Abstract
Background
The increase in bla NDM ‐1 in Enterobacteriaceae has become a major concern worldwide. In previous study, we investigated clonal dissemination and mechanisms of resistance to carbapenem in China.
Methods
We carried out retrospective surveillance for bla NDM ‐1 among carbapenem‐resistant enterobacter strains, which were isolated from patients at our hospital by bacterial strains selection, antimicrobial susceptibility testing, species identification, and molecular detection of resistance gene.
Results
We found three bla NDM ‐1‐positive isolates which were identified as Enterobacter aerogenes in clinical patients in China. The bla NDM ‐1‐positive Enterobacter aerogenes isolates were first found.
Conclusion
It is important to mandate prudent usage of antibiotics and implement infection control measures to control the spread of these resistant bla NDM ‐1‐positive strains.
Keywords: blaNDM‐1, enterobacter aerogenes, New Delhi metallo‐Beta‐Lactamase, resistance
Introduction
The increase of the novel carbapenem resistance mechanism of New Delhi metallo‐beta‐lactamase (bla NDM‐1) in Enterobacteriaceae has become a major concern worldwide 1. The bla NDM‐1 gene was first reported in 2009 in a Klebsiella pneumonia isolated in a Swedish hospital from a patient previously hospitalized in a medical institution in India 2. Since then, the isolation of bacterial species carrying the bla NDM‐1 gene has been reported in several countries, usually from patients who travelled to the Indian subcontinent.
There were several studies which demonstrated the high similarity of the genomic region surrounding bla NDM‐1 gene among members of Enterobacteriacae 1, 2, 3. Moreover, bla NDM‐1‐positive plasmids were revealed by complete genome sequencing in E. coli that was indicated a broad host range and multiple mobile genetic elements, and highlighted the potential for the extensive spread of this determinant of resistance.
There were some previous reports on bla NDM‐1 gene which was focused on its epidemiological dissemination in different countries, including India, China, the United States, Germany, France, Canada, Austria, Africa, and the United Kingdom 1, 4. However, there was no bla NDM‐1–producing Enterobacter aerogenes detected in these studies. In the current study, we investigated clonal dissemination and mechanisms of resistance to carbapenem on tumor patients in China.
Materials and Methods
Bacterial Strains
We carried out retrospective surveillance for bla NDM‐1 among carbapenem‐resistant Enterobacter strains which were isolated from patients at the Affiliated Tumor Hospital of Zhengzhou University in China. There are 2,600 beds in this tumor hospital and a total of 14,825 tumor patients were detected for bla NDM‐1 gene by Kirby–Bauer diffusion test and PCR amplification from January 2010 to June 2014. The research protocol was approved by the institutional ethics committee of the bioscience of Zhengzhou University. All the participants signed an individual informed consent after receiving a detailed explanation of the research. All the data were processed respecting the confidentiality of the participants.
Antimicrobial Susceptibility Testing
MICs of 17 antibiotics, together with the MBL phenotype test, were determined using the E‐test. The susceptibility results were interpreted according to the CLSI guidelines 5. The MIC breakpoint of cefoperazone for Enterobacteriaceae was used for cefoperazone/sulbactam 6. Due to the absence of CLSI criteria, the BSAC breakpoint and the FDA breakpoint for Enterobacteriaceae were applied for aztreonam and tigecycline, respectively 5. Quality controls were included using standard strains (E. coli ATCC 25922).
Species Identification
The bla NDM‐1‐positive isolates were distinguished by the automated bacterial identification susceptibility analyzers. The metalloenzyme phenotype of isolates was determined using the E‐test. Further differentiations within the strains were performed using the 16S rRNA gene intergenic spacer (ITS) sequence to differentiate the strains. The isolates were screened for the species identification by PCR with primers 16s‐F‐(5′‐AGTTTGATCTGGCTCAG‐3′) and 16s‐R‐(5′‐GGACTACAGGGTATCTAAT‐3′).
In order to distinguish the source of the strains, the isolates were detected by PCR with primer ERIC2‐5′‐AAGTAAGTGACTGGGGTGAGCG‐3′. PCR experiments were performed according to standard conditions with an annealing temperature of 45°C.
Test strains were adjusted to the McFarland 0.5 standard and used to inoculate Mueller‐Hinton agar plates. Depending on the test, a 10‐μg imipenem (IPM) disk was placed on the plate, and a blank filter paper disk was placed at a distance of 10 mm (edge to edge). To the blank disk, 10 μl of a 0.5 M EDTA solution (ca. 1,900 μg of disodium salt, dihydrate) was added. After overnight incubation, the presence of even a small synergistic inhibition zone (4 mm) was interpreted as positive.
Molecular Detection of Resistance Genes
The isolates were detected for the presence of bla NDM‐1 by PCR with primers NDM‐1‐U‐(5′TCGCATAAAACGCCTCTG‐3′) and NDM‐1‐L‐(5′‐GAAACTGTCG CACCTCAT‐3′). PCR experiments were performed according to standard conditions with an annealing temperature of 55°C.
Results
Clinical Features of the bla ndm‐1‐Positive Strains
From a total of 14,825 clinical tumor patients, there were 42 patients who were infected with Enterobacter aerogenes. We then found three bla NDM‐1‐positive Enterobacter aerogenes isolates. According to the epidemiological data for the three corresponding patients, there were two isolates, which were found from venous indwelling tube and another was from abdominal drainage. The three patients who only underwent surgical treatment without radiotherapy and chemotherapy drugs were diagnosed with lymphoma, colorectal cancer, and gastric cancer, respectively. The histories of the patients were confirmed as lacking any foreign travel. However, they had lived in the same ward at the same time (Table 1). The patients received empirical antibiotic treatment, including ceftriaxone, meropenem, imipenem, piperacillin/tazobactam, ampicillin/sulbactam, amikacin, levofloxacin, or trimethoprim/sulfamethoxazole, during their hospitalization.
Table 1.
Clinical Features of the bla NDM‐1‐Positive Strains
| Isolate | Sex | Age | Source | Diagnosis | Ward | Admission time | Foreign travel | Antibiotic treatment | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| 0711 | Male | 55 | Venous indwelling catheter | Lymphoma | General Surgery | Feb.21 2011 | No | Yes | Cureda |
| 0807 | Female | 63 | Abdominal drainage | Gastric cancer | General Surgery | Feb.18 2011 | No | Yes | Cured |
| 1311 | Female | 48 | Venous indwelling catheter | Colorectal cancer | General Surgery | Feb.21 2011 | No | Yes | Cured |
Cured: The bla NDM‐1‐positive strains were not detected for 2 weeks.
Antimicrobial Susceptibility Testing
The results of antimicrobial susceptibility testing had been showed that three isolates were sensitive only to LVX (levofloxacin), CIP (ciprofloxacin), and PB (polymyxin B) 5. However, two isolates (0711 and 0807) were intermediary to IPM (imipenem) and MEC (meropenem). The three isolates were resistant to other antibiotics (Table 2).
Table 2.
Antibiotic susceptibilities and β‐lactamase detection of bla NDM‐1‐positive Enterobacter aerogenes (mg/l)
| Isolate | CZa | GEN | AM | CTX | CIP | FEP | FOX | CRO | TZP | PIP | IPM | MEC | AMK | LVX | CAZ | ATM | PB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0711 | >8b | >16 | >32 | >16 | ≤1 | >16 | >16 | >32 | >64/4 | >64 | 2 | 4 | >32 | ≤2 | >16 | >8 | <2 |
| 0807 | >8 | >8 | >16 | >16 | ≤1 | >16 | >16 | >32 | >64/4 | >64 | >8 | 2 | >32 | ≤2 | >16 | >8 | <2 |
| 1311 | >8 | >16 | >32 | >32 | ≤1 | >32 | >16 | >32 | >64/4 | >64 | 4 | 8 | >32 | ≤2 | >16 | >8 | <2 |
CZ, cefazolin; GEN, gentamicin; AM, Ampicillin; CTX, cefotaxime; CIP, ciprofloxacin; FEP, cefepime; FOX, cefoxitin; CRO, ceftriaxone; TZP, piperacillin/tazobactam(E‐test); PIP, piperacillin; IPM, imipenem(E‐test); MEC, meropenem(Etest); AMK, amikacin; LVX, levofloxacin; CAZ, ceftazidime; ATM, aztreonam; PB, polymyxin B(E‐test).
MICs were determined by the BD PhoenixTM‐100 automated bacterial identification susceptibility analyzers system or the E‐test. Where a “>“ sign is present in an entry with two values, the sign applies to both values in the entry. The second MIC refers to the second drug in the compound.
Species Identification
The three bla NDM‐1‐positive isolates were diagnosed as Enterobacter aerogenes using BD PhoenixTM‐100 automated bacterial identification susceptibility analyzers system. The three isolates are consistent with Enterobacter aerogenes by using 16s rRNA sequence alignment. So we believed that three bacteria were Enterobacter aerogenes.
All the metalloenzyme phenotypes of three isolates were positive. The 0711, 0807, and 1311 isolates measured 20 mm, 7 mm, and 10 mm in the IPM E‐test, respectively. After the antimicrobial papers that add EDTA, the inhibition zone diameters were 28 mm, 12 mm, and 16 mm, respectively. Because of the difference of diameter above 4 mm, this indicated that three Enterobacter aerogenes metalloenzyme phenotype experiments were positive.
Every isolate amplified four belts in the ERIC2 typing. The three isolates were of the same molecular weight belts in Marker 2000 belts, Marker 1000 belts, Marker 750 belts, and Marker 150 belts, respectively. So it was indicated that the three Enterobacter aerogenes were of the same clone (Fig. 1).
Figure 1.
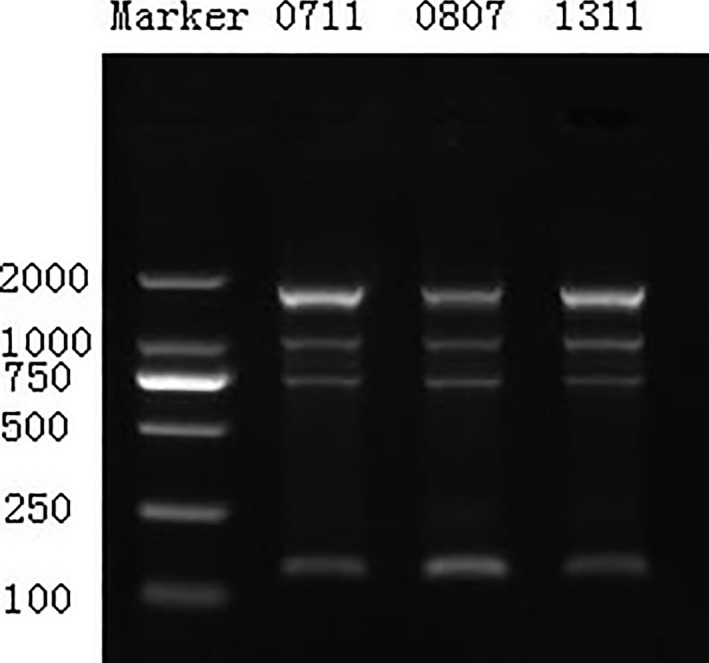
ERIC2‐PCR amplification products of the three Enterobacter aerogenes.
Molecular Detection of Resistance Genes
The isolates were detected for the presence of bla NDM‐1 by PCR. Approximately a 1,000‐bp fragment from DNA of the three Enterobacter aerogenes was obtained (Fig. 2), and the three fragments were of the same DNA sequence by gene sequencing. The fragment was entirely sequenced by primer walking which revealed 99% identity to the sequence reported for A Klebsiella pneumoniae RJF866. The fragment entirely sequenced in our study has already been accepted by GenBank. The accession number is KJ577744.
Figure 2.
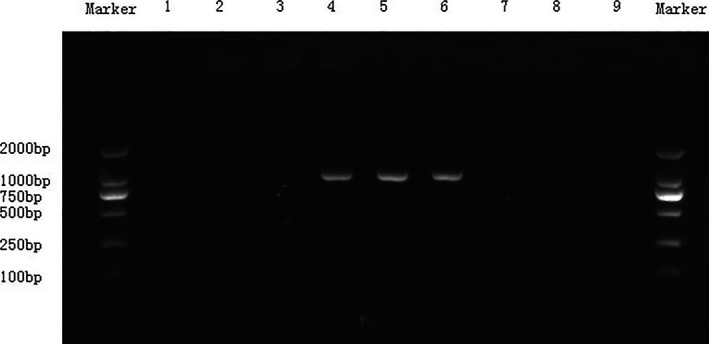
bla NDM ‐1‐PCR products of the three Enterobacter aerogenes; 1–9 isolates are all Enterobacter aerogenes: 1, 0125; 2, 0478; 3, 0684; 4, 0711; 5, 0807; 6, 1311; 7, 1987; 8, 2063; 9, 2794. Approximately a 1000 bp fragment from DNA of 4(0711), 5(0807), and 6(1311) was obtained, respectively. Others were negative.
Discussion
The bla NDM‐1 gene was first reported in 2009 in India. Since then, the global distribution of the bla NDM‐1 gene has been extensively described 1, 7, 8, 9. However, the presence of this gene in Enterobacter aerogenes has not been reported. Here, in our study, we find three bla NDM‐1‐positive Enterobacter aerogenes in clinical cancer patients in China. Based on the detailed clinical information, the surgery of patients may be more susceptible, and contact transmission was the main route of infection.
In addition, we found that the bla NDM‐1‐positive Enterobacter aerogenes were highly resistant to all antibiotics except levofloxacin, ciprofloxacin, imipenem, and meropenem (Table 2). It was different from previous reports in antimicrobial susceptibility results. The antimicrobial susceptibility testing to bla NDM‐1‐positive Acinetobacter and Klebsiella pneumoniae indicated that the bla NDM‐1‐positive strains has been reported in China, which were resistant to most antibiotics, only sensitive to polymyxin and tigecycline 10, 11. In fact, the therapeutic agents prescribed to the patients were followed by empirical antibiotic treatment paradigms rather than the results of antimicrobial susceptibility. Thus, the antibiotics could have been no effective for the control of infections produced by bla NDM‐1‐positive Enterobacter aerogenes.
As a gene resistant to the metallo‐beta‐lactamase, the bla NDM‐1 gene can decompose most antibiotics, including cephalosporins, penicillins, carbapenems, and other beta‐lactam antibiotics. It was reported that the bla NMD‐1 gene was in the coding sequence of plasmid. There were often some other resistance genes inserted around the coding sequence 12. Thus, it was a complex resistant phenotype to bla NDM‐1‐positive isolates.
We detected three bla NDM‐1‐positive Enterobacter aerogenes which had the same molecular weight belt by ERIC2‐PCR and bla NDM‐1‐PCR. So we can realize that the bla NDM‐1 gene was broken out in small scope. The three patients had not gone abroad, and they were the first to be hospitalized. All of them were infected after surgery in hospital. Coincidentally, they almost lived in the same ward of a department in the meantime. Thus, there was the existence of cross time and space to infect.
The isolates, bla NDM‐1‐producing metalloenzymes, had strong ability to spread by previous studies which had been reported the bla NDM‐1‐positive isolates could cause widespread popularity in the world. There were some small outbreaks reported 8, 13. It was found that four of the same ST‐typing Klebsiella pneumoniae isolates producing bla NDM‐1 prevailed in South Korea 14. Same type of ST‐type Acinetobacter isolates producing bla NDM‐1 were reported in French 15. Further studies had shown that bla NDM‐1 gene is typically encoded on the plasmid. There were many movable original copies around the gene (e.g., ISAba125), which could make the bla NDM‐1 gene had strong spread ability. If the patients had underlying disease, it was easy to cause small range of popularity through crossing infection, even large‐scale outbreak of the epidemic. Thus, it is important to mandate prudent usage of antibiotics and implement infection control measures to control the spread of these resistant bla NDM‐1‐positive strains.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by Science and Technology Agency Foundation of Henan (No. 112102310256). The authors thank the skillful technical assistance of Yan‐Min Chang, Zhe Zou, and Ling Zhou.
References
- 1. Gefen‐Halevi S, Hindiyeh MY, Ben‐David D, et al. Isolation of genetically unrelated bla(NDM‐1)‐positive Providencia rettgeri strains in Israel. J Clin Microbiol 2013;51:1642–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010;10:597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rolain JM, Parola P, Cornaglia G. New Delhi metallo‐beta‐lactamase (NDM‐1): Towards a new pandemia? Clin Microbiol Infect 2010;16:1699–1701. [DOI] [PubMed] [Google Scholar]
- 4. Sartor AL, Raza MW, Abbasi SA, et al. Molecular epidemiology of NDM‐1 producing Enterobacteriaceae and Acinetobacter baumannii isolates from Pakistan. Antimicrob Agents Chemother 2014;58:5589–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poulou A, Grivakou E, Vrioni G, et al. Modified CLSI extended‐spectrum beta‐lactamase (ESBL) confirmatory test for phenotypic detection of ESBLs among Enterobacteriaceae producing various beta‐lactamases. J Clin Microbiol 2014;52:1483–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bobenchik AM, Deak E, Hindler JA, Charlton CL, Humphries RM. Performance of Vitek 2 for antimicrobial susceptibility testing of Enterobacteriaceae with Vitek 2 (2009 FDA) and 2014 CLSI breakpoints. J Clin Microbiol 2015;53:816–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shenoy KA, Jyothi EK, Ravikumar R. Phenotypic identification & molecular detection of bla ndm‐1 gene in multidrug resistant Gram‐negative bacilli in a tertiary care centre. Indian J Med Res 2014;139:625–631. [PMC free article] [PubMed] [Google Scholar]
- 8. Rozales FP, Ribeiro VB, Magagnin CM, et al. Emergence of NDM‐1‐producing Enterobacteriaceae in Porto Alegre, Brazil. Int J Infect Dis 2014;25:79–81. [DOI] [PubMed] [Google Scholar]
- 9. Berrazeg M, Diene S, Medjahed L, et al. New Delhi Metallo‐beta‐lactamase around the world: An eReview using Google Maps. Euro Surveill 2014;19:pii: 20809. [DOI] [PubMed] [Google Scholar]
- 10. Fu Y, Du X, Ji J, Chen Y, Jiang Y, Yu Y. Epidemiological characteristics and genetic structure of blaNDM‐1 in non‐baumannii Acinetobacter spp. in China. J Antimicrob Chemother 2012;67:2114–2122. [DOI] [PubMed] [Google Scholar]
- 11. Chen Y, Zhou Z, Jiang Y, Yu Y. Emergence of NDM‐1‐producing Acinetobacter baumannii in China. J Antimicrob Chemother 2011;66:1255–1259. [DOI] [PubMed] [Google Scholar]
- 12. Khajuria A, Praharaj AK, Kumar M, Grover N, Aggarwal A. Multidrug resistant NDM‐1 metallo‐beta‐lactamase producing Klebsiella pneumoniae sepsis outbreak in a neonatal intensive care unit in a tertiary care center at central India. Indian J Pathol Microbiol 2014;57:65–68. [DOI] [PubMed] [Google Scholar]
- 13. Lauderdale TL, Hsu MC, Mu JJ, et al. NDM‐1‐producing Acinetobacter soli from Taiwan. Diagn Microbiol Infect Dis 2014;80:168–169. [DOI] [PubMed] [Google Scholar]
- 14. Kim SY, Shin J, Shin SY, Ko KS. Characteristics of carbapenem‐resistant Enterobacteriaceae isolates from Korea. Diagn Microbiol Infect Dis 2013;76:486–490. [DOI] [PubMed] [Google Scholar]
- 15. Diene SM, Bruder N, Raoult D, Rolain JM. Real‐time PCR assay allows detection of the New Delhi metallo‐beta‐lactamase (NDM‐1)‐encoding gene in France. Int J Antimicrob Agents 2011;37:544–546. [DOI] [PubMed] [Google Scholar]


