Abstract
Background
Cytochrome P4501A1 (CYP1A1) is a member of the cytochrome P450 gene family and plays an important role in the metabolism of exogenous and endogenous material. In recent research, it has been shown that its genetic polymorphisms are associated with many diseases. But the isoschizomers such as the BsrDI enzyme required for the detection of this polymorphism are expensive.
Methods
The study used an improved PCR‐RFLP method with mismatched base for detection of the single‐nucleotide polymorphism rs1048943. A new restriction enzyme cutting site was created by created restriction site PCR (CRS‐PCR), and the restriction enzyme StyI for RFLP was cheaper than other enzymes. A total of 320 samples from Han Chinese were tested to evaluate this novel method. The PCR results were confirmed by DNA sequencing.
Results
After detecting 320 Chinese Han individuals, the genotype frequencies were 63.74% for AA, 31.54% for AG, and 4.72% for GG. The allelic frequencies were 75.48% for A and 24.52% for G. The x 2 test showed the genotype and allele frequencies of CYP1A1 do not deviate from Hardy‐Weinberg equilibrium, and the sequences of amplified products were consistent with the one published in GenBank with the exception of mismatched base.
Conclusions
Based on PCR with mismatched primers we designed, theCYP1A1 polymorphism could be identified effectively with low cost.
Keywords: Cytochrome P4501A1, Single‐nucleotide polymorphism, PCR‐RFLP, created restriction site, rs1048943
1. Introduction
Cytochrome P4501A1 (CYP1A1) is a member of the cytochrome P450 gene family located on chromosome 15, is one of the most important phases I enzymes, and plays an important role in the metabolism of exogenous and endogenous material. In recent research, it has been shown that there have been variants of rs1048943 within the cytochrome P4501A1 (CYP1A1) gene, which have been associated with smooth muscle tumor of the uterus,1 chronic obstructive pulmonary disease2 and esophageal cancer3 and other diseases.
Polymorphism of CYP1A1 gene has four sites, an A/G single‐nucleotide polymorphism (SNP) rs1048943 in exon 7 of the CYP1A1 gene introduces the coding variant of Ile/Val. This mutation enhances the expressive activity of the CYP1A1 or significantly increased. At present, the polymorphism studies associated with the diseases are in progress. For this SNP detection, there are many methods for SNP detection, including dynamic allele‐specific hybridization (DASH),4 fluorescent labeled probes technology,5 and polymerase chain reaction‐restriction fragment length polymorphism6 (PCR‐RFLP). Although the above methods have its own advantages, the complexity and expense of the methods limit their use. PCR‐RFLP is widely used in detection of polymorphism of the human genome owing to its high sensitivity and easy operation and reliability. However, some enzymes like, the enzyme for CYP1A1 gene (rs1048943), BsrDI are very expensive. In this study, we demonstrated an improved PCR‐RFLP assay for the detection of a polymorphism of the CYP1A1 gene.
2. Materials and Methods
2.1. Primer design
The sequence of CYP1A1 gene and polymorphism rs1048943 was abstracted from NCBI website (http://www.ncbi.nlm.nih.gov). The sequences of primer pairs were designed by primer premier 5.0 software as follows:
Forward: 5′CCACTCACTTGACACTTCTGAGCCC 3′
Reverse: 5′AAAGACCTCCCAGCGGGCCA 3′
The natural occurring DNA sequences according to the positions of primers were CCACTCACTTGACACTTCTGAGCCCtgaactgccacttcagctgtctccctctggttacaggaagctatgggtcaacccatctgagttcctacctgaacggtttctcacccctgatggtgctatcgacaaggtgttaagtgagaaggtgattatctttggcatgggcaagcggaagtgtatcggtgagaccrtTGCCCGCTGGGAGGTCTTT.
Among the sequence, the capital bases A, T, C, and G represent for the same sequences as primers. The r represents polymorphism A/G. The length of target DNA sequence is 212 bp. To introduce a recognized site for the StyI restriction endonuclease, the unpaired base “C” (bold italic) in the reverse primer was used in the PCR reaction, and then the natural occurring “T” nucleotide (bold italic) in the DNA sequences was changed to a “G” in the PCR products.
2.2. Genotyping of DNA
Genomic DNA was extracted from peripheral blood leukocytes using genomic DNA isolation kit (Bio Teke Corporation, Beijing, China). PCR reactions were performed in a final volume of 15 μL containing 50 ng of genomic DNA, 0.6 μmol/L of each primer (Sangon Biotech, Shanghai, China), 1× PCR mix (VazymeBiotech, Nanjing, China), and double distilled water as follows: at 94°C for 5 minutes; at 94°C—61°C—72°C for 30 seconds each, 30 cycles; and then at 72°C for 7 minutes.
2.3. RFLP conditions
Enzyme digestion was carried out in a 20 μL final volume reaction mix consisting of 5 units (U) of StyI enzyme (New England Biolabs, Los Angeles, CA, USA), 2 μL 10× Buffer Tango, 7.5 μL double distilled H2O, and 10 μL of PCR reaction mixture. The reaction was performed at 37°C overnight, and the digested products were electrophoresed on 3% agarose gel containing ethidium bromide (0.5 μg/mL) for 50 minutes.
2.4. DNA sequencing
PCR products sequences were detected by Sangon Biotech Company (Shanghai, China), and the forward primer was used in the sequencing process.
3. Results
The prices of the two restriction enzymes that were from New England Biolabs website (NEB) are as Table 1. Compared to using BsrDI enzyme, about 92% the cost was saved using StyI.
Table 1.
Recognition sites and prices of respective endonucleases in NEB Co
| Endonuclease | Recognition sequence | $/500 units |
|---|---|---|
| BsrDI | GCAATGNNˆ | 231 |
| StyI | CˆCWWGG | 19 |
We successfully introduced a StyI restriction enzyme site into the PCR products through CRS‐PCR. As can be seen in the Figure 1, the PCR amplification yielded a product of 212 bp. After digestion with StyI, homozygous allele G had only one 212 bp band. The homozygous allele A showed two bands of 190 and 22 bp, respectively.
Figure 1.
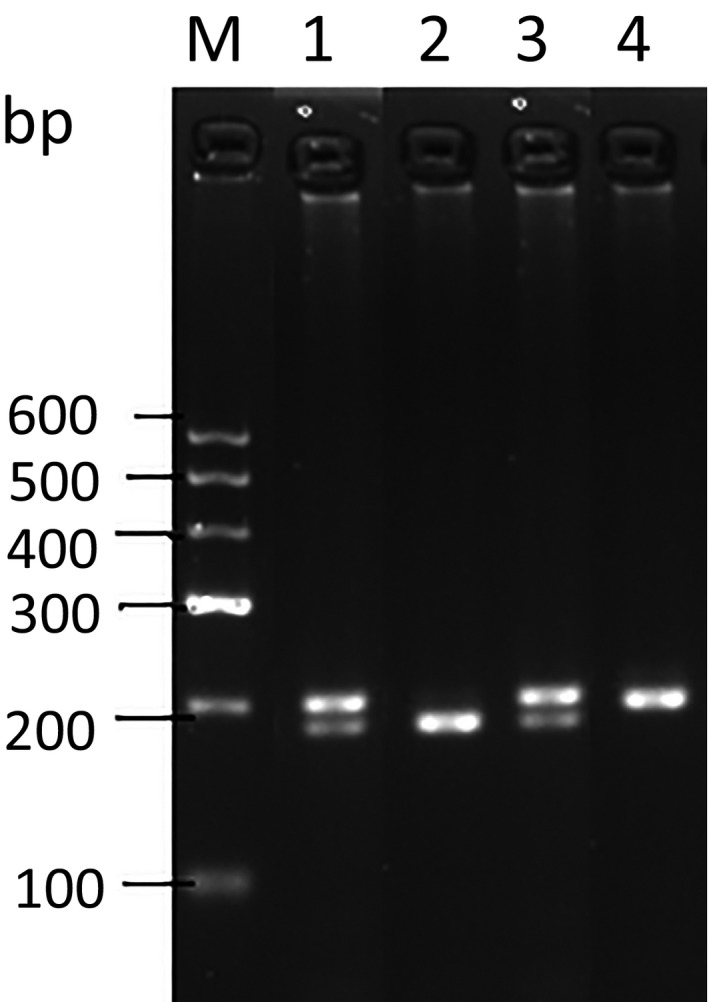
StyI PCR‐RFLP analysis of the CYP1A1 biallelic polymorphism. M, DNA ladder; Lane1, AG; Lane2, GG; Lane4, AA
Polymorphisms of CYP1A1 were detected in a population of 320 Chinese Han individuals who came from one enterprise in Zhengzhou. The genotype frequencies were 63.74% for AA, 31.54% for AG, and 4.72% for GG. The allelic frequencies were 75.48% for A and 24.52% for G. The PCR results were confirmed by DNA sequencing. The χ2 test showed the genotype and allele frequencies of CYP1A1 do not deviate from Hardy‐Weinberg equilibrium (χ2=0.29, P>.05).
Examples of DNA sequencing of the PCR product of the CYP1A1 gene were shown in the Figure 2. As expected, the sequences of amplified products were consistent with the one published in GenBank with the exception of the mismatched base.
Figure 2.
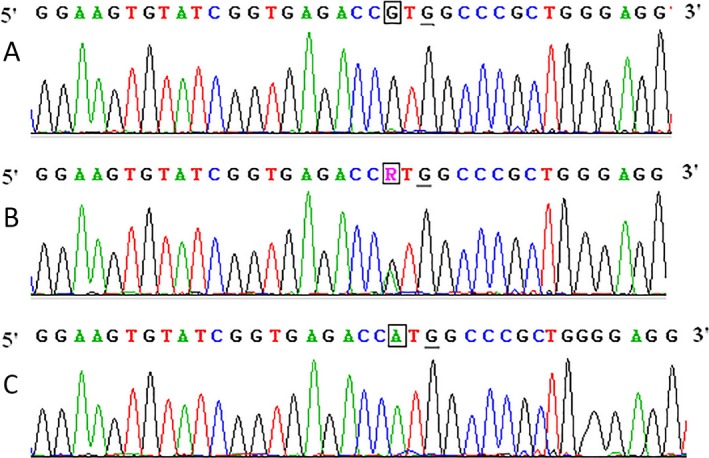
Examples of DNA sequencing of PCR product of CYP1A1 gene. The underlined base G is mismatched base. Three figures representing the genotype of (A) GG, (B) AG, and (C) AA are shown, and the bases R (A and G) representing the polymorphism sites are in the squares
4. Discussion
Li et al.7 studied the association between CYP1A1 gene polymorphism and cervical cancer risk in Chinese women and found that compared to subjects with AA of rs1048943 genotype, subjects with AG or GG of rs1048943 genotype have the highest cervical cancer risk. Research has shown that gene polymorphisms of CYP1A1 were found to contribute significant risk to presbycusis.8 Fan et al.9 studied the association between CYP1A1 Ile462Val polymorphism and endometriosis risk, particularly in Asians. We can by detecting polymorphism loci screening susceptible populations. Therefore, the SNP detection is becoming more and more important.
For SNP detection using CRS‐PCR‐RFLP method, both at home and abroad have reported.10, 11 In this study, we introduced a new restriction enzyme site for StyI to type the CYP1A1 polymorphism rs1048943. Compared to using BsrDI enzyme, about 92% the cost was saved using StyI. By establishing the method of detecting polymorphism of the CYP1A1 gene, polymorphism analysis provides a simple and economical method. This method has gradually served to some tests,12, 13 and related research methods are increasingly mature. CRS‐PCR‐RFLP greatly widened the scope of the PCR‐RFLP and has much flexibility in practical application. It has a good application prospect, especially suitable for the basic unit to carry out the work of knew site SNP. CRS‐PCR‐RFLP method also has a few limitations. This method cannot detect multiple SNPs in the same reaction, and it cannot realize high‐throughput detection.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Acknowledgments
The present study was supported by the Outstanding Youth Grant of Zhengzhou University(1521329035).
Ding M, Duan X, Feng X, Wang P, Wang W. Application of CRS‐PCR‐RFLP to identify CYP1A1 gene polymorphism. J Clin Lab Anal. 2017;31:e22149 10.1002/jcla.22149
References
- 1. Salimi S, Sajadian M, Khodamian M, et al. Combination effect of cytochrome P450 1A1 gene polymorphisms on uterine leiomyoma: a case‐control study. Bosn J Basic Med Sci. 2016;16:209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Korytina GF, Akhmadishina LZ, Kochetova OV, Zagidullin SZ, Viktorova TV. [Association of cytochrome P450 genes polymorphisms (CYP1A1 and CYP1A2) with the development of chronic obstructive pulmonary disease in Bashkortostan]. Mol Biol. 2008;42:32–41. [PubMed] [Google Scholar]
- 3. Gong FF, Lu SS, Hu CY, et al. Cytochrome P450 1A1 (CYP1A1) polymorphism and susceptibility to esophageal cancer: an updated meta‐analysis of 27 studies. Tumour Biol. 2014;35:10351–10361. [DOI] [PubMed] [Google Scholar]
- 4. Russom A, Haasl S, Brookes AJ, Andersson H, Stemme G. Rapid melting curve analysis on monolayered beads for high‐throughput genotyping of single‐nucleotide polymorphisms. Anal Chem. 2006;78:2220–2225. [DOI] [PubMed] [Google Scholar]
- 5. Goni L, Cuervo M, Milagro FI, Martinez JA. A genetic risk tool for obesity predisposition assessment and personalized nutrition implementation based on macronutrient intake. Genes Nutr. 2015;10:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fontecha GA, Garcia K, Rueda MM, Sosa‐Ochoa W, Sanchez AL, Leiva B. A PCR‐RFLP method for the simultaneous differentiation of three Entamoeba species. Exp Parasitol. 2015;151–152:80–83. [DOI] [PubMed] [Google Scholar]
- 7. Li S, Li G, Kong F, et al. The association of CYP1A1 gene with cervical cancer and additional SNP‐SNP interaction in Chinese women. J Clin Lab Anal. 2016;30:1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Manche SK, Jangala M, Putta P, Koralla RM, Akka J. Association of oxidative stress gene polymorphisms with presbycusis. Gene. 2016;593:277–283. [DOI] [PubMed] [Google Scholar]
- 9. Fan W, Huang Z, Xiao Z, Li S, Ma Q. The cytochrome P4501A1 gene polymorphisms and endometriosis: a meta‐analysis. J Assist Reprod Genet. 2016;33:1373–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Viana AC, Kim YJ, Cirelli JA, et al. A novel PCR‐RFLP assay for the detection of the single nucleotide polymorphism at position+1440 in the human CXCR2 gene. Biochem Genet. 2007;45:737–741. [DOI] [PubMed] [Google Scholar]
- 11. Feng X, Wang S, Duan X, et al. An improved PCR‐RFLP assay for the detection of a polymorphism rs2289487 of PLIN1 gene. J Clin Lab Anal. 2016;30:986–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu WW, Hou LL, Zhang WW, et al. Study on heteroplasmic variation and the effect of chicken mitochondrial ND2. Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27:2303–2309. [DOI] [PubMed] [Google Scholar]
- 13. Wang J, Wang C, Tian R, et al. Sequence variants in the bovine PRDM16 gene associated with body weight in Chinese cattle breeds. Genet Mol Res. 2012;11:746–755. [DOI] [PubMed] [Google Scholar]


