Abstract
Background
Based on genetic heterogeneity, hepatitis C virus (HCV) is classified into seven major genotypes and 64 subtypes. In spite of the sequence heterogeneity, all genotypes share an identical complement of colinear genes within the large open reading frame. The genetic interrelationships between these genes are consistent among genotypes. Due to this property, complete sequencing of the HCV genome is not required. HCV genotypes along with subtypes are critical for planning antiviral therapy. Certain genotypes are also associated with higher progression to liver cirrhosis.
Methods
In this study, 100 blood samples were collected from individuals who came for routine HCV genotype identification. These samples were used for the comparison of two different genotyping methods (5′NCR PCR‐RFLP and HCV core type‐specific PCR) with NS5b sequencing.
Results
Of the 100 samples genotyped using 5′NCR PCR‐RFLP and HCV core type‐specific PCR, 90% (κ = 0.913, P < 0.00) and 96% (κ = 0.794, P < 0.00) correlated with NS5b sequencing, respectively. Sixty percent and 75% of discordant samples by 5′NCR PCR‐RFLP and HCV core type‐specific PCR, respectively, belonged to genotype 6. All the HCV genotype 1 subtypes were classified accurately by both the methods.
Conclusion
This study shows that the 5′NCR‐based PCR‐RFLP and the HCV core type‐specific PCR‐based assays correctly identified HCV genotypes except genotype 6 from this region. Direct sequencing of the HCV core region was able to identify all the genotype 6 from this region and serves as an alternative to NS5b sequencing.
Keywords: 5′NCR PCR‐RFLP, HCV core type‐specific PCR, HCV genotype, HCV subtype, NS5b sequencing
Introduction
Hepatitis C virus (HCV) exhibits a great degree of genetic heterogeneity 1. Based on this genetic variability, HCV is classified into genotypes, subtypes, isolates, and quasi‐species. Phylogenetic analysis of HCV genome sequence consisting of > 95% of coding region has led to the classification of HCV into seven major genotypes and 67 confirmed subtypes 2. HCV genotypes differ from each other by 31–33% and subtypes by 20–25% at the nucleotide level 3. Despite the sequence variability of HCV, several subgenomic regions of HCV can be used for genotyping. Partial sequencing of HCV 5′NCR, core, E1, and NS5b are used by researchers to classify HCV genotypes because the sequences from these genomic regions of HCV isolates grouped under the same phylogenetic branch.
Hepatitis C virus genetic diversity occurs due to poor proof‐reading ability of the RNA‐dependant RNA polymerase and the absence of exonuclease activity. The 5′ and 3′ noncoding regions of the HCV genome are the most conserved, while E1 and E2 are the most variable regions. The core and NS5b regions of HCV are also relatively well conserved. HCV genotypes are somewhat geographically restricted with HCV genotypes 1a and 1b most prevalent in northern Europe and United States 4, 5, HCV genotype 2 in Italy 6, HCV genotype 3 in Southern Asia, HCV genotype 4 in North Africa and Middle East 7, 8, 9, HCV genotype 5a in South Africa 10, 11, HCV genotype 6 in South East Asia 12, 13, 14, and HCV genotype 7 from Central Africa 15. The most prevalent genotype in the Indian population is HCV genotype 3 followed by 1 and 4 16.
Certain genotypes of HCV are more refractory to antiviral therapy and can lead to higher level of chronicity after acute infection 17. Accurate genotyping and subtyping of HCV is very important especially for deciding appropriate antiviral therapy 17 as HCV viral load and genotype are the strong prognostic factors for sustained viral response. Earlier, treatment for HCV was done using pegylated‐interferon as monotherapy or in combination with ribavirin. Newer direct‐acting antivirals (DAAs) approved by FDA are recommended in combination with or without ribavirin 18, 19. DAAs are classified into NS3/4a serine protease inhibitors, NS5a inhibitor, and NS5b inhibitor. In phase III clinical trials, a combination of NS3/4a inhibitor and NS5a inhibitor resulted in SVR in 95% of genotype 1a and 99% of genotype 1b patients 20. Various studies have compared NS5b sequencing to commercially available HCV genotyping platforms like Trugene HCV genotyping kit, Versant HCV genotype 2.0 assay, and Real‐time HCV genotype II and reported discrepancies in subtyping 1a and 1b 21, 22.
Hepatitis C virus genotyping is done mainly by the following methods: (a) PCR followed by restriction fragment length polymorphism (RFLP) 23, (b) PCR using genotype‐specific primers 24, (c) PCR followed by molecular hybridization of type‐specific probes 25, (d) NS4 antibody‐based typing 26, (e) nucleotide sequencing of partial regions of HCV genome 27, (f) matrix‐assisted laser desorption ionization time of flight (MALDI‐TOF) 28, (g) melt curve analysis using LightCycler 29, (h) deep sequencing 22, and (i) pyrosequencing 30.
Even though whole‐genome sequencing is the gold standard for identifying HCV genotypes, the similarities within different genomic regions of HCV are conserved within genotypes obliterating the need to do whole genome sequencing which is tedious and expensive. Since the methods used for identifying HCV vary and HCV genotypes are geographically restricted, a need to identify the best method to detect all HCV genotypes prevalent in the different regions is a paramount need. Among the genome regions used for genotyping, HCV NS5b region is considered the gold standard as the region is variable but is still conserved within genotypes. Various studies have compared NS5B sequencing to commercially available HCV genotyping platforms. In this study, we compared two other methods of HCV genotyping from two different regions of the HCV genome, i.e., 5′NCR PCR‐RFLP and core region‐based type‐specific PCR with NS5b sequencing. Also, we compared short sequences of the HCV 5′NCR and core regions with the sequences of NS5b region of the HCV genome to see which sequenced region of the HCV genome is best suited to identify the genotypes prevalent in this region.
Materials and Methods
Samples
Blood samples were collected from 100 HCV RNA‐positive individuals who came to the department of Clinical Virology as referrals for routine HCV viral genotype identification between the years 2004 and 2007. Samples were representative of the most prevalent HCV genotypes in this region 16, 31. All patients were recruited after getting a verbal consent in addition to a general consent received by the institution for all blood collections as part of patient management. Ten milliliters of blood was collected in EDTA tubes from each individual who gave consent. Separated plasma was stored at −70°C until testing. In this study, plasma samples were genotyped using three different methods: 5′NCR region‐based PCR‐RFLP, core region‐based type‐specific PCR, and sequencing of the NS5b region. The HCV genomic regions used for evaluation are depicted in Figure 1. The results were compared to look for the efficacy of these HCV genotyping methods in this population. Discordant results compared with the NS5b sequencing method were resolved by sequencing the specific region (core/5′NCR).
Figure 1.
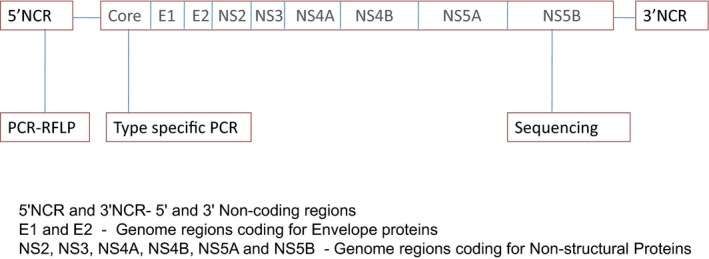
HCV genome region used for genotyping.
Extraction and reverse transcription
For all the three genotyping methods, HCV RNA was extracted using 140 μl of plasma sample using QIAamp Viral RNA mini kit (Qiagen GmbH, Hilden, Germany) as per manufacturer's protocol. Elution was done using 60 μl of AVE buffer. Reverse transcription was done using Moloney‐Murine Leukemia virus reverse transcriptase (Invitrogen, Corp., Carlsbad, CA) and random primers (Roche Diagnostics GmbH, Mannheim, Germany) in a 20‐μl reaction volume using 9 μl of RNA extract. The mix was incubated at 37°C for 1 hr followed by 95°C for 15 min. The cDNA obtained was used for the different genotyping methods.
HCV genotyping methods
HCV core type‐specific PCR
Genotyping of HCV was done by PCR using genotype‐specific primers to the HCV core gene which was earlier standardized in this laboratory 32. This is a nested PCR with the second round of amplification made in two reaction mixes containing primers specific for different HCV genotypes. The first master mix designated “X” had primers (S7, S2a, G1b, S2a, G2a, G2b, G3b) specific for HCV genotypes 1b, 2a, 2b, and 3b. The second master mix designated “Y” had primers (S7, G1a, G3, G4, G5a, G6a) specific for HCV genotypes 1a, 3a, 4, 5a, and 6a. The primer sequences are shown in Table 1. The base pair size of the amplified product was resolved using the gel documentation system (Gel Doc. Bio‐Rad, Hercules, CA). To resolve discrepancies between genotypes identified by the genotype‐specific primer assay in comparison with the NS5b‐based sequencing, the core region was sequenced using sense (S7) and antisense (A5) primers.
Table 1.
Primer Sequence
| Primer Name | Primer Sequence | Reference | Region of HCV | |
|---|---|---|---|---|
| First round | Sc2 | GGGAGGTCTCGTAGACCGTGCACCATG | Ohno et al. 23 | Core |
| First round | Ac2 | GAG(AC)GG(GT)AT(AG)TACCCCATGAG(AG)TCGGC | ||
| Sequencing | S7 | AGACCGTGCACCATGAGCAC | ||
| Sequencing | A5 | TACGCCGGGGGTCA(TG)T(GA)GGGCCCCA | ||
| Mix X | ||||
| Second round | S7 | AGACCGTGCACCATGAGCAC | ||
| Second round | S2a | AACACTAACCGTCGCCCACAA | ||
| Second round | G1b | CCTGCCCTCGGGTTGGCTA(AG) | ||
| Second round | G2a | CACGTGGCTGGGATCGCTCC | ||
| Second round | G2b | GGCCCCAATTAGGACGAGAC | ||
| Second round | G3b | CGCTCGGAAGTCTTACGTAC | ||
| Mix Y | ||||
| Second round | S7 | AGACCGTGCACCATGAGCAC | ||
| Second round | G1a | GGATAGGCTGACGTCTACCT | ||
| Second round | G3a | GCCCAGGACCGGCCTTCGCT | ||
| Second round | G4 | CCCGGGAACTTAACGTCCAT | ||
| Second round | G5a | GAACCTCGGGGGGAGAGCAA | ||
| Second round | G6a | GGTCATTGGGGCCCCAATGT | ||
| First round | P1203 | GGGTTCTCGTATGATACCCGCTGCTTTGACTC | Harris et al. 40 | NS5b |
| First and Second round | P1204 | GGAGGGGCGGAATACCTGGTCATAGCCTCCGTGAA | ||
| Second round | NS5b IP | TGATACCCGCTGCTTTGACTCNACNGTCAC | ||
| First round | GOP1 | AGCGTCTAGCCATGGCGT | Harris et al. 40 | 5′NCR |
| First round | GOP2 | GCACGGTCTACGAGACCT | ||
| Second round | GIP1 | GTGGTCTGCGGAACCGG | ||
| Second round | GIP2 | GGGCACTCGCAAGCACCC |
HCV PCR‐RFLP
Amplification as per the HCV PCR‐RFLP protocol was done utilizing primers specific for the 5′NCR region using 10 μl of cDNA and 5 pmol of each first‐round (outer) primers (GOP 1, GOP 2) (Invitrogen, Carlsbad, CA). First‐round PCR was performed using the following cycling protocol: 94°C for 4 min followed by 94°C for 20 sec, 62°C for 40 sec, 72°C for 35 sec for 35 cycles, and 72°C for 3 min. The second‐round amplification was done using 2 μl of the first‐round product at 94°C for 20 sec, 68°C for 40 sec, and 72°C for 30 sec for 25 cycles followed by 72°C for 5 min using 20 pmol of each second‐round (inner) primers (GIP 1, GIP 2). Primer sequences are shown in Table 1. Amplification of this region produced a 174 base pair product which was visualized by gel electrophoresis. RFLP was performed using four restriction enzymes Mva I (Roche Diagnostics GmbH), ScrF I, Hinf I, and BstU I (New England Biolabs, Beverly, MA) after obtaining the amplified product 23. Restriction endonuclease digestion was done as per manufacturer's protocol. The digested products were analyzed by electrophoresis in a 3% agarose gel by loading equal volume of ScrF I and Mva I digested PCR products together in a single well and Hinf I and BstU I digest together in the adjacent well. The size of the digested product was determined using the gel documentation system (Gel Doc. Bio‐Rad) using the Quantity One software version 4.1.1(Bio‐Rad). The digestion pattern is shown in Figure 2.
Figure 2.
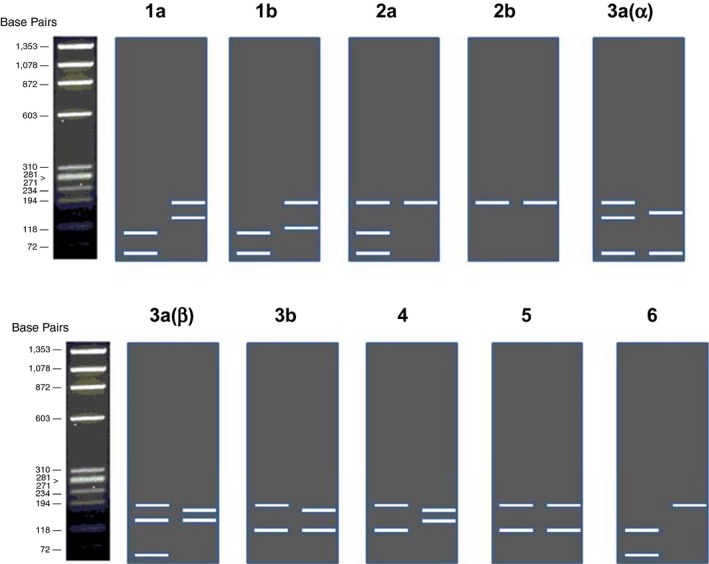
Diagrammatic representation of the digestion pattern of 5′NCR PCR‐RFLP.
Sequencing of the HCV NS5b region
Sequencing of the NS5b region of HCV was a done using a hemi‐nested PCR using 10 μl of cDNA. Amplification was done using the following protocol, 94°C for 4 min followed by 35 cycles at 94°C for 30 sec, 55°C for 40 sec, 72°C for 50 sec followed by 72°C for 180 sec. Second‐round amplification was done using 5 μl of the first‐round product. Second‐round amplification was performed using the following protocol: 94°C for 4 min followed by 94°C for 30 sec, 54°C for 40 sec, 72°C for 30 sec with a final extension of 72°C for 180 sec. The primer sequences are shown in Table 1. The amplified 392 bp product was resolved using gel electrophoresis before sequencing.
All the samples amplified using NS5b region were sequenced after a clean‐up step to remove primer dimers and excess dNTPs. The nucleotide sequences obtained were subjected to BLAST using the HCV BLAST database (http://hcv.lanl.gov).
Results
Evaluation of genotyping methods
Of the 100 samples genotyped using core type‐specific PCR, 96 (96%) samples correlated with the genotype results of NS5b sequencing (Table 2). The correlation between both the genotyping methods was excellent (κ = 0.913, P < 0.000). Of the four discordant samples, three were genotype 6 and the remaining was genotype 1a as per NS5b sequencing. As per the core type‐specific PCR assay, the three of the seven genotype 6 samples were identified as genotype 4 and one of the 15 genotype 1 was identified as genotype 3 (Table 2). Two of these three discordant genotype 6 and four concordant genotype 6 were further identified correctly by sequencing of this region. The remaining discordant genotype 6 could not be sequenced due to paucity of sample. The single discordant genotype 1 sample was also correctly identified by sequencing of this HCV core region.
Table 2.
Correlation of HCV Genotypes Between HCV Core Type‐Specific PCR and NS5b Sequencing
| HCV genotypes determined by core type‐specific PCR | HCV genotypes determined by NS5b sequencing | Total | |||
|---|---|---|---|---|---|
| 1 | 3 | 4 | 6 | ||
| 1 | 14 | 14 | |||
| 3 | 1a | 71 | 72 | ||
| 4 | 7 | 3b | 10 | ||
| 6 | 4 | 4 | |||
| Total | 15 | 71 | 7 | 7 | 100 |
Kappa coefficient = 0.913 (P < 0.0001).
Sequencing of the HCV core region classified this strain as HCV genotype 1.
Sequencing of the HCV core region classified these three strains as HCV genotype 6.
Ninety (90%) of the 100 samples genotyped using 5′NCR PCR‐RFLP correlated with NS5b sequencing (κ = 0.794, P < 0.000) (Table 3). Of the ten discordant samples, six were genotype 6 and the remaining four were HCV genotype 3 by NS5b sequencing. All the discordant genotype 6 samples (n = 6) were identified as genotype 1 and the discordant genotype 3 samples (n = 4) were identified as genotype 4 by the 5′NCR assay. In all, six (85.7%) of the seven genotype 6 samples as identified by the gold standard were classified as genotype 1 by 5′NCR PCR‐RFLP method. By sequencing the 5′NCR region, two of the four discordant genotype 3 samples were correctly identified as genotype 3. There was insufficient plasma to sequence the remaining two samples. Five of six discordant and one concordant genotype 6 samples were sequenced. The 5′NCR region sequencing of five of the six discordant genotype 6 samples also identified them as genotype 1. One of the six discordant genotype 6 samples could not be sequenced due to paucity of sample. Further analysis of the genotype 6 sequences revealed that all the discordant genotype 6 samples had a restriction site for the restriction enzyme BstU 1, which is used to differentiate genotype 1a from 1b. This was also observed in some GenBank HCV genotype 6 samples (Fig. 3). This was observed only with nongenotype 6a samples.
Table 3.
Correlation of HCV genotypes between HCV 5′NCR PCR‐RFLP and NS5b sequencing
| HCV genotypes determined by 5′NCR PCR‐RFLP | HCV genotypes determined by NS5b sequencing | Total | |||
|---|---|---|---|---|---|
| 1 | 3 | 4 | 6 | ||
| 1 | 15 | 6a | 21 | ||
| 3 | 67 | 67 | |||
| 4 | 4b | 7 | 11 | ||
| 6 | 1 | 1 | |||
| Total | 15 | 71 | 7 | 7 | 100 |
Kappa coefficient = 0.794 (P < 0.0001).
Sequencing of the HCV 5′NCR region in 5 of 6 of these samples classified these strains as HCV genotype 1. One sample could not be sequenced due to insufficient sample.
Sequencing 2 of the four samples by HCV 5′NCR classified these strains as HCV genotype 3 (two samples could not be sequenced because the sample was insufficient).
Figure 3.

Nucleic acid sequence alignment of the HCV genotype 6 from position 143 to 316 of the consensus HCV genotype 6 genome compared with the strains identified from this study.
Overall, 89% (n = 100) of the samples concurred across all the three genotyping protocols (Table 4).
Table 4.
Correlation of HCV genotyping methods: Core type‐specific PCR, 5′NCR PCR‐RFLP, and NS5b sequencing
| HCV genotypes | NS5b sequencing | 5′NCR PCR‐RFLP (%) | Core type‐specific PCR (%) |
|---|---|---|---|
| 1 | 15 | 15 (100) | 14 (93.3) |
| 3 | 71 | 67 (94.4) | 71 (100) |
| 4 | 7 | 7 (100) | 7 (100) |
| 6 | 7 | 1 (14.3) | 4 (57.1) |
89% correlation between all genotyping methods.
90% correlation between 5′NCR PCR‐RFLP and NS5b sequencing.
96% correlation between core type‐specific PCR and NS5b sequencing.
HCV subtype
Of the 71 genotype 3 samples, 22 (31%) samples belonged to subtype 3a, 37 (52.1%) to 3b, 4 (5.6%) classified as 3f, 5 (7.0%) as 3i, and 3 (4.2%) as 3k as classified by the NS5b sequencing PCR (Fig. 4). Of the 15 genotype 1 samples, 5 belonged to 1a and 10 were subtype 1b as classified by NS5b sequencing. All the subtypes of genotype 1 were correctly identified by both the 5′NCR PCR‐RFLP and the core type‐specific PCR when compared with NS5b sequencing.
Figure 4.
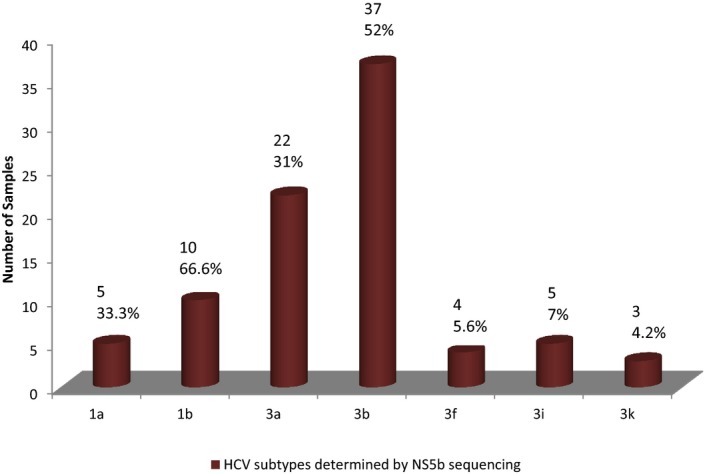
Bar diagram depicting the subtype distribution of HCV determined by NS5b sequencing.
Of the 22 subtype 3a samples, two samples were identified by core type‐specific PCR to have mixed infection (3a & 3b), all the other 20 samples correlated with the NS5b sequencing method. Of the 37 genotype 3b samples, 24 (64.9%) samples correlated in both methods. In the remaining 13 discordant samples, 11 (29.7%) showed mixed infection with 3a & 3b (9 samples) and 3b & 3g (2 samples). Two (5.4%) of the 13 discordant samples were subtyped as 3a by the core type‐specific PCR.
Hepatitis C virus 5′NCR PCR‐RFLP showed a 100% correlation with NS5b sequencing for HCV subtype 3a. Of the 37 subtype 3b samples, 27 (73.0%) were correctly subtyped as 3b, 7 (18.9%) samples were subtyped as 3a, and remaining 3 (8.1%) samples were classified as genotype 4 by the PCR‐RFLP method.
Discussion
The gold standard for genotyping of HCV is nucleotide sequencing. But in a diagnostic laboratory, a simple, faster, and an inexpensive method of HCV genotyping is required. Several methods of genotyping have been developed for HCV of which, hybridization of amplified products with specific probes and real‐time HCV genotyping assay are commercially available 33, 34, 35. Accurate genotyping of HCV RNA‐positive samples is a must as it has important clinical and therapeutic implications 36. HCV genotyping is also essential for epidemiological studies. This study was done using three “in‐house” genotyping assays on a panel of 100 HCV RNA‐positive plasma samples. The three “in‐house” assays amplified three different genomic regions of HCV: (a) Ohno's genotype‐specific reverse transcription‐nested polymerase chain reaction (RT‐nPCR) which amplifies the core region of the HCV genome, (b) a restriction fragment length polymorphism (RFLP) analysis of PCR products amplified from the 5′ noncoding region (5′NCR) of the viral genome, and (c) sequencing of the NS5b region of the HCV genome. The 5′NCR PCR‐RFLP and the core type‐specific PCR technique were compared with NS5b sequencing technique which is currently considered the gold standard for HCV genotyping 37.
The core type‐specific PCR utilizes primers specific for each HCV genotype and can differentiate subtypes 1a, 1b, 2a, 2b, 3a, and 3b but cannot differentiate subtypes for genotypes 4, 5, and 6 32. The core region of the HCV genome is well conserved with sequence similarity between genotypes ranging from 81% to 88% and between subtypes from 88% to 93% 27. In our study, core type‐specific PCR showed excellent correlation (96%) (κ = 0.913, P < 0.001) with the genotype results of NS5b sequencing. Seventy‐five percent (n = 3) of the discordant samples were genotype 6 and the remaining was genotype 1 (n = 1) as classified by NS5b sequencing method. However, sequencing results of the core region of these discordant samples correlated 100% with NS5b sequencing results. This suggests that the core region used in this study is variable enough to classify all six genotypes prevalent in this region.
The 5′NCR is one of the most conserved regions in the HCV genome. But a set of well‐characterized polymorphisms in this genomic region is used to classify HCV genotype by PCR‐RFLP. The PCR‐RFLP method can differentiate subtypes 1a, 1b, 2a, 2b, 3a(α), 3a(β), 3b but cannot differentiate subtypes of 4, 5, and 6 23. This method of genotyping is also widely used as 5′NCR is the region of choice for diagnosis of HCV. In our study, 90% of HCV genotypes, as classified by 5′NCR PCR‐RFLP, correlated with NS5b sequencing (κ = 0.794, P < 0.001). In another study, which compared two methods of core type‐specific PCR and 5′NCR PCR‐RFLP with sequencing of the core region, an overall genotyping sensitivity of 96.2% for 5′NCR PCR‐RFLP38 was observed, which is similar to our study findings where we used NS5b sequencing to compare 5′NCR PCR‐RFLP results. In our study, core sequencing and NS5b sequencing yielded 100% correlation.
In addition to the six major genotypes of HCV, an additional genotype (genotype 7) has been identified which is restricted to Central Africa. In this study, approximately 86% of HCV genotype 6 as classified using NS5b sequencing was genotyped as genotype 1 by the 5′NCR PCR‐RFLP method. Also, sequencing of the same 5′NCR region used for PCR‐RFLP could not identify the genotype 6 (except 6a) samples from this region. Hence, the region of 5′NCR of the HCV genome used in this study is not variable enough to differentiate most of the genotype 6 prevalent in this region. This is in concordance with another study done in Thailand and Vietnam, where the HCV isolates, which were identified as genotype 6 by phylogenetic analysis of core region of the genome, were identified as genotype 1 by 5′NCR PCR‐RFLP. Sequencing of the 5′NCR region in that study also was not able to classify the isolates as genotype 6 39.
The 5′NCR PCR‐RFLP method differentiates genotype 6 from genotype 1 by the absence of BstU1 restriction site. We compared the sequences from our isolates with the database and found that except for one isolate (genotype 6a) which was genotyped correctly by the PCR‐RFLP, all the other strains had BstU1 restriction site and hence will be classified as genotype 1. This is of relevance since HCV genotype 6 is found frequently in Southeast Asia 12. All our patients in this study who had genotype 6 were from Northeastern region of India from where there is much travel to other parts of southeast Asia. This region is geographically closest to Thailand and Vietnam, and hence, it is possible that the genotype 6 strains in this study could have originated from these countries.
Hepatitis C virus subtype identification is significant especially for the identification of 1a and 1b for treatment using the recently licensed DAAs 18, 19. Also, subtype identification has a greater significance in epidemiological studies. In our study, there was 100% concordance between subtypes of genotype 1. But in contrast, other studies which compared direct sequencing of the NS5b region with a 5′NCR‐based commercial reverse line probe assay (Inno‐LiPA HCV II; Innogenetics, Ghent, Belgium) identified only 80% of the subtypes accurately where a significant percent of genotype 1a isolates were misclassified by LiPA as genotype 1b. The region of 5′NCR used by Inno‐LiPA HCV II has a A/G sequence polymorphism at position ‐99 that does not allow differentiation of subtype 1a from subtype 1b isolates using reverse line probe assay 35. Sequencing of the respective regions of HCV is required for the accurate identification of HCV subtypes.
In this study, we have genotyped HCV using the core region type‐specific PCR and the 5′NCR PCR‐RFLP method and compared each with NS5b sequencing as the gold standard. We found good overall agreement between the different methods of HCV genotyping when compared to the NS5b‐based sequencing. But when the core type‐specific PCR was compared with NS5b sequencing for specific genotypes, genotype 3 and 4 correlated 100%, while genotype 6 showed only 57.1% correlation with NS5b sequencing results. Similarly, the 5′NCR PCR‐RFLP was able to identify only 14% of the HCV genotype 6 samples in this study, the remaining 86% were classified as genotype 1. This study suggests that core type‐specific PCR and 5′NCR PCR‐RFLP can be underreporting genotype 6 prevalent in this region. The core type‐specific PCR classified genotype 6 as genotype 4 and the 5′NCR PCR‐RFLP classified genotype 6 as genotype 1. Sequencing of the discordant genotype 6 samples using core region‐specific primers were able to identify all the HCV genotype 6, whereas sequencing of the 5′NCR region was not able to identify the genotype 6 prevalent in this region. Based on this study, compared with the NS5b region‐based sequencing, core type‐specific PCR and 5′NCR PCR‐RFLP fell short in identifying HCV genotype 6 from this region.
Our study has limitations: our sample size for some HCV genotypes especially genotype 1 is very low and may not have the discriminative power to draw conclusions on the subtype classification. Despite this limitation, our results indicate that 5′NCR PCR‐RFLP and core type‐specific PCR failed to identify all the HCV genotype 6. Also, HCV 5′NCR region used for this genotyping method failed to identify genotype 6 after sequencing, indicating that the region may not be able to differentiate HCV genotype 6 from HCV genotype 1. In this study, we did not test the samples against any commercial platforms as they are not yet available in our setting.
Laboratories with expertise in PCR and sequencing would be able to produce reliable genotyping results without having to rely on other more expensive commercial platforms not easily accessible in India. Participation in a quality assurance programs would ascertain the suitability of a genotyping assay to correctly identify HCV genotypes and subtypes. It would also give confidence in the genotyping strategy of the participating laboratory. Use of any one of these cost‐effective genotyping assays would enhance the effective management of HCV infected individuals.
Acknowledgments
This study was carried out using intramural funds from departmental resources. We acknowledge Dr. Solomon for analyzing the results.
References
- 1. Ogata N, Alter HJ, Miller RH, Purcell RH. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci U S A 1991;88:3392–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith DB, Bukh J, Kuiken JC, et al. Expanded Classification of Hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatology 2014;59:318–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simmonds P, Bukh J, Combet C, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 2005;42:962–973. [DOI] [PubMed] [Google Scholar]
- 4. Nousbaum JB, Pol S, Nalpas B, Landais P, Berthelot P, Brechot C. Hepatitis C virus type 1b (II) infection in France and Italy. Collaborative Study Group. Ann Intern Med 1995;122:161–168. [DOI] [PubMed] [Google Scholar]
- 5. Zein NN, Rakela J, Krawitt EL, Reddy KR, Tominaga T, Persing DH. Hepatitis C virus genotypes in the United States: Epidemiology, pathogenicity, and response to interferon therapy. Collaborative Study Group. Ann Intern Med 1996;125:634–639. [DOI] [PubMed] [Google Scholar]
- 6. Maggi F, Vatteroni ML, Fornai C, et al. Subtype 2c of hepatitis C virus is highly prevalent in Italy and is heterogeneous in the NS5A region. J Clin Microbiol 1997;35:161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abdulkarim AS, Zein NN, Germer JJ, et al. Hepatitis C virus genotypes and hepatitis G virus in hemodialysis patients from Syria: Identification of two novel hepatitis C virus subtypes. Am J Trop Med Hyg 1998;59:571–576. [DOI] [PubMed] [Google Scholar]
- 8. Chamberlain RW, Adams N, Saeed AA, Simmonds P, Elliott RM. Complete nucleotide sequence of a type 4 hepatitis C virus variant, the predominant genotype in the Middle East. J Gen Virol 1997;1997(78):1341–1347. [DOI] [PubMed] [Google Scholar]
- 9. Shobokshi OA, Serebour FE, Skakni L, Al‐Saffy YH, Ahdal MN. Hepatitis C genotypes and subtypes in Saudi Arabia. J Med Virol 1999;58:44–48. [PubMed] [Google Scholar]
- 10. Gededzha MP, Selabe SG, Blackard JT, Kyaw T, Mphahlele MJ. Near full‐length genome analysis of HCV genotype 5 strains from South Africa. Infect Genet Evol 2014;21:118–123. [DOI] [PubMed] [Google Scholar]
- 11. Smuts HE, Kannemeyer J. Genotyping of hepatitis C virus in South Africa. J Clin Microbiol 1995;33:1679–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mellor J, Walsh EA, Prescott LE, et al. Survey of type 6 group variants of hepatitis C virus in Southeast Asia by using a core‐based genotyping assay. J Clin Microbiol 1996;34:417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tokita H, Okamoto H, Luengrojanakul P, et al. Hepatitis C virus variants from Thailand classifiable into five novel genotypes in the sixth (6b), seventh (7c, 7d) and ninth (9b, (c) major genetic groups. J Gen Virol 1995;76:2329–2335. [DOI] [PubMed] [Google Scholar]
- 14. Tokita H, Okamoto H, Tsuda F, et al. Hepatitis C virus variants from Vietnam are classifiable into the seventh, eighth, and ninth major genetic groups. Proc Natl Acad Sci U S A 1994;91:11022–11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murphy DG, Sablon E, Chamberland J, Fournier E, Dandavino R, Tremblay CL. Hepatitis C virus genotype 7, a new genotype originating from Central Africa. J Clin Microbiol 2015;53:967–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Christdas J, Sivakumar J, David J, Daniel HD, Raghuraman S, Abraham P. Genotypes of hepatitis C virus in the Indian sub‐continent: A decade‐long experience from a tertiary care hospital in South India. Indian J Med Microbiol 2013;31:349–353. [DOI] [PubMed] [Google Scholar]
- 17. Hnatyszyn HJ. Chronic hepatitis C and genotyping: The clinical significance of determining HCV genotypes. Antivir Ther 2005;10:1–11. [PubMed] [Google Scholar]
- 18. Recommendation for testing, managing, and treating hepatitis C. AASLD, 2014, http://www.hcvguidelines.org.
- 19. European Association for the Study of the Liver . EASL recommendation on treatment of hepatitis C 2015. Clinical Practice Guidelines. J Hepatol 2015; 63: 199–236.25911336 [Google Scholar]
- 20. Zeuzem S, Ghalib R, Reddy KR, et al. Grazoprevir‐elbasvir combination therapy for treatment‐naive cirrhotic and noncirrhotic patients with chronic HCV genotype 1, 4, or 6 infection: A randomized trial. Ann Intern Med 2015;163:1–13. [DOI] [PubMed] [Google Scholar]
- 21. Chueca N, Rivadulla I, Lovatti R, et al. Using NS5B Sequencing for Hepatitis C Virus Genotyping Reveals Discordances with Commercial Platforms. PLoS ONE 2016;11:e0153754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Quer J, Gregori J, Rodríguez‐Frias F, et al. High‐Resolution Hepatitis C Virus Subtyping Using NS5B Deep Sequencing and Phylogeny, an Alternative to Current Methods. J Clin Microbiol 2015;53:219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pohjanpelto P, Lappalainen M, Widell A, Asikainen K, Paunio M. Hepatitis C genotypes in Finland determined by RFLP. Clin Diagn Virol 1996;7:7–16. [DOI] [PubMed] [Google Scholar]
- 24. Ohno O, Mizokami M, Wu RR, et al. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microbiol 1997;35:201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stuyver L, Wyseur A, van Arnhem W, Hernandez F, Maertens G. Second‐generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol 1996;34:2259–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simmonds P, Rose KA, Graham S, et al. Mapping of serotype‐specific, immunodominant epitopes in the NS‐4 region of hepatitis C virus (HCV): Use of type‐specific peptides to serologically differentiate infections with HCV types 1, 2, and 3. J Clin Microbiol 1993;31:1493–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simmonds P, Smith DB, McOmish F, et al. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS‐5 regions. J Gen Virol 1994;75:1053–1061. [DOI] [PubMed] [Google Scholar]
- 28. Ilina EN, Malakhova MV, Generozov EV, Nikolaev EN, Govorun VM. Matrix‐assisted laser desorption ionization‐time of flight (mass spectrometry) for hepatitis C virus genotyping. J Clin Microbiol 2005;43:2810–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takemura M, Fujigaki H, Takahashi K, et al. Genotyping and the quantification of hepatitis C virus by the melting curves in light cycler. Rinsho Byori 2004;52:167–171. [PubMed] [Google Scholar]
- 30. Trémeaux P, Caporossi A, Ramière C, et al. Amplification and pyrosequencing of near‐full‐length hepatitis C virus for typing and monitoring antiviral resistant strains. Clin Microbiol Infect 2016;22:460.e1–460.e10. [DOI] [PubMed] [Google Scholar]
- 31. Raghuraman S, Abraham P, Sridharan G. Hepatitis C virus genotypes: Special reference to the Indian scene. Indian J Gastroenterol 2003;22:180–186. [PubMed] [Google Scholar]
- 32. Raghuraman S, Shaji RV, Sridharan G, et al. Distribution of the different genotypes of HCV among patients attending a tertiary care hospital in south India. J Clin Virol 2003;26:61–69. [DOI] [PubMed] [Google Scholar]
- 33. McCormick AL, Macartney MJ, Abdi‐Abshir I, et al. Evaluation of sequencing of HCV core/E1, NS5A and NS5B as a genotype predictive tool in comparison with commercial assays targeting 5'UTR. J Clin Virol 2015;66:56–59. [DOI] [PubMed] [Google Scholar]
- 34. Chinchai T, Labout J, Noppornpanth S, et al. Comparative study of different methods to genotype hepatitis C virus type 6 variants. J Virol Methods 2003;109:195–201. [DOI] [PubMed] [Google Scholar]
- 35. Chen Z, Weck KE. Hepatitis C virus genotyping: Interrogation of the 5’ untranslated region cannot accurately distinguish genotypes 1a and 1b. J Clin Microbiol 2002;40:3127–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hino K, Sainokami S, Shimoda K, et al. Genotypes and titers of hepatitis C virus for predicting response to interferon in patients with chronic hepatitis C. J Med Virol 1994;42:299–305. [DOI] [PubMed] [Google Scholar]
- 37. Simmonds P, Holmes EC, Cha TA, et al. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS‐5 region. J Gen Virol 1993;74:2391–2399. [DOI] [PubMed] [Google Scholar]
- 38. Furione M, Simoncini L, Gatti M, Baldanti F, Grazia Revello M, Gerna G. HCV genotyping by three methods: Analysis of discordant results based on sequencing. J Clin Virol 1999;13:121–130. [DOI] [PubMed] [Google Scholar]
- 39. Nolte FS, Green AM, Fiebelkorn KR, et al. Clinical evaluation of two methods for genotyping hepatitis C virus based on analysis of the 5’ noncoding region. J Clin Microbiol 2003;41:1558–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harris KA, Teo CG. Diversity of hepatitis C virus quasispecies evaluated by denaturing gradient gel electrophoresis. Clin Diagn Lab Immunol 2001;8:62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]


