Abstract
Accurate classification and localization of anatomical structures in images is a precursor for fully automatic image-based diagnosis of placental abnormalities. For placental ultrasound images, typically acquired in clinical screening and risk assessment clinics, these structures can have quite indistinct boundaries and low contrast, and image-level interpretation is a challenging and time-consuming task even for experienced clinicians. In this paper, we propose an automatic classification model for anatomy recognition in placental ultrasound images. We employ deep residual networks to effectively learn discriminative features in an end-to-end fashion. Experimental results on a large placental ultra-sound image database (10,808 distinct 2D image patches from 60 placental ultrasound volumes) demonstrate that the proposed network architecture design achieves a very high recognition accuracy (0.086 top-1 error rate) and provides good localization for complex anatomical structures around the placenta in a weakly supervised fashion. To our knowledge this is the first successful demonstration of multi-structure detection in placental ultrasound images.
1. Introduction
Ultrasonography is a low-cost, non-invasive and non-radiative technique used worldwide for clinical assessment of the human placenta. Expertise is required to both acquire placental ultrasound images and to perform clinical diagnosis from them. These images are particularly challenging for automated biomedical image analysis as the contrast between the textured areas of interest is often low.
Abnormally invasive placentation (AIP) is a general term that covers conditions where the human placenta adheres to the uterus in an invasive fashion. Various diagnostic criteria based on placental ultrasound imaging have been reported or suggested in the literature to characterise this condition [3]. The general approach is to first detect and localise anatomical structures such as the placenta itself, the utero-placental interface and the myometrium within grayscale ultrasound images (B-Mode). Vascular examination using Doppler ultrasound imaging can provide further evidence to support diagnosis (analysis of Doppler is beyond the scope of the current paper). However, interpretation of the criteria by sonographers is quite inclined to subjectivity [4]. Moreover, manual search for visual evidence among sequences of 2D or 3D placental ultrasound data is sometimes too time-consuming to be considered in clinical workflow.
The contributions in this paper are twofold. First, we propose a deep convolutional neural network (CNN) model for describing anatomical structures present in a 2D placental ultrasound image. This image-level model achieves accurate classification (0.086 top-1 error rate) of the four multi-anatomical structure combinations typically observed in a 2D placental image (as illustrated in Fig. 1), namely (1) placenta only (PL); (2) placenta and myometrium (PL+MY); (3) placenta and subcutaneous tissue (PL+ST); (4) placenta, myometrium and subcutaneous tissue (PL+MY+ST). Second, we show that the proposed model achieves good localization of anatomical structures (placenta, myometrium, sub-cutaneous tissue) based on our multi-structure classification formulation. This is achieved by incorporating a global average pooling (GAP) layer before the fully-connected layer. Thus we demonstrate that image-level classification suffices for localization of anatomical structures in a weakly supervised fashion without any additional training.
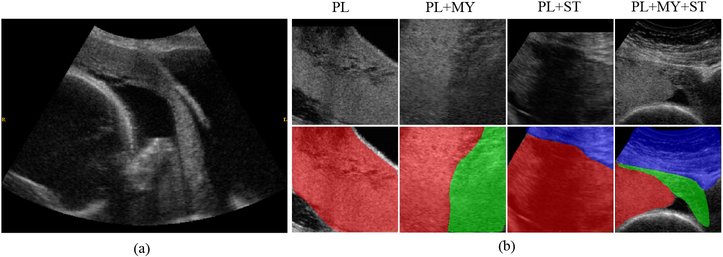
Fig. 1.
(a) a placental ultrasound image taken from the sagittal plane. (b) samples from four image categories cropped from sagittal planes, the bottom row shows the reference segmentation mask of the follow anatomical structures: placenta (PL), subcutaneous tissue (ST) and myometrium (MY). Please note that all segmentation masks that appear in this paper are used solely for illustration purpose rather than training models.
2. Related Work
Weakly-supervised object localization:
CNN-based weakly-supervised object localization has been a popular research topic in computer vision in recent years and applications are starting to appear in the medical image analysis literature [10], though not to our knowledge for placental ultrasound image analysis. It relies only on image-level labels, rather than annotations in a fully-supervised setting (e.g. manually-annotated bounding box or dense pixel-level annotation), to learn from cluttered scenes with multiple objects. It is of great research interest to develop weakly-supervised localization models that perform comparably to its fully-supervised counterparts due to the fact that the former saves a considerable amount of time in annotation and is less prone to subjectivity. Recent work has further demonstrated that CNNs originally trained for image classification can be used to localize objects via analysis of representative features across layers [1, 11, 16, 13]. For instance, Simonya et al. proposed a visualization technique by computing the gradient of the class score with respect to the input image [13]. The resulting saliency map pinpoints the location of objects correlated with the class label. Oquad et al. proposed a method to transfer mid-level image representations and explicitly search for high-score regions [11]. Zhou et al. recently proposed class activation mapping (CAM) to localize regions with discriminative features in an end-to-end fashion [16]. In general, weakly-supervised localization relies only on image-level classification, which is a desirable property that makes object localization a preferable by-product of classification without additional training.
3. Learning to Classify and Localize with Residual Units
Problem formulation:
Our approach is built on an observation that there are four local anatomical scenarios, which clinicians observe in routine placenta scans, namely PL, PL+MY, PL+ST, PL+MY+ST. Thus we have designed a CNN to distinguish between these classes. Further, since the placenta (PL) is shared in all classes it acts as a distractor for localization. To be discriminative, the other three categories are forced to activate their unique regions, which can then be visualized by CAM as is described later. First, however, we describe the general CNN architecture we use.
Deep residual learning for placental ultrasound images:
Deep residual networks (Res-Net [7, 8]) have shown impressive representative ability and good convergence behaviours in recent large-scale natural image classification tasks (e.g. ImageNet ILSVRC 2015 [12]), yielding state-of-the-art performance. In a recent work [8], a simple and effective identity mapping structure was proposed to enable smooth information propagation through the entire network. In addition, large-scale data experiments reveal that the full pre-activation residual unit, as shown in the top-left corner of Fig. 2, consistently outperforms the original design by putting batch normalization [9] and rectified linear unit (ReLU) before convolution. This network design modification has been found to accelerate learning and improves global regularization. In general, a Res-Net typically contains a number of basic residual units. Each unit performs the following computation: , where xl refers to the input feature to the l-th residual unit and is a set of weights and biases associated with the l-th residual unit. Here denotes the residual function which is learnt with respect to the input feature xl. Such a design allows a recursive derivation:
for any deeper unit L and any shallow unit l, which implies the smooth information propagation through the network.
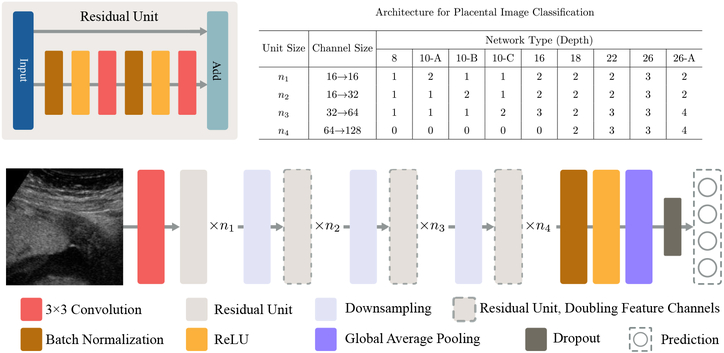
Fig. 2.
Proposed deep residual network architecture with different sizes of residual units, as listed in the table.
We adopted the pre-activation residual unit in [8] and designed a series of multi-layer Res-Nets with various representative abilities. The table in Fig. 2 shows different network architectures. In general, the model contains four groups of residual modules, each of which further consists of ni stacked residual units for i = 1, 2, 3, 4. Here ni is an architecture hyper-parameter that controls the entire depth D of the Res-Net, where D = 2(n1 + n2 + n3) + 2 denotes the number of convolutional layer. We follow the principle adopted in recent recognition and segmentation researches [14, 2, 7] to employ a small convolutional kernel of size 3 × 3 for all the convolutional layers in Res-Nets. At the beginning of the second, third and fourth residual modules, convolutional layers with stride 2 are used to downsample the feature map. Meanwhile, the convolution doubles the feature channel size (also by two), yielding the change of channel sizes: 16 → 32 → 64 → 128. It is followed by a global average pooling (GAP) layer and a fully-connected layer to generate the final prediction. To increase regularization, we use a dropout layer [15] with dropout probability of p = 0.3, which demonstrates a good regularization performance across various architectures according to experiments. The use of GAP is described in the following subsection to boost discriminative localization. The Res-Nets are trained in an end-to-end fashion by stochastic gradient descent with a momentum of 0.9. We tested different combinations of hyper-parameter ni and report results in the following section.
Global average pooling (GAP) for localization:
As shown in [16], the use of GAP encourages the network to identify the extent of the object, rather than pinpointing the object on a specific location as global max pooling (GMP) does [11]. It is intuitive that this global averaging operation should boost identification of a local discriminative region in order to reach a lower global loss, while a global maximum operation only influences the maximal value of a feature map. We take advantage of GAP to generate CAM [16]. Each class has its corresponding CAM which visualizes discriminative image regions used by the network to identify this specific class. As displayed in the bottom of Fig. 3, the CAM for class i is generated by computing a weighted sum of all feature maps from the rectified activation of the last convolutional layer. Here the weights refer to the corresponding weight vector learnt in the fully-connected layer. A simple up-sampling would suffice to map the CAM heat-map (28 × 28) back to the input size (224 × 224).
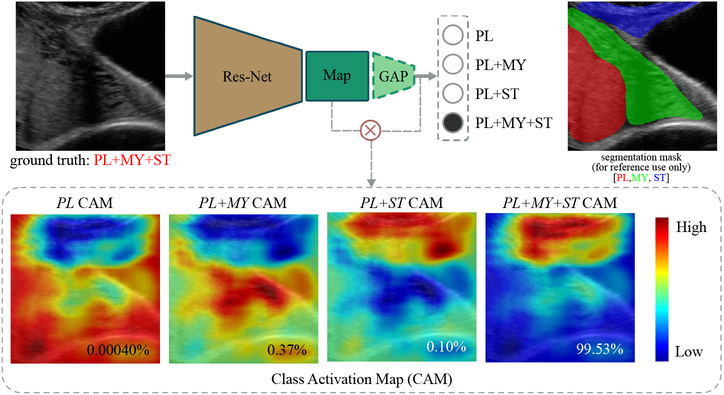
Fig. 3.
Proposed pipeline for placental ultrasound image detection and localization. Given an unseen image patch, the network can predict its label, which indicates the multi-anatomical structure combination within. By computing the class activation maps for each category, the network can provide reasonable localization on the detected anatomical structures.
For PL+MY, its CAM is expected to visualize the myometrium region (MY). For the same reason, PL+ST CAM would illustrate the region of subcutaneous tissue (ST) and PL+MY+ST would ideally highlight the joint region of myometrium and subcutaneous tissue. There are potentially many ways to formulate this classification problem. The most intuitive way is probably to build a multi-label learning model by training a group of one-verses-all binary classifiers to identify these anatomical structures respectively. However, this type of model suffers from over-fitting and generalization problems in our data experiments. One possible explanation is that there are strong correlations among these anatomical structures (e.g. myometrium is almost always co-localized with placenta), thus it may not be appropriate to model them separately. Moreover, the multi-label learning does not contribute to the generation of CAM due to the removal of softmax normalization. In this paper we formulate the problem with the intention to both achieve high classification accuracy and boost weakly supervised localization. As shown in Fig. 4, experimental results confirm the validity of our formulation. CAM demonstrates reasonable localization ability for the corresponding anatomical structures. More details will be discussed in the following sections. In this section, we present the two major parts of the proposed model for placental ultrasound image classification and anatomy localization. An overview of our pipeline is given in Fig. 3. We exploit residual units, GAP and CAM to classify ultrasound images and to localize corresponding anatomical structures.
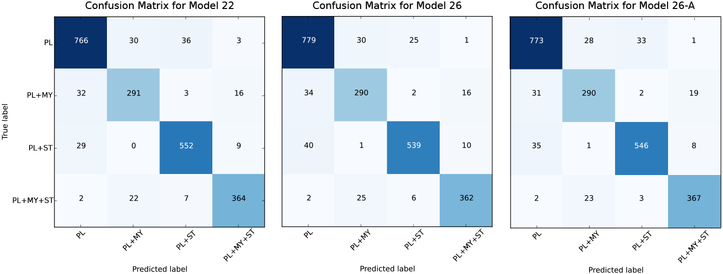
Fig. 4.
Confusion Matrices in the test stage for best three models.
4. Experiments
To evaluate our proposed method, we conducted data experiments on a placental ultrasound image dataset, which was collected as part of a large placenta clinical study. Classification performance of different CNN models are presented. Results of the weakly-supervised localization are also displayed. All the images used in this work are obtained from the sagittal plane, annotated by H.Q. under the guidance of S.C., who is a consultant obstetrician and subspecialist in maternal and fetal medicine of John Radcliffe Hospital in Oxford. This method is implemented in Torch 7 [6] on a 64-bit Ubuntu 15.04 machine with a NVIDIA graphics card.
Dataset:
All placental ultrasound data (N = 60) used in this experiment were obtained as part of a large obstetrics research project [5]. Written informed consent was obtained with local research ethics approval. Static, transabdominal 3D gray-scale ultrasound volumes of the placental bed were obtained according to a predefined protocol with the participant in a semi-recumbent position and a full bladder using a 3D curved array abdominal transducer on a GE Voluson E8 machine. Each 3D volume was then sliced along the sagittal plane into 2D placental ultrasound images, as shown in Fig. 1(a). We then randomly cropped 224 × 224 image patches from these sagittal planes and formed a training and testing dataset by annotating the patches into the four categories described in Sect. 3. In total, the dataset contains 10,808 placental image patches, which is then randomly divided into a training set (64%), a validation set (16%) and a test set (20%), as shown in Table 1. Here the validation set is used to tune CNN hyper-parameters such as the learning rate, weight decay and architecture parameters {ni}.
Table 1.
Statistics of Placental Ultrasound Image Dataset
| Training | Val. | Test | Total | |
|---|---|---|---|---|
| PL | 2,764 | 711 | 835 | 4,310 |
| PL+MY | 1,064 | 283 | 342 | 1,689 |
| PL+ST | 1,834 | 422 | 590 | 2,846 |
| PL+MY+ST | 1,256 | 312 | 395 | 1,963 |
| Total | 6,918 | 1,728 | 2,162 | 10,808 |
Evaluation metrics:
We used top-1 error rate to evaluate the classification performance of our proposed method. Experimental results are presented in Table 2. For reference, we also list the top-1 error rate for the validation set and the classification error for individual image categories during the test. It is worth noting that PL+MY presents the worst classification performance as expected. In placental ultrasound imaging, it is generally difficult to identify the myometrium for various reasons. First, it may not exist at all due to certain placental abnormality (such as AIP). Second, it appears but the texture or intensity is not sufficiently discriminative to be identified. Third, ultrasound signal dropout may hinder a clear visualization. We also present confusion matrices in the test stage for our best three models in Fig. 4. They all suffer from the same problem of identifying PL+MY.
Table 2.
Classification Performance of Various Architectures
| Network type | Mean error (val., %) | Mean error (test, %) | Class error (test, %) | |||
|---|---|---|---|---|---|---|
| PL | PL+MY | PL+ST | PL+MY+ST | |||
| 8 | 18.23 | 17.81 | 13.05 | 26.61 | 20.85 | 15.70 |
| 10-A | 14.53 | 14.29 | 9.82 | 29.53 | 12.71 | 12.91 |
| 10-B | 12.85 | 13.55 | 8.74 | 30.41 | 11.36 | 12.41 |
| 10-C | 10.82 | 11.29 | 7.07 | 23.98 | 9.15 | 12.41 |
| 16 | 11.40 | 10.78 | 8.02 | 22.22 | 9.15 | 9.11 |
| 18 | 9.95 | 9.20 | 8.62 | 19.59 | 6.10 | 6.08 |
| 22 | 8.56 | 8.74 | 8.26 | 14.91 | 6.44 | 7.85 |
| 26 | 9.84 | 8.88 | 6.71 | 15.21 | 8.64 | 8.35 |
| 26-A | 8.08 | 8.60 | 7.43 | 15.21 | 7.46 | 7.09 |
Architecture hyper-parameter:
In Section 3, we introduced four architecture hyper-parameters n1, n2, n3, n4, corresponding to the number of stacked residual units in each residual module. By altering these hyper-parameters, we can investigate how network depth casts impact on the generalization ability of our classification problem at different abstraction levels. As shown in Fig. 2, nine architectures were evaluated. Here A,B,C denotes variants for models of the same depth. Referring to classification performance in Table 2 we see that: (1) more residual units should be put in deeper modules that have larger channel sizes, as demonstrated by the better performance of 10-C and 26-A compared to their counterparts of the same depth; (2) the identity mapping structure of Res-Net indeed boosts the propagation of information, yielding better performance for deeper networks without causing degradation problems described in [7].
Weakly-supervised localization:
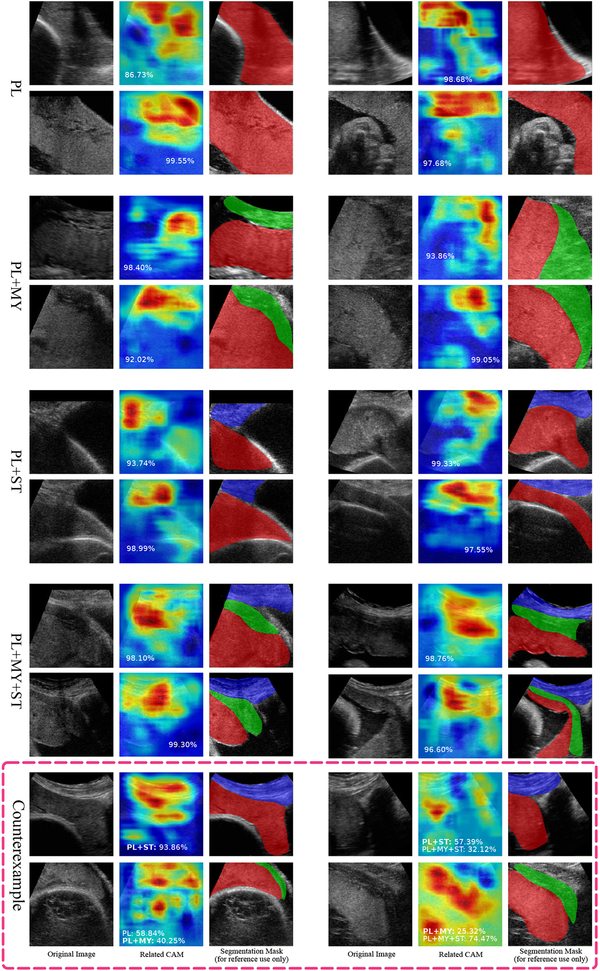
Here we show results of the weakly supervised localization of placental anatomical structures based on the learnt classification model (model 26-A). An input image was first classified into one of the four categories. After this, we generated its CAM for the predicted category, which highlights the discriminative regions of this image. Some results are shown in Fig. 5 for each category, where we also provide softmax scores as well as segmentation masks for illustration. For example in the first triple set, the input image is correctly classified as PL with a score of 0.8673. CAM heat-map for PL highlights the approximate position of the placenta, as verified by the reference segmentation mask. We also present some counterexamples in the bottom of Fig. 5, which are either mis-classified (the correct class is labelled in bold font), mis-localized or both. Good weakly-supervised localization tends to be achieved based on an accurate classification.
Fig. 5.
Results of weakly supervised localization with reference segmentation mask of the follow anatomical structures: placenta (PL), subcutaneous tissue (ST) and myometrium (MY), all images are from the test set.
Discussions:
In the CAM heat-maps of Fig. 5, we observe that the network appears to use texture as well as boundary information as discriminative features. For example, the PL CAM hot zone typically covers a partition of the placenta as well as the placenta-background boundaries. Similarly, the PL+MY CAM hot zone covers a part of the myometrium and the myometrium-placenta boundaries. The PL+ST CAM hot zone contains the subcutaneous tissue as well as the tissue-placenta interface. Finally, the PL+MY+ST CAM hot zone contains regions of both myometrium and subcutaneous tissue, as well as their interface. This is observed across the test set. Joint analysis of PL+MY+ST CAM and PL+MY CAM will be carried out in the future, which may provide some insight to help further refine weakly-supervised localization of the myometrium since they both contain a partition of the anatomical structure.
5. Conclusion
In this paper, we have formulated automatic placental ultrasound image structure detection and localization as a multi-structure classification problem. The proposed model is based on deep residual networks. Experimental results show good detection accuracy using our approach. Moreover, we demonstrate that reasonable localization of placental anatomical structures can be achieved, without explicit training to perform localization.
References
- [1].Bazzani L, Bergamo A, Anguelov D, Torresani L: Self-taught object localization with deep networks. In: Applications of Computer Vision (WACV), 2016 IEEE Winter Conference on. pp. 1–9. IEEE; (2016) [Google Scholar]
- [2].Chen H, Dou Q, Yu L, Heng PA: Voxresnet: Deep voxelwise residual networks for volumetric brain segmentation. arXiv preprint arXiv:1608.05895 (2016) [DOI] [PubMed] [Google Scholar]
- [3].Collins SL, Ashcroft A, Braun T, Calda P, Langho-Roos J, Morel O, Stefanovic V, Tutschek B, Chantraine F: Proposal for standardized ultrasound descriptors of abnormally invasive placenta (aip). Ultrasound in Obstetrics & Gynecology (2016) [DOI] [PubMed] [Google Scholar]
- [4].Collins SL, Stevenson GN, Al-Khan A, Illsley NP, Impey L, Pappas L, Zamudio S: Three-dimensional power doppler ultrasonography for diagnosing abnormally invasive placenta and quantifying the risk. Obstetrics & Gynecology (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Collins S, Stevenson G, Noble J, Impey L, Welsh A: Influence of power doppler gain setting on virtual organ computer-aided analysis indices in vivo: can use of the individual sub-noise gain level optimize information? Ultrasound in Obstetrics & Gynecology (2012) [DOI] [PubMed] [Google Scholar]
- [6].Collobert R, Kavukcuoglu K, Farabet C: Torch7: A matlab-like environment for machine learning. In: BigLearn, NIPS Workshop (2011) [Google Scholar]
- [7].He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 770–778 (2016) [Google Scholar]
- [8].He K, Zhang X, Ren S, Sun J: Identity mappings in deep residual networks. In: European Conference on Computer Vision. pp. 630–645. Springer; (2016) [Google Scholar]
- [9].Io e S, Szegedy C: Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167 (2015) [Google Scholar]
- [10].Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B: Efficient multi-scale 3d cnn with fully connected crf for accurate brain lesion segmentation. Medical Image Analysis 36, 61–78 (2017) [DOI] [PubMed] [Google Scholar]
- [11].Oquab M, Bottou L, Laptev I, Sivic J: Is object localization for free?-weakly-supervised learning with convolutional neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 685–694 (2015) [Google Scholar]
- [12].Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L: ImageNet Large Scale Visual Recognition Challenge. International Journal of Computer Vision (IJCV) 115(3), 211–252 (2015) [Google Scholar]
- [13].Simonyan K, Vedaldi A, Zisserman A: Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv preprint arXiv:1312.6034 (2013) [Google Scholar]
- [14].Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014) [Google Scholar]
- [15].Srivastava N, Hinton GE, Krizhevsky A, Sutskever I, Salakhutdinov R: Dropout: a simple way to prevent neural networks from overfitting. Journal of Machine Learning Research 15(1), 1929–1958 (2014) [Google Scholar]
- [16].Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A: Learning deep features for discriminative localization. In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (June 2016) [Google Scholar]