Abstract
Introduction
Over the last decade, atomic force microscopy (AFM) has played an important role in understanding nanomechanical properties of various cancer cell lines. This study is focused on Lewis lung carcinoma cell tumours as 3D multicellular spheroid (MS). Not much is know about the mechanical properties of the cells and the surrounding extracellular matrix (ECM) in rapidly growing tumours.
Methods
Depth-dependent indentation measurements were conducted with the AFM. Force-vs.-indentation curves were used to create stiffness profiles as a function of depth. Here studies were focused on the outer most layer, i.e., proliferation zone of the spheroid.
Results
Both surface and sub-surface stiffness profiles of MS were created. This study revealed three nanomechanical topographies, Type A-high modulus due to collagen fibers, Type B-high stiffness at cell membrane and ECM interface and Type C-increased modulus due to cell lying deep inside matrix at a depth of 1.35 μm. Both Type and Type-B topographies result from collagen-based structures in ECM.
Conclusion
This study has first time revealed mechanical constitution of an MS. Depth-dependent indentation studies have the revealed role of various molecular and cellular components responsible for providing mechanical stability to MS. Nanomechanical heterogeneities revealed in this investigation can shed new light in developing correct dosage regime for collagenase treatment of tumours and designing better controlled artificial extracellular matrix systems for replicating tissue growth in-vitro.
Electronic supplementary material
The online version of this article (10.1007/s12195-019-00577-0) contains supplementary material, which is available to authorized users.
Keywords: AFM, Nanomechanics, Spheroids, Lewis lung carcinoma, Extracellular matrix
Introduction
Tissues in higher living organisms have multilayered cell–cell and cell–extracellular matrix (ECM) interactions. Three dimensional (3D) in-vitro cell cultures used for investigating various kinds of tumours, also known as multicellular spheroids (MS), have become a preferred means to study and simulate an in-vivo environment.27 3D cell cultures with resemblance of structural and functional properties to those of real tissues have helped in understanding the growth of tumours, the effects and penetration of toxins within a tissue, as well as developing targeted drug-delivery systems.18,31,32
It was the discovery of ECM in MS that gave us better insight towards developing 3D cell cultures.34 The matrix acts as both mechanical and biochemical shield within a MS. ECM also works as an information gateway system that allows selective passage of signals via cell membrane. Here, the composition of ECM was found to be similar to that of tumours in-vivo. Fibronectin, laminins, proteoglycans and collagen form major structural components within the matrix.33,34 Structure forming elements like collagen and laminins play a major role in giving stability to MS.
MS are a tumour analog with an inherent gradient of nutrients, oxygen and metabolites. Rapidly growing cultures have central necrotic core, which is surrounded by a layer of quiescent viable cells and an outer most layer of rapidly proliferating cells. The central necrotic core and the region of hypoxia are critical for testing anti-cancer therapeutics. Hypoxia has been identified as one of the causes of drug resistance in tumours.1,22,45 Here, penetration of drug through various layers of MS becomes a limiting factor towards drug efficacy. Therefore, this investigation is aimed towards understanding the nanomechanical properties at the surface of the spheroid where along with the cells, large volume of ECM is present that encapsulates the multicellular structure. Various groups have extensively investigated mechanical characteristic of cancer cells, including metastatic cells and cells isolated from the tumours. However, little is know about their growth mechanics and the impact of ECM forming elements within a compact multicellular structure such as a tumour or spheroid.
In this study, spheroidal cell cultures were developed using lewis lung carcinoma (LLC) cell line. LLC cell lines were derived from carcinoma of the lung of a C57BL mouse. LLC cells are well studied and are being used to determine outcome of drug induced toxicity by chemotherapeutic agents, developing methods to suppress metastasis, etc..4,35 There are a number of techniques like the spinner flask method, the hanging drop method, etc. that are commonly being used for the development of spheroidal cell cultures.27 For the current investigation LLC cell spheroids were developed with the liquid overlay method.5
Understanding nanomechanical properties of spheroids via depth dependent indentation profiling (DDIP) of cells and the surrounding ECM revealed minute changes in stiffness as the function of depth.53 Indentation-dependent profiling methods have been explored by a number of researchers in the field of AFM biology. Measurements conducted by Rico et al. and other groups primarily estimated the modulus corresponding to one data point at a time along the length of the force curve.10,25,41 In this investigation, however, successive segments of Force-vs.-distance (FD) curve were analyzed that provided ‘step-wise-modulus’, which helped in detecting variations in stiffness in 3-dimensions.
ECM is equally distributed within the proliferation zone and the quiescent zone. Hence, the stiffness (corresponding to high density of ECM components) at the cell surface can be a controlling factor for metastasis in tumours, drug penetration as well as growth and proliferation of tumours. To map differential nanomechanics at the MS surface, force-volume measurements were conducted with help of atomic force microscope (AFM). Other applications of AFM in the field biomaterials and cell biology include probing of biocompatible nature of metal oxide surfaces,17,21,52 nanostethoscopy of cardiomyocytes and other living organisms,20,46,51 cell nanomechanics,7,9,30 etc.
Earlier AFM measurements by Plodinec et al. on rat spheroids were only focused on differential nanomechanical properties at the core and periphery of the tumorous structure.38 This study further explores the nanomechanics of MS by mapping the stiffness of cells and the surrounding ECM. Surface scans were executed over 20 μm sized area with high resolution of 64 × 64 pixels, that brought forth three nanomechanical topographies into observation, Type-A, i.e., stiffness of long range Collagen I fibers, Type-B, i.e., high stiffness at the interface of cell membrane and ECM, and Type-C, i.e., the indentation mapping up to depth 1.35 µm has also revealed stiffness of cells lying deep inside ECM. Modulus mapping at the surface was complemented with multidimensional histograms and depth-wise error maps that gave crucial insight into mechanical diversity that constitutes an active 3D cell culture/tumour in in-vitro environment.
Materials and methods
Data Analysis
DDIP measurements were conducted using a pyramidal AFM probe. The probe indents directly into the viscous mass of ECM surrounding the cells. In some places after an indentation of a few nanometers, the tip comes into contact with cells embedded in the ECM. Due to the square pyramidal geometry of the probe, the modulus for developing the indentation profile was calculated using Sneddon’s model.
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
A simplified equation of measured force (Fmodel) vs. indentation to a model-dependent power (δn) curve is shown as Eq. (1). Other terms of the equation include a model dependent constant term (Cmodel), and the local reduced modulus (E*). Equation for Sneddon’s mechanics and reduced modulus (E*) are presented as Eq. (2) and (3), respectively. For calculating reduced modulus, Poisson ratio (ν) of 0.3 was employed for all samples. Modulus (E) of the tip is > 100 GPa as compared to sample’s modulus, which is < 1 MPa.8,14 Hence, the first term in Eq. (3) becomes negligible and is dropped from the final equation.
The local reduced modulus for any given Force-vs.-indentation curve was calculated from the slope of the nth root of the curve, all of which is taken to the nth power and is divided by the appropriate constant (Eq. 4). This approach towards calculating the modulus also provides the standard deviation of the modulus. Here, the standard deviation of the slope is calculated by simple linear regression in Eq. (5) and is substituted by standard deviation of the actual slope in Eq. (4)(braces). The standard deviation (error) is presented as percentage of measured stiffness and was used for developing error maps. Effective local modulus and its standard deviation at varying depths and at every pixel were calculated using Eq. (3). Error maps developed here were used to interpret indentation data from novel perspective.
In this communication our objectives are first, to highlight various nanomechanical heterogeneities within a MS and investigate the interaction at cell-matrix interface. Second, to demonstrate the application of multidimensional histograms along with the stiffness profiles in the identification of such heterogeneities. Error parameters associated with local mechanical properties were also used in the analysis. It helped in identifying minute fluctuations in stiffness as measured by AFM probe during indentation into a cell having a well defined membrane as compared to ECM, which is a highly viscous mass of proteins, glycoproteins, etc. In our previous investigation with agarose gels, one could observe that stiffer gels had a narrow error distribution when compared with agarose gels having lower stiffness.53
Production of spheroids
Lewis lung carcinoma (LLC) cells were obtained from ATCC (Manassas, VA). LLC cells were maintained in Dulbecco’s modified Eagle media (DMEM, Corning cellgro®, Manassas, VA) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Spheroids were produced using a liquid-overlay technique on agar surfaces. Briefly, 1% agar was coated in each well of a 96-well plate. Following the solidification of agar, 1000 LLC cells were plated in each well and the plate was centrifuged using a plate centrifuge (ThermoFisher Marathon 3000R) at 250×g for 15 min. The plate was placed in 5% CO2, 37 °C atmosphere and the spheroids were allowed to grow for 6 days.
After 6 day of incubation spheroids were fixed on the glass coverslip using agarose gel. 0.1% of warm agarose gel was allowed to cool down at room temperature and is gently poured over the coverslip. Before complete solidification of gel, a spheroid is carefully pipetted on its surface and lower half of the spherical mass of cell gets immobilized over the agar surface.
Nanomechanical Measurements
AFM based force mapping measurements were conducted with an Asylum Research MFP-3D AFM with integrated inverted optics (Nikon TE-2000), in a liquid cell for complete immersion during the experiments. A fluid cell heater was used for maintaining the temperature at 37 °C. Arrays of 64 × 64 force indentations were acquired over a 20 μm × 20 μm area. All force maps were collected with indentation velocities of 32 μm/s, amounting to individual indentations at a rate of ~ 2 Hz. Spring constants (0.3–1.3 nN/nm) of the Olympus TR800PB probes used throughout were calibrated following the widely applied Sader method.43 Sneddon’s model was used for estimating modulus as a function of depth. Percentile Error calculated from the standard deviation of the slope was used to develop error maps. Customized analytical and visualization routines were written with Matlab software.
Results
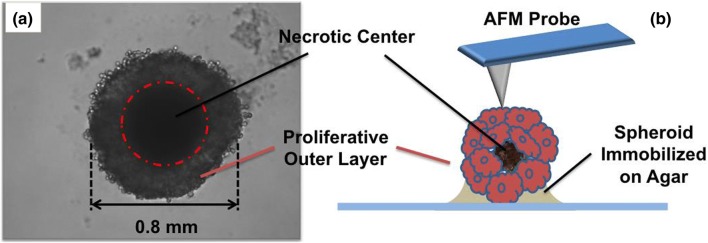
MS were initially regarded as simple aggregates of cells with intimate cell-to-cell contacts. It was with the discovery of ECM, that it became clear how closely MS resembled tumours in-vivo.34 ECM is a mucilaginous and sticky of mass proteo-glycan, collagen and other structure forming elements. These membrane free extracellular components can introduce large local variations in the mechanical properties across the ECM. This study is focused on such nanomechanical characteristics of the MS as a model system consisting of cells embedded within the matrix and forming a 3D aggregate of rapidly growing mass of cells. The lower half of the LLC spheroids was partially immobilized in agarose gel. This provided stability to the spherical structure of the spheroid and prevented lateral motion during long duration measurements via AFM probes (Fig. 1b). The top view of spheroid fixed on the gel’s surface is shown in Fig. 1a. A single spheroid was 0.8 mm in diameter with a dark necrotic center marked by the dotted red circle. The outer proliferative layer was distinguished by lighter shades of grey clearly visible with few loosely bound cells in its periphery.
Figure 1.
(a) Top view of a Spheroid partially embedded in agarose gel. Spheroid has diameter of 0.8 mm with a central necrotic core marked by dotted red circle. Outer proliferative layer having relatively lesser density of cells was used for nanomechanical investigations. (b) Schematic portraying experimental setup, lower half of the spheroid is immobilized in agarose gel. Upper half exposed to aqueous media is probed with AFM cantilevers.
AFM measurements were conducted over 20 μm sized areas. 20 spheroids were examined to ascertain which regions present regions of both ECM and cells adjacent to each other. Each data set contains 4096 force-vs.-indentation curves, i.e., 64 × 64 pixels. Measurements were conducted at an indentation rate of ~ 2 Hz. It is a known fact that the measured mechanical stiffness depends on the loading rate.10,30 This investigation required examining large number of samples, which in turn produced large volumes of data for analysis. Thus, in order to accelerate the experimental procedures and maintain viability of a large number of cells under the AFM probe, experiments were performed at a high indentation rate (~ 2 Hz). At lower rates, the duration of each scan becomes too long, this may kill or damage structural integrity MS under investigation. Therefore, the stiffness maps presented in this article, should be used to appreciate relative differences in stiffness between cells and ECM within a single data set.
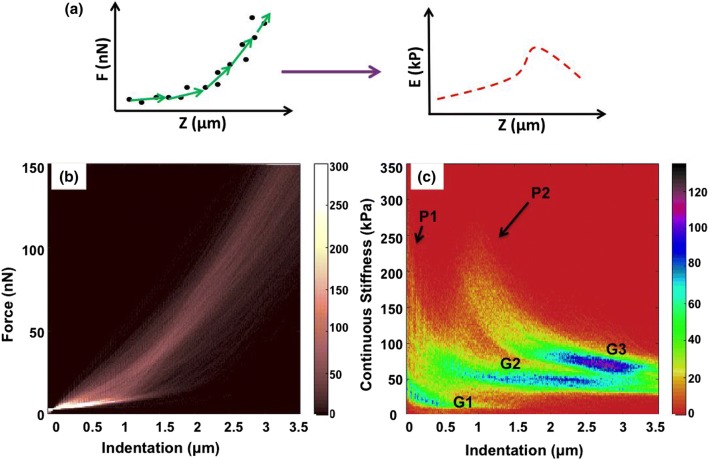
It is important to note that a sharp conical probe can provide unrealistically high values of modulus. The Sneddon’s model is based on the assumption of a constant modulus with a linear, isotropic, elastic material, but it may it may still be used in an attempt to parameterize the measurement results. Data from each indentation curve is split into small fragments and each fragment is fitted with Sneddon’s model to estimate the stiffness at certain depth, as illustrated in Fig. 2a. All the force spectra from 64 × 64 indentations were plotted in a single multi-dimensional histogram (color contrast indicates histograms counts), shown as Fig. 1b. Due to the gelatinous nature of the cells and the surrounding ECM, repulsive forces rise gradually over a distance of 2–3 µm. In regions having relatively lower compliance, one can observe much slower increase in forces curves (0–10 nN/µm). To determine the point of contact with the sample, relatively high contact force of 4 nN was considered. At such high contact force there is sufficient deformation of cell around the AFM probe which has been predicted16,24 and observed2 by various research groups. This point of contact was later used for developing topography of the apical surface of the MS which otherwise cannot be observed using optical microscopy.
Figure 2.
(a) Sketch shows stepwise fitting of Sneddon’s model into force curve data which gets converted into modulus vs. indentation curve. Modulus data from each pixel is plotted as stiffness map at various depths. (b) Multidimensional histogram (color contrast indicates the counts) of all 64 × 64 force (nN) vs. distance (µm) curves in 20 × 20 µm scan of the surface of a spheroid (c) Multidimensional histogram of continuous stiffness (kPa) vs. indentation (µm) of the force volume data shown in (b). Differential color contrast indicates density of the counts at different indentation depths in x axis, corresponding with the stiffness values in y axis.
As described in the illustration in Fig. 2a, FD curves were fitted with Sneddon’s model and as a result, continuous stiffness-vs.-indentation multi-dimensional histograms were developed. A colored histogram (color contrast indicates histograms counts) with depth-wise modulus is shown in Fig. 2c. Plotting data from all continuous stiffness-vs.-indentation curves in a multi-dimensional histogram, produces differential color contrast based on the density of counts at different depths of indentation. Here, two peaks P1 and P2 are marked with black arrows. High density of counts creates three different groups with blue cores (counts in the range of 100–120), which are labeled as G1, G2 and G3. This contrast can only be appreciated with high-resolution force volume measurements. Here 4096 curves were collected over a scan area of 20 µm × 20 µm, which gives a pixel size 312 nm. Such closely packed probing of the surface gives a relatively higher number of data points from the regions having similar nanomechanical characteristics. Therefore, heterogeneities in stiffness as a function of depth can be observed both from the histograms and from the modulus profiles. The correlation between the histogram and the modulus profiles will be discussed latter in the text.
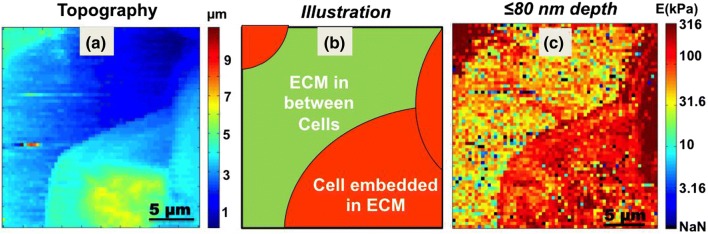
Surface topography is created from all the FD curve is shown in Fig. 3a. Positioning of cells and ECM is explained with help of an illustration in Fig. 3b. The bluish central region in Fig. 3a marks the ECM, which is schematically illustrated with green color in Fig. 3b. There are 3 cells (top left, right and bottom right) with light bluish green tone in the topography image (Fig. 3a). The same are represented with red color in the illustration in Fig. 3b. Here LLC cells seem to have a spherical morphology. This was expected due their growth in a tumour like environment and the presence of a large volume of ECM in their immediate vicinity.
Figure 3.
(a) Surface topography of LLC-spheroid cell created from 128 × 128 force curves. (b) Illustration explaining various nanomechanical features visible in stiffness profile. Large central green colored region is ECM in between cells and cells embedded in the matrix are colored red. (c) Indentation modulus profile of MS up to depth of 80 nm. Scan area of 20 × 20 µm having 64 × 64 pixels.
Data collected from the measured stiffness as a function of depth (Fig. 2c) is plotted in form of a stiffness map shown in Fig. 3c. The stiffness map presented here corresponds to a depth of ≤ 80 nm. The ECM in the center of the Fig. 3c at ≤ 80 nm depth, has large variations in modulus ranging from 10 kPa to 100 kPa. One can observe a large gradient in color contrast, from bluish green to red in color. To get the maximum contrast, the color bar for the modulus is presented in log scale. Presentation of the modulus in logarithmic scale can facilitate the identification of minute features having stiffness unlike the rest of the matrix. During the measurement, MS is suspended in growth medium (DMEM). Continuous poking with the AFM probe can dissociate some of its matrix components into the surrounding aqueous environment. This may create large variations in stiffness at depths closer to the surface of the spheroid since ECM is a mix of various molecular and mechanical compositions can introduce large variation in modulus observed close to the surface of MS.
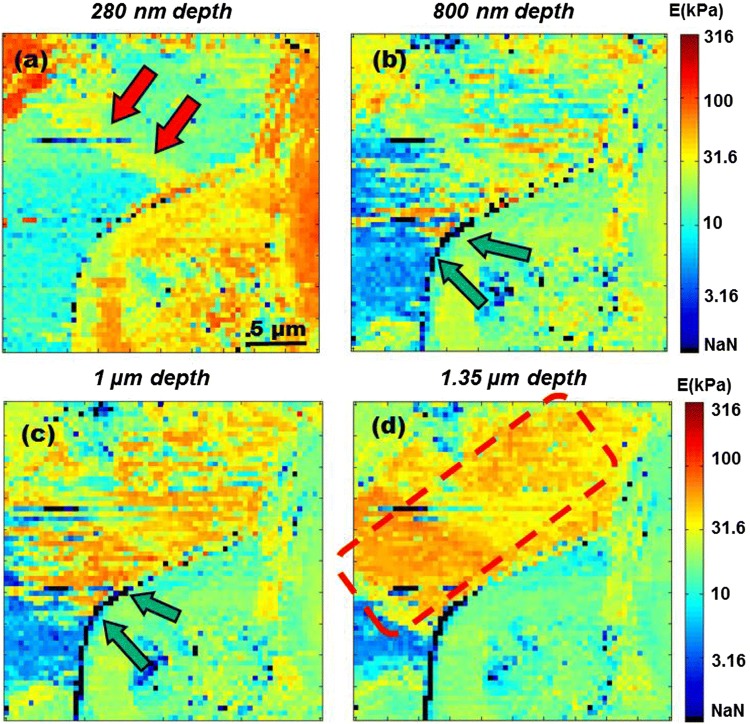
The cells discussed earlier in Figs. 3a and 3c on top left, right and bottom right corner of the image, have a very high modulus when compared with the adjacent ECM. The modulus of all these cells is higher than 100 kPa and goes upto 300 kPa. These high modulus values within the first 100 nm of indentation are also observed in the multi-dimensional histogram and are labeled as peak P1 in Fig. 2c. Increasing the indentation depth beyond 100 nm, there is a gradually decrease in stiffness as the probe indents deeper into the cell. This decrease in modulus within the regions of the cell can be observed from stiffness profiles at various depths of 280 nm, 800 nm, 1 µm and 1.35 µm in Figs. 4a–4d respectively. Similar decrease of modulus can also be observed in other MS samples. The modulus profile of sample MS-2 is discussed as supplementary Fig. 1. Meanwhile, the modulus of ECM at depth of 0.17 µm is around 100 kPa which gradually goes down to 10 kPa at depth of 1.44 µm.
Figure 4.
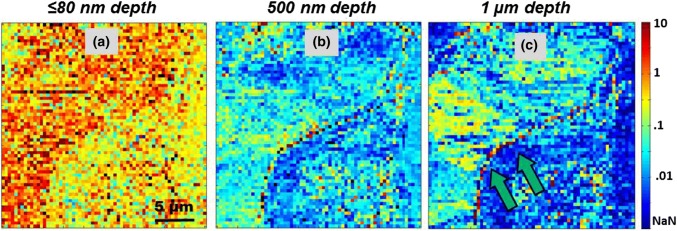
Stiffness maps of MS at various depths (a) At depth of 280 nm, modulus of ECM is at 10 kPa and cells have stiffness < 100 kPa. Double red arrows mark interconnecting collagen type I stress fiber. (b) Interface of cell membrane and ECM at depth of 800 nm is marked by double green arrows, has stiffness greater than 316 kPa, hence observed as string of black pixels. (c) Double green arrows mark regions of high stiffness at cell membrane-ECM interface at 1 µm depth. (d) At 1.35 µm depth, cell embedded deep inside the ECM can be observed with stiffness > 31.6 kPa is marked by dotted red square.
Sub-Surface Nanomechanics
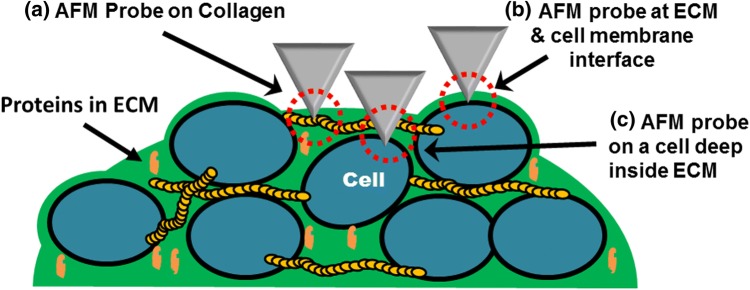
A closer inspection of the maps derived from indentation depths within the range of ≤ 80 nm up to 1.35 µm revealed three types of nanomechanical topographies, Type-A, Type-B and Type-C. These features are explained by illustration in Fig. 5. The illustration highlights the point of contact of the AFM probe with various sub-surface structural components (collagen—long elongated yellow fibers) in the surrounding ECM (green in color) and the cells (oval shaped) embedded in the matrix. Type-A nanomechanical features look like elongated fibrous structures; they can be observed in Fig. 4a (marked by two red arrows). One can also observe similar elongated fiber-like structures in sample MS-2 described in supplementary Fig. 1c. It is important to note that these structural collagen fibers will always appear straight. A collagen fiber may hold on to different cells or similar interconnected fiber, however this will introduce some tension along its length. Hence, they should appear as straightened-out elongated structures within the matrix. They are possibly load bearing collagen fibers that maintain the structural integrity of the spheroid and can also help cells to mechanically sense and respond to the presence of other cells over long distances (~ 100 µm). Collagen of types I, II III, V and XI are fibril-forming collagens.11 Studies show that when hMSCs are grown on type-1 collagen gels, they respond to the stiffness of a glass slide via 1000 µm of collagen gel fibers.26 Similarly, when fibroblast are grown on fibrin gels, they can sense the presence of other cells up to 250 µm away and are also known to respond by aligning themselves relative to each other.55 Therefore, the stiffness and the stability of a MS structure as a whole can be directly correlated to the modulus of these load-bearing fibers. Collagen fibers are also known to have modulus greater than 0.3 GPa but can have lower stiffness depending on the substrate and the source of the biomaterial.15,29,54,56
Figure 5.
Sketch describes three types of nanomechanical topographies identified with DDIP. Type (A) is long range collagen type I stress fibers. They have considerable high stiffness as compared with surrounding ECM. Type (B) is region of high stiffness at interface of cell membrane and ECM. Type (C) is a cell embedded deep inside the ECM. They are identified by gradual increase of stiffness as AFM probe indents deeper into the ECM.
The stiffness of bulk ECM was measured up to few tens of kPa in Fig. 4a. Hence, the presence of type I collagen fibers should introduce stiffness greater than that of the rest of the biological material forming a MS. In Fig. 3c, stress fiber observed in Fig. 4a looks like a smear in the center of the matrix. Its stiffness up to depth of 80 nm is ≥ 100 kPa. Collagen fibers have high stiffness but due to their location close to the surface of spheroid (top layer of ECM), they get masked by a high degree of variability in the modulus of the surrounding matrix. At depth of 280 nm (Fig. 4a), a collagen fiber can be observed as an interconnecting fiber between cells separated by distance of 10 µm. In the supplementary Figs. 1b and 1c one can also observe a collagen fiber attached to the cell on top right corner. Its stiffness is greater than 20 kPa and it is ~15 μm in length. Collagen fibers are known to have diameter greater than 250 nm whereas, sometimes they might be as thin as 30 nm.3,50 Accurate estimation of modulus of these structural fibers in MS will also depend on the loading rate, plus fibers are spread across a matrix of biomolecules. Fibers will get pulled and pushed around as the probe indents into the spheroids. Hence, stress and strain factors must be taken into consideration for a more precise estimation of modulus of the collagen fibers in 3D cell cultures.42
Type-B nanomechanical features illustrated in Fig. 5 arise due to interactions taking place between the ECM and the cell membranes. When cells are grown as monolayered cultures, their apical surfaces are exposed to aqueous media whereas in-vivo most cells are embedded within the ECM. Cells associated with fluid connective tissues like blood and lymph can be found floating or exposed to aqueous environment. In that case, there is hardly any interaction with any viscous mass, such as that of the ECM, other than their immediate aqueous environment. Therefore, stiffness of a cell might be influenced by the interaction between the cell membrane and the surrounding extracellular matrix. In this investigation it was interesting to observe that in regions where the cell membrane was in direct contact with ECM, a very high modulus ≥ 300 kPa can be observed. As the probe indents into membrane-matrix interface (MMI) there is a sharp increase in stiffness. Regions of high stiffness at MMI are marked by green arrows in Figs. 4b and 4c at the depth of 800 nm and 1 µm respectively and in the supplementary Fig. 1b, where the stiffness at the MMI goes up to 300 kPa. Here, pixels having modulus greater than 316 kPa are shown with a black contrast, labeled as NaN in the color bar. For the sake of visual presentation of sub-surface nanomechanical topographies and to accommodate the maximum number of nanomechanical heterogeneities, logarithmic scale was used for the stiffness values in the range of 1 kPa to 316 kPa. Stiffness for most of the soft biological material falls within this range. Hence, modulus greater than the cut-off value of 316 kPa is denoted by black contrast pixels which are shown in color bar below 1 kPa, hence labeled NaN.
The cells present on the outer-most later of the MS also have a thin layer of ECM over them. As the AFM probe indents into the MS surface, those cells with a coating of ECM should have higher stiffness as compared to the rest of the cell body. This effect was earlier observed in Fig. 3c (depth ≤ 80 nm) where cells on the lower right and top left corner have modulus up to 300 kPa. Hence, the cells in top layer of MS also fall into Type-B category. This explains why there is significantly higher stiffness up to indentation depth of 100 nm. Thereafter, there is gradual decrease in the modulus of cells as the AFM probe indents deeper into the cell, which has a low stiffness due to its gelatinous nature.
This increase in stiffness at MMI can be attributed to the formation of a basal lamina assembly at the cell surface.57 ECM architecture near the cell surface is composed of interconnected polymers of collagen IV and VII.58 They are bound to laminin sheets, which are organized at the cell membrane by integrins and dystroglycan. Binding of collagen IV network to laminin takes place via nidogen molecules.47 This assembly is further strengthen by binding to collagen VII dimers.19,33,39 A conclave of structural proteins over the cell membrane strengthens the basal lamina. The combined effect of these tightly packed structures can considerably increase the modulus of the cell membrane that may go above 300 kPa. Therefore, cells embedded within the ECM in spheroids have shell like structure around them that provides them with considerable mechanical stability.
The last nanomechanical topography is Type-C, which is illustrated in Fig. 5 and is marked by a red square in Fig. 4d. The outer surface of the spheroid doesn’t have an even distribution of cells. There are gaps filled with viscous ECM. As the probe indents into the regions filled with ECM there is a decrease in stiffness from the depth of ≤ 80 nm (Fig. 1b) to 280 nm (Fig. 2a). Average stiffness of the ECM goes down from 31.6 kPa to 10 kPa. Only a collagen fiber marked by double red arrows has a relatively higher modulus as compared to the surrounding ECM. Furthermore at indentation depths of 800 nm (Fig. 4b), 1 µm (Fig. 4c) and 1.35 µm (Fig. 4d), one can observe a gradual increase in stiffness that goes up to 100 kPa. As illustrated in Fig. 5, Type-C nanomechanical features could be due to an underlying cell positioned deep inside the ECM. This effect can also be observed in multi-dimensional histograms. This second rise in modulus, labeled as Peak P2 can be observed in histogram in Fig. 2c. This increase of modulus can be observed from the depth of 0.5 µm (indentation) onwards and a significant number of counts having stiffness greater than 100 kPa can be observed up to depths of 1.5 µm. This observation of Type-C features is limited by the length of the AFM probe. Olympus TR800 PB probes have a height of 3 ± 0.5 µm, hence the analysis of the histogram data is considered up to depth of 3.5 µm. Being highly viscous, ECM tends to adhere to the AFM probe. Thus, any further investigating of nanomechanical properties of cells deepeer into the proliferation zone of the spheroid will require the development of specialized AFM probes.
Referring back to the histogram in Fig. 2c, the grouping of the counts can also be correlated with the number of pixels with similar values of stiffness. Group G1 is primarily contribution of modulus values from ECM up to depths of 500 nm. As the probe indents deeper, modulus averages out at all depths hence the grouping of counts is labeled as G2. The highest density of counts is observed beyond 1.5 µm. A downward trend from peak P2 merges into group G3. This third grouping of modulus is observed from indentations between 2 and 3 µm. Here higher density of counts can be due to Type-C nanomechanical topographies. They have a relatively higher average stiffness of 100 kPa. Cells that are deep inside the spheroid are surrounded by ECM and other cells in their vicinity. This would put more mechanical load on to the cells present in the sub-surface regions. Hence, they might have a higher modulus as compared to the cells that are present in the outer periphery.
Indentation Error Maps
Standard deviation from slopes of each FD curve was used for developing error profile at different depths. Error profiles at depths of ≤ 80 nm, 500 nm and 1 µm are shown in Fig. 6. Error profiles are interrelated with the previously discussed stiffness maps at different indentations. At depths ≤ 80 nm (Fig. 6a), the ECM has higher error values as compared to the adjacent cells in top left, right and lower right corner of the image. For ECM, observed standard deviation of the calculated modulus (percentage error) was greater than 1. This might be due to the lack of clearly defined structures or membrane on the outer-most layer of the spheroid. The mucilaginous nature of the ECM will also have significant contribution towards higher percentile error, especially in regions or spaces between the cells filled with the ECM.
Figure 6.
Standard deviation of calculated modulus is shown as percentage error maps at various depths (a) Error profile of stiffness up to depth of 80 nm. ECM has higher percentile error as compared to cells forming MS. (b) At 500 nm depth, error is < 1 for both cells and ECM. (c) Probe indents deeper to a depth of 1 µm. Edge of the cell interacting with ECM has higher error. Region of cell membrane—ECM interface is marked by two green arrows has percentile error > 1.
On closer examination of the error profile in Figs. 6a and 6b, i.e., corresponding to indentaion depths from ≤ 80 to 500 nm there is a decrease in the error to 0.01 units. At 1 micron depth in Fig. 6c regions with higher modulus have errors greater than 0.1 unit. At depth of 1 µm, MMI regions have higher error values that go up to 10 units and creating a reddish parabolic streak across the edge of the cell, which is marked by two green arrows. This high error could be the result of resistance experienced by the probe as it indents deeper at the edges of the cell. The stiffness at MMI is greater that 300 kPa, and as the probe penetrates through the interface it can experience number of minute fluctuations, thereby introducing relatively higher errors. These errors are mildly visible in the error map at the depth of 500 nm but get significantly masked at ≤ 80 nm due to the squishiness of the ECM.
Discussion
The main objective of culturing MS is to replicate the tumorous environment in-vitro for developing therapeutics that can penetrate and kill any cancerous growth inside the human body. Due to layered structuring and compactness of tumours, breaching the outer most layers of cells for targeted drug delivery becomes a limiting factor towards curing a cancer patient. ECM in tumours is known for creating a transport barrier that restricts drug penetration inside a solid tumour. Treatment of collagenase prior to delivery of 100 nm sized nanoparticles has found to increase particle penetration by 4-fold.13 Both Type-A and Type-B topographies originate due to the presence of collagen-based structures. Hence, collagenase treatment, when coupled with nanomechanical studies, could help in building the correct dosage regime. Understanding tumour micro environment, can therefore play a crucial role in the development of effective anticancer drugs.32 By measuring MS stiffness one can look for better means to dissolve the outer mass of cells and to eventually develop methods for the deliver of therapeutics into the quiescent zone for the complete dissolution of the multicellular structure.31
The composition of ECM in a MS is considerably different when compared with monolayer cultures.12 Previous studies have also suggested increased stiffness of ECM in malignant phenotypes/cultures, which is associated with increased concentration of collagen fibers in the matrix.6,37,40 Additionally, collagen IV being the most abundant constituent of the basement membrane, is associated with tumour fibrosis and accumulates in tumour interstitium.19,23,49 Therefore, detection of higher stiffness at the MMI by AFM probe is consistent with studies conducted on ECM and tumourous tissues up to date.
Another interesting application of depth dependent investigations would be to investigate development of artificial ECM. With DDIP, scanning surfaces of the cultures with artificial ECM would help fine-tuning the nanomechanical properties that are most appropriate for growing 3D cell cultures. Some groups have started to develop permissive hydrogels that mimic specifically the ECM in MS. They have substituted protein based ECM components with poly(ethylene glycol) (PEG).28,44,48 Such artificial matrices can only act as a physical scaffold to hold cells proliferating in a 3-dimension (3D). In contrast, the exact nanomechanical properties in a native ECM environment are controlled by collagen, laminins and other components. Tissues engineered with an artificial ECM may have a different mechanism towards the delivery of therapeutics and nutrients in 3D cultures. Therefore, information gathered via nanomechanical investigation of the extracellular matrix and cells in a MS can help in improving hydrogel-based replacements for ECM.
The main limitation in conducting force measurements on MS is its opaque nature. A significant number of hit and trial efforts are required to find the optimum region in its outer periphery. Hence, it was not possible to conduct simultaneous fluorescence or optical measurements. However, with the development of Light Sheet Fluorescence Microscopy (LSFM),36 in the near future it might be possible to combine force measurements with LSFM that would give us a sufficiently fine control to probe and selectively study the mechanical properties of cells located in the proliferation zone and the quiescent zone in MS.
Conclusion
Some groups have reported extensively on the analysis of force curves to determine the stiffness of living tissues; however most of the investigations are focused on measuring the modulus to a depth of only a few nanometers. Here we report for the first time on how the depth dependent indentation profiling (DDIP) can give us crucial insights into the nanomechanical diversity within a complex agglomerate of cells and extra-cellular matrix. Other investigators have conducted similar studies, yet they have worked with spherical probes having diameter greater than 5 micron.14 Due to the poor resolution and the relatively greater area of contact with the spherical probe, the depth dependent investigations won’t reveal any significant sub-surface heterogeneities. This study puts particular emphasis on minute changes in stiffness, which result from interactions between various components present within rapidly proliferating mass of cells. This novel approach of stiffness profiling can have profound application in twenty-first century as we move towards synthetic bio-mechanical platforms. Knowledge regarding mechanics of interactions of cells with their surrounding environment is essential for developing different types of artificial 3D cell cultures. The same can also give significant insight towards targeting tumorous tissues, i.e., the mechanical stiffness at MMI can have a significant impact on the bio-availability of a drug towards the treatment of the concerned biological disorder.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Author Contributions
Conceived and designed the experiments: VV BDH. Performed the experiments: VV. Analyzed the data: VV BDH. Contributed biological material & cells: MS GGMD. Wrote the paper: VV.
Conflict of interest
Varun Vyas, Melani Solomon, Gerard G.M. D’Souza and Bryan D. Huey declare that they have no conflicts of interest.
Ethical Approval
No human studies were carried out by the authors for this article. No animal studies were carried out by the authors for this article.
Funding
Provided by the University of Connecticut’s Provosts Fund and NSF Nano-Bio-Mechanics grant 0626231.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Varun Vyas, Email: varun.shiva@gmail.com.
Melani Solomon, Email: melani.the.piscean@gmail.com.
Gerard G. M. D’Souza, Email: gerard.dsouza@mcphs.edu
Bryan D. Huey, Email: bryan.huey@uconn.edu
References
- 1.Andre F, Berrada N, Desmedt C. Implication of tumor microenvironment in the resistance to chemotherapy in breast cancer patients. Curr. Opin. Oncol. LWW. 2010;22:547–551. doi: 10.1097/CCO.0b013e32833fb384. [DOI] [PubMed] [Google Scholar]
- 2.Bagnoli, K. L. Influence of EGF on HaCaT cells as measured by AFM with a comparison of AFM and 3D optical deconvolution using MH-S cells. M.S. Thesis, University of Connecticut, 2007.
- 3.Bancelin S, et al. Determination of collagen fibril size via absolute measurements of second-harmonic generation signals. Nat. Commun. 2014;5:4920. doi: 10.1038/ncomms5920. [DOI] [PubMed] [Google Scholar]
- 4.Bertram JS, Janik P. Establishment of a cloned line of Lewis lung carcinoma cells adapted to cell culture. Cancer Lett. 2017;11:63–73. doi: 10.1016/0304-3835(80)90130-5. [DOI] [PubMed] [Google Scholar]
- 5.Carlsson J, Yuhas JM. Liquid-overlay culture of cellular spheroids. Recent Results Cancer Res. 1984;95:1–23. doi: 10.1007/978-3-642-82340-4_1. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhuri O, et al. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 2014;13:970–978. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 7.Cross SE, Jin Y-S, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 8.Dokukin ME, Sokolov I. On the Measurements of Rigidity Modulus of Soft Materials in Nanoindentation Experiments at Small Depth. Macromolecules. 2012;45:4277–4288. doi: 10.1021/ma202600b. [DOI] [Google Scholar]
- 9.Dufrene YF, Martinez-Martin D, Medalsy I, Alsteens D, Muller DJ. Multiparametric imaging of biological systems by force-distance curve-based AFM. Nat. Meth. 2013;10:847–854. doi: 10.1038/nmeth.2602. [DOI] [PubMed] [Google Scholar]
- 10.Efremov YM, Dokrunova AA, Bagrov DV, Kudryashova KS, Sokolova OS, Shaitan KV. The effects of confluency on cell mechanical properties. J. Biomech. 2013;46:1081–1087. doi: 10.1016/j.jbiomech.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 11.Gelse K, Pöschl E, Aigner T. Collagens—structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003;55:1531–1546. doi: 10.1016/j.addr.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Glimelius B, Norling B, Nederman T, Carlsson J. Extracellular matrices in multicellular spheroids of human glioma origin: increased incorporation of proteoglycans and fibronectin as compared to monolayer cultures. APMIS. 1988;96:433–444. doi: 10.1111/j.1699-0463.1988.tb05327.x. [DOI] [PubMed] [Google Scholar]
- 13.Goodman TT, Olive PL, Pun SH. Increased nanoparticle penetration in collagenase-treated multicellular spheroids. Int. J. Nanomed. 2007;2:265–274. doi: 10.2217/17435889.2.3.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guz N, Dokukin M, Kalaparthi V, Sokolov I. If cell mechanics can be described by elastic modulus: study of different models and probes used in indentation experiments. Biophys. J. 2014;107:564–575. doi: 10.1016/j.bpj.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heim AJ, Matthews WG, Koob TJ. Determination of the elastic modulus of native collagen fibrils via radial indentation. Appl. Phys. Lett. 2006;89:181902. doi: 10.1063/1.2367660. [DOI] [Google Scholar]
- 16.Hoh JH, Schoenenberger CA. Surface morphology and mechanical properties of MDCK monolayers by atomic force microscopy. J. Cell Sci. 1994;107:1105–1114. doi: 10.1242/jcs.107.5.1105. [DOI] [PubMed] [Google Scholar]
- 17.Jandt KD, Finke M, Cacciafesta P. Aspects of the physical chemistry of polymers, biomaterials and mineralised tissues investigated with atomic force microscopy (AFM) Colloids Surf. B Biointerfaces. 2000;19:301–314. doi: 10.1016/S0927-7765(00)00139-9. [DOI] [PubMed] [Google Scholar]
- 18.Jang SH, Wientjes MG, Lu D, Au JL-S. Drug delivery and transport to solid tumors. Pharm. Res. 2003;20:1337–1350. doi: 10.1023/A:1025785505977. [DOI] [PubMed] [Google Scholar]
- 19.Kalluri R. Angiogenesis: basement membranes: structure, assembly and role in tumour angiogenesis. Nat. Rev. Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 20.Keaton A, Holzrichter JF, Balhorn R, Siekhaus WJ. Nanostethoscopy: a new mode of operation of the atomic force microscope BT. In: Güntherodt HJ, Anselmetti D, Meyer E, editors. Forces in Scanning Probe Methods. Dordrecht: Springer; 1995. pp. 91–97. [Google Scholar]
- 21.Kessel S, et al. Thermoresponsive PEG-based polymer layers: surface characterization with AFM force measurements. Langmuir. 2010;26:3462–3467. doi: 10.1021/la903007v. [DOI] [PubMed] [Google Scholar]
- 22.Kim S-H, Kuh H-J, Dass CR. The reciprocal interaction: chemotherapy and tumor microenvironment. Curr. Drug Discov. Technol. 2011;8:102–106. doi: 10.2174/157016311795563875. [DOI] [PubMed] [Google Scholar]
- 23.Kühn K, et al. Macromolecular structure of basement membrane collagens. FEBS Lett. 1981;125:123–128. doi: 10.1016/0014-5793(81)81012-5. [DOI] [PubMed] [Google Scholar]
- 24.Kwon E-Y, Kim Y-T, Kim D-E. Investigation of penetration force of living cell using an atomic force microscope. J. Mech. Sci. Technol. 2009;23:1932–1938. doi: 10.1007/s12206-009-0508-z. [DOI] [Google Scholar]
- 25.Lekka M, et al. Cancer cell detection in tissue sections using AFM. Arch. Biochem. Biophys. 2012;18:151–156. doi: 10.1016/j.abb.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 26.Leong WS, et al. Thickness sensing of hMSCs on collagen gel directs stem cell fate. Biochem. Biophys. Res. Commun. 2010;401:287–292. doi: 10.1016/j.bbrc.2010.09.052. [DOI] [PubMed] [Google Scholar]
- 27.Lin R-Z, Chang H-Y. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. 2008;3:1172–1184. doi: 10.1002/biot.200700228. [DOI] [PubMed] [Google Scholar]
- 28.Lutolf MP. Integration column: artificial ECM: expanding the cell biology toolbox in 3D. Integr. Biol. 2009;1:235–241. doi: 10.1039/b902243k. [DOI] [PubMed] [Google Scholar]
- 29.Ma X, et al. Fibers in the extracellular matrix enable long-range stress transmission between cells. Biophys. J. 2013;104:1410–1418. doi: 10.1016/j.bpj.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medalsy ID, Müller DJ. Nanomechanical properties of proteins and membranes depend on loading rate and electrostatic interactions. ACS Nano. 2013;7:2642–2650. doi: 10.1021/nn400015z. [DOI] [PubMed] [Google Scholar]
- 31.Mehta G, Hsiao AY, Ingram M, Luker GD, Takayama S. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J. Control Release. 2012;164:192–204. doi: 10.1016/j.jconrel.2012.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat. Rev. Cancer. 2006;6:583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 33.Mouw JK, Ou G, Weaver VM. Extracellular matrix assembly: a multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 2014;15:771–785. doi: 10.1038/nrm3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nederman T, Norling B, Glimelius B, Carlsson J, Brunk U. Demonstration of an extracellular matrix in multicellular tumor spheroids. Cancer Res. 1984;44:3090–3097. [PubMed] [Google Scholar]
- 35.O’Reilly MS, et al. Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by a lewis lung carcinoma. Cell. 1994;79:315–328. doi: 10.1016/0092-8674(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 36.Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 37.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 38.Plodinec M, Schweighauser G, Sütterlin R, Oberman E, Aebi U. Morphology and cytoarchitecture regulate nanomechanical properties of tumor spheroids. AFM Cancer Diagn. 2012;7:116–145. [Google Scholar]
- 39.Pöschl E, Schlötzer-Schrehardt U, Brachvogel B, Saito K, Ninomiya Y, Mayer U. Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development. 2004;131:1619–1628. doi: 10.1242/dev.01037. [DOI] [PubMed] [Google Scholar]
- 40.Provenzano PP, Inman DR, Eliceiri KW, Keely PJ. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 2009;28:4326–4343. doi: 10.1038/onc.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rico F, Roca-Cusachs P, Gavara N, Farré R, Rotger M, Navajas D. Probing mechanical properties of living cells by atomic force microscopy with blunted pyramidal cantilever tips. Phys. Rev. E. 2005;72:21914. doi: 10.1103/PhysRevE.72.021914. [DOI] [PubMed] [Google Scholar]
- 42.Roeder BA, Kokini K, Sturgis JE, Robinson JP, Voytik-Harbin SL. Tensile mechanical properties of three-dimensional type i collagen extracellular matrices with varied microstructure. J. Biomech. Eng. 2002;124:214–222. doi: 10.1115/1.1449904. [DOI] [PubMed] [Google Scholar]
- 43.Sader JE, Larson I, Mulvaney P, White LR. Method for the calibration of atomic force microscope cantilevers. Rev. Sci. Instrum. 1995;66:3789–3798. doi: 10.1063/1.1145439. [DOI] [Google Scholar]
- 44.Sawhney A, Pathak C, Hubbell J. Bioerodible hydrogels based on photopolymerized poly (ethylene glycol)-co-poly (alpha-hydroxy acid) diacrylate macromers. Macromolecules. 1993;26:518–587. doi: 10.1021/ma00056a005. [DOI] [Google Scholar]
- 45.Shekhar MPV. Drug resistance: challenges to effective therapy. Curr. Cancer Drug Targets. 2011;11:613–623. doi: 10.2174/156800911795655921. [DOI] [PubMed] [Google Scholar]
- 46.Sokolov I. Toward the nanoscale study of insect physiology using an atomic force microscopy-based nanostethoscope. MRS Bull. 2012;37:522–527. doi: 10.1557/mrs.2012.91. [DOI] [Google Scholar]
- 47.Takagi J, Yang Y, Liu J, Wang J, Springer TA. Complex between nidogen and laminin fragments reveals a paradigmatic β-propeller interface. Nature. 2003;424:969–974. doi: 10.1038/nature01873. [DOI] [PubMed] [Google Scholar]
- 48.Tibbitt MW, Anseth KS. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009;103:655–663. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timpl R, Wiedemann H, van Delden V, Furthmayr H, Kühn K. A network model for the organization of type IV collagen molecules in basement membranes. Eur. J. Biochem. 1981;120:203–211. doi: 10.1111/j.1432-1033.1981.tb05690.x. [DOI] [PubMed] [Google Scholar]
- 50.Ushiki T. Collagen fibers, reticular fibers and elastic fibers. A comprehensive understanding from a morphological viewpoint. Arch. Histol. Cytol. 2002;65:109–126. doi: 10.1679/aohc.65.109. [DOI] [PubMed] [Google Scholar]
- 51.Vyas V, Nagarajan N, Zorlutuna P, Huey BD. Nanostethoscopy: atomic force microscopy probe contact force versus measured amplitude of cardiomyocytic contractions. J. Bionanosci. 2017;11:319–322. doi: 10.1166/jbns.2017.1441. [DOI] [Google Scholar]
- 52.Vyas V, Podestà A, Milani P. Probing nanoscale interactions on biocompatible cluster-assembled titanium oxide surfaces by atomic force microscopy. J. Nanosci. Nanotechnol. 2011;11:4739–4748. doi: 10.1166/jnn.2011.4113. [DOI] [PubMed] [Google Scholar]
- 53.Vyas V, Solomon M, D’Souza GGM, Huey BD. Dynamic and depth dependent nanomechanical properties of dorsal ruffles in live cells and biopolymeric hydrogels. J. Nanosci. Nanotechnol. 2018;18:1557–1567. doi: 10.1166/jnn.2018.14219. [DOI] [PubMed] [Google Scholar]
- 54.Wenger MP, Bozec L, Horton MA, Mesquida P. Mechanical properties of collagen fibrils. Biophys. J. 2007;93:1255–1263. doi: 10.1529/biophysj.106.103192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winer JP, Oake S, Janmey PA. Non-linear elasticity of extracellular matrices enables contractile cells to communicate local position and orientation. PLoS ONE. 2009;4:e6382. doi: 10.1371/journal.pone.0006382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang YL, Leone LM, Kaufman LJ. Elastic moduli of collagen gels can be predicted from two-dimensional confocal microscopy. Biophys. J. 2009;97:2051–2060. doi: 10.1016/j.bpj.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yurchenco PD, Patton BL. Developmental and pathogenic mechanisms of basement membrane assembly. Curr. Pharm. Des. 2009;15:1277–1294. doi: 10.2174/138161209787846766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yurchenco PD, Schittny JC. Molecular architecture of basement membranes. FASEB J. 1990;4:1577–1590. doi: 10.1096/fasebj.4.6.2180767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.