Abstract
Background
Protein arginine N‐methyltransferase 6 (PRMT6) was deemed to be indispensable in the variety of biological processes. Upregulated PRMT6 was found in various human diseases including cancer. Herein, we investigated the performance of PRMT6 methylation in the diagnosis for CRC.
Methods
A quantitative methylation‐specific polymerase chain reaction (qMSP) method was used to measure PRMT6 promoter methylation. The percentage of methylated reference (PMR) was applied to represent gene methylation level.
Results
Our data indicated that PRMT6 promoter methylation levels were significantly lower in CRC tissues than those in paired nontumor tissues (median PMR: 36.93% vs 63.12%, P = 1E‐6) and normal intestinal tissues (median PMR: 36.93% vs 506.55%, P = 8E‐12). We further examined the potential role of PRMT6 hypomethylation by the receiver operating characteristic (ROC) curve. Our results showed that the area under the curve (AUC) was 0.644 (95% CI = 0.596‐0.733) between CRC tissues and paired nontumor tissues, 0.958 (95% CI = 0.919‐0.998) between CRC tissues and normal intestinal tissues, and 0.899 (95% CI = 0.825‐0.972) between paired nontumor tissues and normal intestinal tissues.
Conclusion
Our study firstly indicated that the hypomethylation of PRMT6 promoter could be a novel diagnostic biomarker for CRC.
Keywords: colorectal cancer, DNA methylation, protein arginine N‐methyltransferase 6, quantitative methylation‐specific polymerase chain reaction
Abbreviations
- CpG
cytosine‐phosphate‐guanine
- CRC
colorectal cancer
- GEO
gene expression omnibus
- PMR
percentage of methylated reference
- PRMT6
protein arginine methyltransferases 6 genes
- qMSP
quantitative methylation‐specific polymerase chain reaction
- ROC
receiver operating characteristic
- TCGA
The Cancer Genome Atlas
1. INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer and the fourth most universal cause of cancer‐related death globally.1, 2 CRC is a complex disease influenced both by genetic factors and by environmental factors.3 As for bridging factors of genetics and environment, epigenetic modifications modulate gene expression.4, 5 DNA methylation is one of the widely studied epigenetic mechanisms, and it often occurs in CpG dinucleotide‐rich regions. Aberrant gene methylation 6, 7, 8 was deemed as one of the most promising diagnostic tools for cancer,9, 10, 11 including CRC.12
PRMT6 encodes protein arginine methyltransferase 6 which could methylate protein on arginine residue.13 The arginine N‐methyltransferase family members often bind to chromatin and act as transcriptional coactivators or corepressors. PRMTs were often found to be aberrantly regulated in various cancer types, such as prostate cancer,14 breast cancer,15, 16 and lung cancer.17 However, no article was reported on whether PRMT6 hypomethylation was associated with CRC.
Here, we assessed the association between PRMT6 promoter methylation and CRC. The goal of our study was to determine whether PRMT6 hypomethylation could be used as a diagnostic biomarker for CRC.
2. MATERIALS AND METHODS
2.1. Tissue samples
A total 121 CRC patients (mean age, 61.62 ± 11.55 years) were recruited from Zhejiang Tumor Hospital (Zhejiang, China), Shaoxing First People's Hospital (Zhejiang, China), and Third Affiliated Hospital of Nanjing University of Traditional Chinese Medicine (Nanjing, China) between August 2011 and January 2015. Tumor tissues were obtained from the central parts of tumor, and paired nontumor tissues were recruited at least 5 cm away from tumor lesion. In addition, normal intestinal tissues from 22 healthy participants were collected at Zhejiang Tumor Hospital (Zhejiang, China) at the same time. All the individuals were diagnosed by colonoscopy and pathological examination. None of the participants had a history of preoperative chemotherapy or radiation therapy before sampling. Microscopic examination showed at least 80% of cancer cells in each tumor tissue sample and no tumor cells in paired nontumor tissues. Normal intestinal tissues were collected from healthy participants. Tumor tissues and their paired nontumor tissues were taken in the same block and layer.18 All the clinical information was extracted from the medical records (Table 1). Written informed consent form was obtained from each participant. Permission for the study was given by Human Research Ethics Committees in Ningbo University and the above three hospitals.
Table 1.
The qMSP primer sequences
| Gene (product length) | Forward primer sequences (5′→3′) | Reverse primer sequences (5′→3′) |
|---|---|---|
| PRMT6 (64 bp) | AGCGATTAGATGTTGGAATG | CCACACCATAATACTACTTCAC |
| ACTB (133 bp) | TGGTGATGGAGGAGGTTTAGTAAGT | AACCAATAAAACCTACTCCTCCCTTAA |
2.2. DNA extraction, bisulfite conversion, and qMSP
The details of DNA extraction from tissue samples and bisulfite conversion were as previously described.6 The qMSP was applied to measure the methylation level, and the details of qMSP were as shown in our previous publications.7, 19 The detail of PCR was conducted as before 6and the qMSP primer sequences were shown in Table 1. Percentage of methylated reference (PMR) was calculated by 2−ΔΔCt quantification approach, in which ΔΔCt = sample DNA(CtPRMT6 ‐ CtACTBcontrol) ‐ fully methylated DNA(CtPRMT6 ‐ CtACTBcontrol) to represent PRMT6 methylation level.
2.3. GEO and TCGA datasets
To evaluate the association between PRMT6 methylation and PRMT6 expression, the data of 372 samples in TCGA colorectal adenocarcinoma cohort was downloaded from cBioPortal (http://www.cbioportal.org/). Meanwhile, data with PRMT6 expression changes in several cancer cell lines before and after 5′‐AZA‐deoxycytidine (5‐AZA) treatment was retrieved from GEO DataSets (GSE38823, https://www.ncbi.nlm.nih.gov/pubmed).
2.4. Statistical analysis
Nonparametric rank test was used to assess the methylation differences between two groups. Spearman correlation test was used to assess the correlation between PRMT6 methylation and clinical characteristics. ROC curve analysis was used to assess the diagnostic value of PRMT6 promoter methylation for CRC. A two‐sided P < .05 was defined to be statistically significant.
3. RESULTS
The target fragment in promoter was used to represent the methylation level of PRMT6 gene (GRCh37/hg19 assembly, chr1:107599926‐107599989, Figure 1A). In addition, Sanger sequencing manifested that the amplified fragment matched the target sequence and the bisulfite conversion was complete (Figure 1B). Capillary electrophoresis showed that the length of PRMT6 target fragment was 64 bp as expected (Figure 1C).
Figure 1.
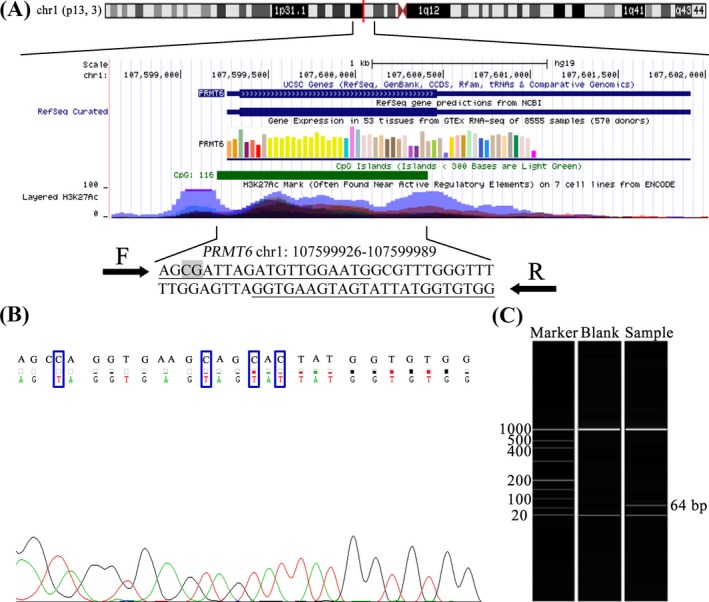
The target sequence of PRMT6 methylation assay. A, The genomic position and functional annotation of amplified fragment were based on UCSC genome browser GRCh37 assembly. The qMSP primers are underlined, and one CpG sites are in gray. F denotes forward primer; R denotes reverse primer. B, The picture on the left is the sequencing result of PRMT6 qMSP product. The top row of the sequence is the original DNA sequence; the second row of the sequence is the converted sequence. C, The results of capillary electrophoresis. The first column is the marker band; the second column is the blank control; the third column is 64‐bp qMSP product as expected
To evaluate the value of PRMT6 methylation in the diagnosis of CRC, we recruited tumor tissues and paired nontumor tissues of 121 CRC patients and normal intestinal tissues of 22 healthy participations. Our study showed that the methylation levels of PRMT6 promoter were significantly lower in CRC tissues than those in the paired nontumor tissues (P = 1E‐6, Figure 2), and than those in the normal intestinal tissues (P = 8E‐12, Figure 2). In addition, we also observed significantly lower PRMT6 methylation levels in the paired nontumor tissues compared to those in normal intestinal tissues (P = 3E‐9, Figure 2).
Figure 2.
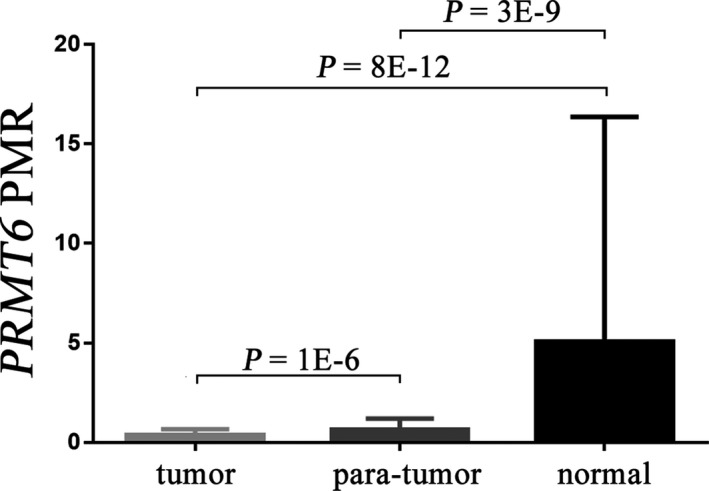
Comparisons of percentage of methylated reference (PMR) of PRMT6 between tumor tissues, paired nontumor tissues, and normal intestinal tissues. PMR denotes the percentage of methylated reference, and data are presented as median (interquartile range). P value is calculated by a nonparametric test
Further ROC curve analysis showed that the AUC was 0.644 (95% CI = 0.596‐0.733) with a specificity of 53.7% and a sensitivity of 74.4% between tumor tissues and paired nontumor tissues (Figure 3A). Between tumor tissues and normal intestinal tissues, PRMT6 hypomethylation yielded a significant AUC of 0.958 (95% CI = 0.919‐0.998) with a specificity of 78.5% and a sensitivity of 95.5% (Figure 3B). Furthermore, PRMT6 hypomethylation yielded a significant AUC of 0.899 (95% CI = 0.825‐0.972) with a specificity of 78.5% and a sensitivity of 81.8% to distinguish the difference between adjacent nontumor tissues and normal intestinal tissues (Figure 3C). To further evaluate the diagnostic value of PRMT6 hypomethylation in CRC, we compared the positive predictive value (PPV) of PRMT6 hypomethylation and CEA in 39 samples. Then, we used ROC best value (73.0%) as cutoff value to divide the PRMT6 methylation levels into either hypermethylation or hypomethylation, and we found that the PPV of PRMT6 hypomethylation in tumor was 64.1%, which was higher than the PPV of CEA (33.3%).
Figure 3.
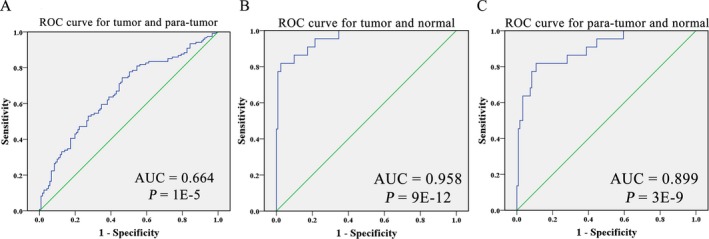
ROC curves of PRMT6 hypomethylation as a diagnostic biomarker for colorectal cancer (CRC). ROC curve for the diagnostic value of PRMT6 hypomethylation between CRC tumor tissues and para‐tumor tissues A, between CRC tumor tissues and normal intestinal tissues B, between para‐tumor tissues, and normal intestinal tissues C, ROC: receiver operating characteristic. AUC: area under the curve
Subsequently, we examined the correlation between PRMT6 methylation and the clinicopathological features of CRC patients. Our results showed that PRMT6 methylation in tumor was not significantly associated with gender, age, clinical stage, differentiation, tumor size or lymph node metastasis of the patients (all P > .05, Table 1). There was a significant correlation between PRMT6 hypomethylation and differentiation in paired nontumor tissues (P = .032, Table 1), and there was no significant correlation of PRMT6 hypomethylation with other clinical features (all P > .05, Table 2).
Table 2.
Association of PRMT6 methylation with clinical characteristics in CRC patients
| Variables | N | Tumor PMR (%) | P value | Para‐tumor PMR (%) | P value |
|---|---|---|---|---|---|
| Total | 121 | 36.93 (19.89, 65.95) | 63.12 (37.01, 119.50) | ||
| Gendera | |||||
| Male | 80 | 41.37 (20.43, 70.80) | .553 | 67.08 (41.24, 120.30) | .198 |
| Female | 39 | 34.97 (18.96, 56.38) | 61.56 (16.52, 120.50) | ||
| Age (y)a | |||||
| ≤65 | 78 | 33.15 (19.38, 59.63) | .097 | 64.33 (41.41, 146.58) | .114 |
| >65 | 41 | 51.77 (21.84, 92.10) | 55.05 (20.19, 101.24) | ||
| Differentiationa | |||||
| poorly | 17 | 40.55 (12.73, 66.66) | .653 | 51.97 (38.80, 108.48) | .032 |
| moderate + well | 101 | 36.93 (21.81, 69.09) | 64.12 (36.81, 120.70) | ||
| Tumor sizea | |||||
| ≤5 cm | 80 | 36.92 (23.52, 67.80) | .563 | 55.29 (41.24, 106.53) | .210 |
| >5 cm | 40 | 38.52 (14.6, 65.04) | 87.96 (33.9, 128.32) | ||
| Lymphatic metastasisa | |||||
| Yes | 56 | 31.49 (13.95, 56.61) | .235 | 67.53 (41.80, 146.12) | .442 |
| No | 63 | 47.24 (21.96, 77.17) | 57.21 (35.77, 111.7) | ||
PMR, the percentage of methylated reference.
Data are presented as median (interquartile range).
P value is calculated by Spearman test. Two‐sided P value < .05 (in bold).
A small amount (<1%) of patients did not have the information.
In addition, an inverse correlation between PRMT6 methylation and PRMT6 expression was observed according to our analysis of data from 372 TCGA colorectal adenocarcinoma samples (r = −.378, P = 4E‐14, Figure 4A). In addition, GEO data analysis showed that PRMT6 expression was significantly increased when cell lines (SAS and HCS3) were treated with demethylation agent (5‐AZA, fold changes (FC) = 1.30 and 1.16, Figure 4B).
Figure 4.
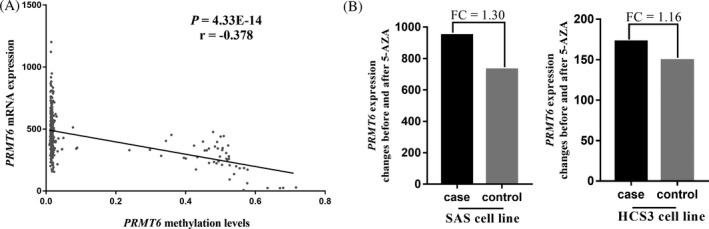
Association between PRMT6 methylation status and expression. A, There was an inverse correlation between DNA methylation level and mRNA expression in TCGA colorectal adenocarcinoma cohort (http://www.cbioportal.org/); B, expression status of several cell lines before and after 5‐AZA treated in GEO database. P value is calculated by Spearman rank correlation test. FC: fold change
4. DISCUSSION
In this study, we analyzed PRMT6 methylation levels of tumor tissues with those in the paired nontumor tissues and normal intestinal tissues. Moreover, we evaluated the correlations between PRMT6 methylation and a series of clinical features. Our results showed that PRMT6 methylation levels were significantly lower in tumor tissues than those in nontumor and normal intestinal tissues. These findings led us to speculate that PRMT6 hypomethylation might be used as a diagnostic biomarker of CRC.
PRMTs can methylate the arginine residues of proteins,13 including p16 and p21 which are tumor suppressor proteins.16, 20, 21 Methylation of p16 usually resulted in decreased protein function and promoted tumorigenesis.22 The p21 gene encodes a cyclin‐dependent kinase inhibitory protein with tumor suppressor activity.23 The p21 plays an important role in the development and progression of cancer,24 and hypermethylation of p21 leads to downregulation of p21 gene expression.24 Downregulated expression of PRMT6 in CRC cells can induce p21 overexpression and inhibit cell growth and colony formation capacity.25 PRMT6 is often found to be overexpressed in cancers, including CRC,12, 25 bladder cancer,17 lung cancer,17 prostate cancer,15, 26 and breast cancer.15 In the present study, we found PRMT6 hypomethylation was significantly associated with CRC. Meanwhile, TCGA and GEO data analyses showed that PRMT6 methylation was inversely correlated with mRNA expression. We hypothesized that PRMT6 hypomethylation might increase the expression of PRMT6 protein and thus promotes cell proliferation by downregulating tumor suppressor genes.27
As a noninvasive method widely used in the detection of CRC, fecal occult blood test (FOBT) comprises guaiac (gFOBT) and immunochemical (iFOBT). However, the gFOBT test was easy to be effected by other factors, resulting in false‐positive results. The detection rate of iFOBT decreased rapidly once the time from collection to laboratory inspection was deferred for over 5 days.28, 29, 30 As for colonoscopy, despite its high sensitivity and specificity, there were still some shortcomings, such as complications (intestinal gastrointestinal bleeding, intestinal perforation,) and low compliance.31 Carcinoembryonic antigen (CEA) in serum is one of the biomarkers widely used in the diagnosis of cancer;32 however, it is still defective because increased CEA levels were not only found in cancer patients but also in smokers, which reduced its specificity in cancer diagnosis.33, 34 Meanwhile, CEA lacks ideal sensitivity in the early diagnosis of CRC.35, 36 A review based on 9834 CEA test results indicated that the sensitivity of CEA ranged from 50% to 80%.37 In addition, the sensitivity of serum CEA in detecting the risk of CRC recurrence was only 64%.38 Here, our study found that PRMT6 hypomethylation had a specificity of 78.5% and a sensitivity of 95.5% for the detection of CRC, and we also found the PPV of PRMT6 hypomethylation was higher than that of serum CEA. However, future investigation is needed to access the combined diagnosis of PRMT6 methylation and CEA levels for CRC.
The benign colorectal disease can gradually progress to advanced adenoma and invasive adenocarcinoma.39, 40, 41 Abnormal gene methylation was found in hyperplastic polyps with cancerous potential.42 Previous studies identified that there was aberrant methylation of multiple genes between CRC tissues and colorectal benign polyp tissues and between colorectal benign polyp tissues and healthy intestinal tissues.43, 44, 45, 46, 47, 48 In the present study, we did not perform the similar comparison due to a lack of colorectal benign polyp tissues. Future study is needed to investigate the role of PRMT6 methylation in colorectal benign polyp tissues.
However, our study has some limitations. Firstly, although this is the first study to investigate the methylation of PRMT6 in CRC, we did not evaluate the role of PRMT6 hypomethylation in the benign colorectal tissues, which usually have a high risk of CRC. Secondly, we did not collect serum protein markers, such as CEA, in healthy individuals to compare the diagnostic value of PRMT6 methylation with the conventional biomarkers. Future analysis is needed to evaluate the joint role of methylation biomarkers and protein biomarkers for the diagnosis of CRC. Lastly but not least, our study did not have enough amount of samples to evaluate the correlation of PRMT6 methylation with PRMT6 expression. However, our TCGA data analysis showed an inverse correlation between PRMT6 methylation and PRMT6 expression.
In summary, we found a significant association of PRMT6 hypomethylation with CRC. Our results suggested that PRMT6 hypomethylation might be used as a diagnostic biomarker for CRC, although further validation with large samples is needed in the future.
AUTHORS CONTRIBUTIONS
SD and RP conceived and designed the experiments. HY, XY, CZ, JZ, JD, YZ, BW, and YM performed the experiments. RP analyzed the data. RP and SD contributed to completion of figures, tables, and the writing of this manuscript.
ACKNOWLEDGMENTS
We thank TCGA and GEO databases for their open access. The research is supported by K. C. Wong Magna Fund in Ningbo University.
Pan R, Yu H, Dai J, et al. Significant association of PRMT6 hypomethylation with colorectal cancer. J Clin Lab Anal. 2018;32:e22590 10.1002/jcla.22590
REFERENCES
- 1. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490‐1502. [DOI] [PubMed] [Google Scholar]
- 2. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359‐E386. [DOI] [PubMed] [Google Scholar]
- 3. Bishak YK, Payahoo L, Osatdrahimi A, et al. Mechanisms of cadmium carcinogenicity in the gastrointestinal tract. Asian Pac J Cancer Prev. 2015;16:9‐21. [DOI] [PubMed] [Google Scholar]
- 4. Toporcov TN, Znaor A, Zhang ZF, et al. Risk factors for head and neck cancer in young adults: a pooled analysis in the INHANCE consortium. Int J Epidemiol. 2015;44:169‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matsubara N. Epigenetic regulation and colorectal cancer. Dis Colon Rectum. 2012;55:96‐104. [DOI] [PubMed] [Google Scholar]
- 6. Li J, Chen C, Bi X, et al. DNA methylation of CMTM3, SSTR2, and MDFI genes in colorectal cancer. Gene. 2017;630:1‐7. [DOI] [PubMed] [Google Scholar]
- 7. Li B, Chen X, Jiang Y, et al. CCL2 promoter hypomethylation is associated with gout risk in Chinese Han male population. Immunol Lett. 2017;190:15‐19. [DOI] [PubMed] [Google Scholar]
- 8. Wu D, Chen X, Xu Y, et al. Prognostic value of MLH1 promoter methylation in male patients with esophageal squamous cell carcinoma. Oncol Lett. 2017;13:2745‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen X, Hu H, Liu J, et al. FOXF2 promoter methylation is associated with prognosis in esophageal squamous cell carcinoma. Tumour Biol. 2017;39:1010428317692230. [DOI] [PubMed] [Google Scholar]
- 11. Chen X, Yang Y, Liu J, et al. NDRG4 hypermethylation is a potential biomarker for diagnosis and prognosis of gastric cancer in Chinese population. Oncotarget. 2017;8:8105‐8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ravegnini G, Zolezzi Moraga JM, Maffei F, et al. Simultaneous analysis of SEPT9 promoter methylation status, micronuclei frequency, and folate‐related gene polymorphisms: the potential for a novel blood‐based colorectal cancer biomarker. Int J Mol Sci. 2015;16:28486‐28497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Migliori V, Phalke S, Bezzi M, et al. Arginine/lysine‐methyl/methyl switches: biochemical role of histone arginine methylation in transcriptional regulation. Epigenomics. 2010;2:119‐137. [DOI] [PubMed] [Google Scholar]
- 14. Almeida‐Rios D, Graca I, Vieira FQ, et al. Histone methyltransferase PRMT6 plays an oncogenic role of in prostate cancer. Oncotarget. 2016;7:53018‐53028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim NH, Kim SN, Seo DW, et al. PRMT6 overexpression upregulates TSP‐1 and downregulates MMPs: its implication in motility and invasion. Biochem Biophys Res Comm. 2013;432:60‐65. [DOI] [PubMed] [Google Scholar]
- 16. Phalke S, Mzoughi S, Bezzi M, et al. p53‐Independent regulation of p21Waf1/Cip1 expression and senescence by PRMT6. Nucleic Acids Res. 2012;40:9534‐9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoshimatsu M, Toyokawa G, Hayami S, et al. Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int J Cancer. 2011;128:562‐573. [DOI] [PubMed] [Google Scholar]
- 18. Rauscher GH, Kresovich JK, Poulin M, et al. Exploring DNA methylation changes in promoter, intragenic, and intergenic regions as early and late events in breast cancer formation. BMC Cancer. 2015;15:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Y, Chen X, Hu H, et al. Elevated UMOD methylation level in peripheral blood is associated with gout risk. Sci Rep. 2017;7:11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X, Huang Y, Zhao J, et al. Suppression of PRMT6‐mediated arginine methylation of p16 protein potentiates its ability to arrest A549 cell proliferation. Int J Biochem Cell Biol. 2012;44:2333‐2341. [DOI] [PubMed] [Google Scholar]
- 21. Kleinschmidt MA, de Graaf P, van Teeffelen HA, et al. Cell cycle regulation by the PRMT6 arginine methyltransferase through repression of cyclin‐dependent kinase inhibitors. PLoS One. 2012;7:e41446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma WL, Wang L, Liu LX, et al. Effect of phosphorylation and methylation on the function of the p16INK4a protein in non‐small cell lung cancer A549 cells. Oncol Lett. 2015;10:2277‐2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li YJ, Hoang‐Xuan K, Zhou XP, et al. Analysis of the p21 gene in gliomas. J Neurooncol. 1998;40:107‐111. [DOI] [PubMed] [Google Scholar]
- 24. Akhter N, Akhtar MS, Ahmad MM, et al. Association of mutation and hypermethylation of p21 gene with susceptibility to breast cancer: a study from north India. Mol Biol Rep. 2014;41:2999‐3007. [DOI] [PubMed] [Google Scholar]
- 25. Lim Y, Yu S, Yun JA, et al. The prognostic significance of protein arginine methyltransferase 6 expression in colon cancer. Oncotarget. 2018;9:9010‐9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vieira FQ, Costa‐Pinheiro P, Ramalho‐Carvalho J, et al. Deregulated expression of selected histone methylases and demethylases in prostate carcinoma. Endocr Relat Cancer. 2014;21:51‐61. [DOI] [PubMed] [Google Scholar]
- 27. Stein C, Riedl S, Ruthnick D, et al. The arginine methyltransferase PRMT6 regulates cell proliferation and senescence through transcriptional repression of tumor suppressor genes. Nucleic Acids Res. 2012;40:9522‐9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oono Y, Iriguchi Y, Doi Y, et al. A retrospective study of immunochemical fecal occult blood testing for colorectal cancer detection. Clin Chim Acta. 2010;411:802‐805. [DOI] [PubMed] [Google Scholar]
- 29. van Rossum LG, van Rijn AF, van Oijen MG, et al. False negative fecal occult blood tests due to delayed sample return in colorectal cancer screening. Int J Cancer. 2009;125:746‐750. [DOI] [PubMed] [Google Scholar]
- 30. Li Y, Song L, Gong Y, et al. Detection of colorectal cancer by DNA methylation biomarker SEPT9: past, present and future. Biomark Med. 2014;8:755‐769. [DOI] [PubMed] [Google Scholar]
- 31. Committee ASoP, Fisher DA, Maple JT, et al. Complications of colonoscopy. Gastrointest Endosc. 2011;74:745‐752. [DOI] [PubMed] [Google Scholar]
- 32. Jessup JM, Thomas P. Carcinoembryonic antigen: function in metastasis by human colorectal carcinoma. Cancer Metastasis Rev. 1989;8:263‐280. [DOI] [PubMed] [Google Scholar]
- 33. Lee H, Song M, Shin N, et al. Diagnostic significance of serum HMGB1 in colorectal carcinomas. PLoS One. 2012;7:e34318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qi J, Qian C, Shi W, et al. Alu‐based cell‐free DNA: a potential complementary biomarker for diagnosis of colorectal cancer. Clin Biochem. 2013;46:64‐69. [DOI] [PubMed] [Google Scholar]
- 35. Fletcher RH. Carcinoembryonic antigen. Ann Intern Med. 1986;104:66‐73. [DOI] [PubMed] [Google Scholar]
- 36. Peng HX, Yang L, He BS, et al. Combination of preoperative NLR, PLR and CEA could increase the diagnostic efficacy for I‐III stage CRC. J Clin Lab Anal. 2017;31 10.1002/jcla.22075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sorensen CG, Karlsson WK, Pommergaard HC, et al. The diagnostic accuracy of carcinoembryonic antigen to detect colorectal cancer recurrence – a systematic review. Int J Surg. 2016;25:134‐144. [DOI] [PubMed] [Google Scholar]
- 38. Tan E, Gouvas N, Nicholls RJ, et al. Diagnostic precision of carcinoembryonic antigen in the detection of recurrence of colorectal cancer. Surg Oncol. 2009;18:15‐24. [DOI] [PubMed] [Google Scholar]
- 39. Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159‐170. [DOI] [PubMed] [Google Scholar]
- 40. Stella S, Bruzzese A, Chiarini S. [Biological bases of the colorectal adenoma‐carcinoma sequence]. Il Giornale di Chirurgia. 1996;17:473‐476. [PubMed] [Google Scholar]
- 41. Takami K, Yana I, Kurahashi H, et al. Multistep carcinogenesis in colorectal cancers. Southeast Asian J Trop Med Public Health. 1995;26(Suppl 1):190‐196. [PubMed] [Google Scholar]
- 42. Wynter CV, Walsh MD, Higuchi T, et al. Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut. 2004;53:573‐580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang HL, Liu P, Zhou PY, et al. Promoter methylation of the RASSF1A gene may contribute to colorectal cancer susceptibility: a meta‐analysis of cohort studies. Ann Hum Genet. 2014;78:208‐216. [DOI] [PubMed] [Google Scholar]
- 44. Wang YC, Yu ZH, Liu C, et al. Detection of RASSF1A promoter hypermethylation in serum from gastric and colorectal adenocarcinoma patients. World J Gastroenterol. 2008;14:3074‐3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li YW, Kong FM, Zhou JP, et al. Aberrant promoter methylation of the vimentin gene may contribute to colorectal carcinogenesis: a meta‐analysis. Tumour Biol. 2014;35:6783‐6790. [DOI] [PubMed] [Google Scholar]
- 46. Suzumura K, Okada T, Satake M, et al. Laparoscopic splenectomy for inflammatory pseudotumor of the spleen. Hepatogastroenterology. 2011;58:1909‐1911. [DOI] [PubMed] [Google Scholar]
- 47. Abouzeid HE, Kassem AM, Abdel Wahab AH, et al. Promoter hypermethylation of RASSF1A, MGMT, and HIC‐1 genes in benign and malignant colorectal tumors. Tumour Biol. 2011;32:845‐852. [DOI] [PubMed] [Google Scholar]
- 48. Wu PP, Zou JH, Tang RN, et al. Detection and clinical significance of DLC1 gene methylation in serum DNA from colorectal cancer patients. Chin J Cancer Res. 2011;23:283‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]


