [Google Scholar]
- 21.Kim JS, Yoon TJ, Yu KN, Noh MS, Woo M, Kim BG, Lee KH, Sohn BH, Park SB, Lee JK, Cho MH. Selective targeting of gold nanorods at the mitochondria of cancer cells: implications of cancer therapy. J. Vet. Sci. 2006;11:772. [Google Scholar]
- 22.Kirchhausen T, Macia E, Pelish HE. Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol. 2008;438:77. doi: 10.1016/S0076-6879(07)38006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kostarelos K, Lacerda L, Pastorin G, Wi W, Wieckowski S, Luangsivilay J, Godefroy S, Pantarotto D, Briand JP, Muller S, Prato M, Bianco A. Cellular uptake of functionalized carbon nanotubes is independent of functional group and cell type. Nat. Nanotechnol. 2007;2:108. doi: 10.1038/nnano.2006.209. [DOI] [PubMed] [Google Scholar]
- 24.Kumari S, Swetha MG, Mayor S. Endocytosis unplugged: multiple ways to enter the cell. Cell Res. 2010;20:256. doi: 10.1038/cr.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang M, Lin IC, Whittaker MR, Minchin RF, Monteiro MJ, Toth I. Cellular uptake of densely packed polymer coatings on gold nanoparticles. ACS Nano. 2010;4:403. doi: 10.1021/nn9011237. [DOI] [PubMed] [Google Scholar]
- 26.Liu Z, Liang XJ. Nano-carbons as theranostics. Theranostic. 2012;2(3):235. doi: 10.7150/thno.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson AK. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. USA. 2008;105:14265. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macia E, Ehrlich M, Massol R, Boucrot E, Bruneer C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell. 2006;10(6):839. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Madani SY, Naderi N, Dissanayake O, Tan A, Seifalian AM. A new era of cancer treatmemt: carbon nanotubes as drug delivery tools. Int. J. Nanomed. 2011;6:2963–2979. doi: 10.2147/IJN.S16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agents smancs. Cancer Res. 1986;46:6387. [PubMed] [Google Scholar]
- 31.Mc Mahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2011;12:517. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 32.Michael MD, Christopher BC, Jessica B, Kelly S, Linda W, Gary SK, Vita F, David G, Robert G, Chris H. Optimized high-throughtput microRNA expression profiling provides novel biomarker assessment of clinical prostrate and breast cancer biopsies. Mol. Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Misra SK, Chang H-H, Mukherjee P, Tiwari S, Ohoka A, Pan D. Regulating biocompatibility of carbon spheres via defined nanoscale chemistry and a careful selection of surface functionalites. Sci. Rep. 2015;5:14986. doi: 10.1038/srep14986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misra SK, Kus J, Kim S, Pan D. Nanoscopic poly-DNA-cleaver for breast cancer regression with induced oxidative damage. Mol. Pharm. 1976;2014:33. doi: 10.1021/mp500444c. [DOI] [PubMed] [Google Scholar]
- 35.Misra SK, Mukherjee P, Chang HH, Tiwari S, Gryka M, Bhargava R, Pan D. Multi-functionality redefined with colloidal carotene carbon nanoparticles for synchronized chemical imaging, enriched cellular uptake and therapy. Sci Rep. 2016;6:29299. doi: 10.1038/srep29299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misra SK, Ostadhossein F, Daza E, Johnson EV, Pan D. Hyperspectral imaging offers visual and quantitative evidence of drug release from Zwitterionic-Phospholipid-Nanocarbon when concurrently tracked in 3D intracellular space. Adv. Funct. Mater. 2016;26:8031. doi: 10.1002/adfm.201602966. [DOI] [Google Scholar]
- 37.Misra SK, Srivastava I, Tripathi I, Daza E, Ostadhossein F, Pan D. Macromolecularly “caged” carbon nanoparticle for intracellular trafficking via switchable photoluminescence. J. Am. Chem. Soc. 2017;139(5):1746–1749. doi: 10.1021/jacs.6b11595. [DOI] [PubMed] [Google Scholar]
- 38.Misra SK, Wang X, Srivastava I, Imgruet MK, Graff RW, Ohoka A, Kampert TL, Gao H, Pan D. Combinatorial therapy for triple negative breast cancer using hyperstar polymer-based nanoparticles. Chem. Commun. 2005;51:16710. doi: 10.1039/C5CC07709E. [DOI] [PubMed] [Google Scholar]
- 39.Misra SK, Ye M, Kim S, Pan D. Highly efficient anti-cancer therapy using scorpion ‘NanoVenin’. Chem. Commun. 2014;50:13220. doi: 10.1039/C4CC04748F. [DOI] [PubMed] [Google Scholar]
- 40.Mukherjee P, Misra SK, Gryka MC, Chang H-H, Tiwari S, Wilson WL, Scott JW, Bhargava R, Pan D. Tunable luminescent carbon nanospheres with well-defined nanoscale chemistry for synchronized imaging and therapy. Small. 2016;36:4691. doi: 10.1002/smll.201500728. [DOI] [PubMed] [Google Scholar]
- 41.Ostadhossein F, Pan D. Functional carbon nanodots for multiscale imaging and therapy. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017;9(3):e1436. doi: 10.1002/wnan.1436. [DOI] [PubMed] [Google Scholar]
- 42.Papakonstanti EA, Stournaras C. Cell responses regulated by early reorganization of actin cytoskeleton. FEBS Lett. 2008;582(14):2120. doi: 10.1016/j.febslet.2008.02.064. [DOI] [PubMed] [Google Scholar]
- 43.Qin W, Ding D, Liu J, Yuan WZ, Hu Y, Liu B, Tang BZ. Biocompatible nanoparticles with aggregation-induced emission characteristics as far-red/near-infrared fluorescent bioprobes for in vitro and in vivo imaging applications. Adv. Funct. Mater. 2011;22(4):771. doi: 10.1002/adfm.201102191. [DOI] [Google Scholar]
- 44.Rakha EA, El-Sayed ME, Green AR, Lee AHS, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25. doi: 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- 45.Ren X, Duan L, He Q, Zhang Z, Zhou Y, Wu D, Pan J, Pei D, Ding K. Identification of niclosamide as a new small-molecule inhibitor of the STAT3 signalling pathway. ACS Med. Chem. Lett. 2010;1:454. doi: 10.1021/ml100146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saha K, Kim ST, Yan B, Miranda OR, Alfonso FS, Shlosman D, Rotello VM. Surface functionality of nanoparticles determines cellular uptake mechanisms in mammalian cells. Small. 2013;9(2):300. doi: 10.1002/smll.201201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sigismund JS, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, DiFiore PP, Polo S. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. USA. 2005;102:2760. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srivastava I, Misra SK, Ostadhossein F, Daza E, Singh J, Pan D. Surface chemistry of carbon nanoparticles functionally select their uptake in various stages of cancer cells. Nano Res. 2017 [Google Scholar]
- 49.Stuart AD, Brown TDK. Entry of feline calicivirus is dependent on clathrin-mediated endocytosis and acidification in endosomes. J. Cell Virol. 2006;80(15):7500. doi: 10.1128/JVI.02452-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson JA, Watts C. Macropinocytosis. Trends Cell Biol. 1995;5:424. doi: 10.1016/S0962-8924(00)89101-1. [DOI] [PubMed] [Google Scholar]
- 51.Vercauteren D, Vandenbroucke RE, Jones AT, Rejman J, Demeester J, DeSmedt SC, Sanders NN, Braeckmans K. The use of inhibitors to study endocytic pathways of gene carriers: optimization and pitfalls. Mol. Therapy. 2010;18(3):561. doi: 10.1038/mt.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walkey CD, Olsen JB, Guo H, Emill A, Chan WCW. Nanoparticle size and surface chemistry determine serum proteins adsorption and macrophage uptake. J. Am. Chem. Soc. 2012;134(4):2139. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 53.Wang T, Bai J, Jiang X, Nienhaus GU. Cellular uptake of nanoparticles by membrane penetration: a study combining confocal microscopy with FTIR spectroelectrochemistry. ACS Nano. 2012;6(2):1251. doi: 10.1021/nn203892h. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y-C, Chao T-K, Chang C-C, Yo Y-T, Yu M-H, Lai H-C. Drug screening identifies niclosamide as an inhibitor of breast cancer stem-like cells. PLoS ONE. 2013;8:74538. doi: 10.1371/journal.pone.0074538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Liu Y, Li W, Jiang X, Ji Y, Wu X, Xu L, Qiu Y, Zhao K, Wei T, Li Y, Zhao Y, Chen C. Selective targeting of gold nanorods at the mitochondria of cancer cells: implications of cancer therapy. Nano Lett. 2011;11(2):772. doi: 10.1021/nl103992v. [DOI] [PubMed] [Google Scholar]
- 56.Wang LH, Rothberg KG, Anderson RG. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J. Cell Biol. 1993;123(5):1107. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xin-Sheng D, Shuiliang W, Anlong D, Bolin L, Susan EM, Stuart LE, Reema W-A, Ann TD. Metformin targets Stat3 to inhibit cell growth and induce apoptosis in triple-negative breast cancers. Ann. Cell Cycle. 2012;11:367. doi: 10.4161/cc.11.2.18813. [DOI] [PubMed] [Google Scholar]
- 58.Ye T, Xiong Y, Yan Y, Xia Y, Song X, Liu L, Li D, Wang N, Zhang L, Zhu Y, Zeng J, Wei Y, Yu L. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS ONE. 2014;9:P1. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Youan BB. Impact of nanoscience and nanotechnology on controlled drug delivery. Nanomedicine. 2008;3(4):401. doi: 10.2217/17435889.3.4.401. [DOI] [PubMed] [Google Scholar]
- 60.Zhang SL, Li J, Lykotrafitis G, Bao G, Suresh S. Size-dependent endocytosis of nanoparticles. Adv. Mater. 2009;21:419. doi: 10.1002/adma.200801393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.