Abstract
Objective
Genetic polymorphisms in ALDH2 and C12orf30 genes have been reported to increase the risk of developing esophageal squamous cell carcinoma (ESCC). This study aims to investigate the relationship between ALDH2 rs671 and c12orf30 rs4767364 polymorphisms in the chromosome 12q24 gene, and risk and prognosis of individuals developing esophageal cancer (ESCC) in Xinjiang Kazak and Han populations.
Methods
The case group consisted of 127 ESCC patients. The control group comprised of 125 healthy individuals. Subjects that were recruited all come from Xinjiang province. TaqMan and the Hardy‐Weinberg equilibrium were the main methods employed to detect and examine the distribution of genotypes of rs671 and rs4767364.
Results
The genotype frequencies of ALDH2 rs671 between the Kazak case and control groups were statistically significant, while no significant difference was observed between the Han case and control groups (P>.05). Moreover, ALDH2 rs671 (G>A) was associated with poor prognosis of ESCC in both Kazak and Han populations, and c12orf30 rs4767364 (A>G) was also connected with poor prognosis of ESCC in Kazak but not in Han population.
Conclusion
In the chromosome 12q24 locus, ALDH2 rs671 (G>A) is related to the susceptibility to ESCC in Kazak populations, and it is also associated with poor prognosis of EC in Kazak and Han populations. Furthermore, c12orf30 rs4767364 (A>G) may be correlated with poor ESCC prognosis in Kazak population.
Keywords: ALDH2, c12orf30, esophageal squamous cell carcinoma, gene polymorphism, Han, Kazak, prognosis, susceptibility
1. Introduction
Originating from the gastrointestinal tract, esophageal cancer (EC) is considered to be one of the most invasive malignant tumors and is the sixth major cause of cancer‐related death worldwide, contributing to more than 400 000 deaths each year.1 The two major subtypes of EC are: esophageal squamous‐cell carcinoma (ESCC) and esophageal adenocarcinoma, both of which may arise due to excessive cigarette smoking, alcohol consumption, obesity, as well as gastroesophageal reflux disease (GERD).2 In China, the highest mortality rates of people diagnosed with ESCC are found in areas of northwestern Xinjiang. The majority of the population were of Kazak origin whereby the age‐adjusted mortality of ESCC have reached as many as 150/100 000 individuals.3 Behavioral and environmental risk factors such as tobacco smoking, alcohol consumption, obesity, and GERD, have all been associated with the carcinogenesis and development of ESCC.2 However, only a small portion of people exposed to the aforementioned risk factors will go on to develop ESCC. In addition, studies of familial aggregation and its significant to EC have also been reported, highlighting a genetic contribution to the pathogenesis of ESCC.4 Interestingly, multiple gene polymorphisms, such as oncogene activation coupled with the loss of tumor suppressor genes, are reported to affect the progression and prognosis of ESCC.5, 6
The Aldehyde dehydrogenase‐2 (ALDH2) gene is composed of 12 coding exons that are located on chromosome 12q24.7 It has been shown that the varied ALDH2 rs671 gene in different Asian populations may contribute to the susceptibility to ESCC.8 The A allele at the ALDH2 rs671 gene locus has been linked with an increase in susceptibility in developing ESCC in Chinese populations, implying that abnormally high levels of acetaldehyde may be an indicator for ESCC development.9 Another gene of particular interest is the cl2orf30 gene. The c12orf30 gene is also located on chromosome 12q24 and is found to play a role in the type 1 diabetes due to their presumed significance in immune signaling.10 Furthermore, a study has shown that variants in c12orf30 regions are associated with autoimmune diseases such as rheumatoid arthritis as well as systemic lupus erythematosus.11 More importantly to us, c12orf30 rs4767364 is identified to be related to upper aero‐digestive tract cancers including ESCC.12 Thus, we conduct this study in order to gain a better understanding between the associations of ALDH2 rs671 and c12orf30 rs4767364 polymorphisms with the risk and prognosis of ESCC in Xinjiang Han and Kazakh populations in China.
2. Materials and Methods
2.1. Ethics statement
This study was approved by the Ethical Committee of the First Affiliated Hospital of Xinjiang Medical University. Written informed consents were obtained from all study subjects.
2.2. Subject selection
A total of 127 ESCC patients (designated as the case group) in the First Affiliated Hospital of Xinjiang Medical University between 2006 and 2010 were randomly recruited. The subjects that were recruited included 65 Kazak cases and 62 Han cases. Patients who can partake in the case group have to fulfill the following criteria: live as local residents of Xinjiang for more than 20 years,; patients diagnosed with ESCC from tumor tissue obtained from surgical resection and with confirmation by pathological examination of endoscopic biopsy according to the diagnostic criteria of ESCC13; and patients who have not undergone radiotherapy or chemotherapy to treat their tumors.
The exclusion criteria for patients who are not eligible for this study include: patients with a history of malignant tumors or malignant neoplasms of esophagus metastasis; patients with incomplete clinical data. The control group comprised of 125 healthy individuals (75 cases of Kazak origin and 50 cases of Han origin) that lived in the same area of Xinjiang province for more than 20 years. Finally, the patients’ personal information and clinical data of smoking, drinking, and family history of each subject was obtained by questioning the subjects and their family members.
2.3. Sample preparation and single nucleotide polymorphism (SNP) detection
Participants were first subjected to 5 mL of venous blood collection which were anticoagulated with ethylene diamine tetraacetic acid (EDTA) and then stored at −80°C for preservation. The extraction of DNA was carried out using a genomic DNA blood extraction kit (TianGen Biotech Co., Ltd, Beijing, China). TaqMan probes were applied to detect the genotypes of targeted gene polymorphism. The SNPs of ALDH2 rs671 and c12orf30 rs4767364 were synthesized by polymerase chain reaction (PCR) amplifications and fluorescent probes (ABI Company, Oyster Bay, NY, USA). The TaqMan genotyping reaction was prepared in the 96‐well plate following operating instructions. PCR amplification for targeted fragments was performed using GeneAmp PCR System 9700 thermal cycler (Applied Biosystems, Life Technologies Ltd, Paisley, UK). The scanning and analysis of the amplified fragments were performed using ABI PRISM 7900 HT real‐time quantitative real‐time polymerase chain reaction (qRT‐PCR), and genotyping results were analyzed using SDS 2.4 software (Applied Biosystems, Inc., Foster City, CA, USA). A random set of three reactions without DNA isolation and two replicates were used as controls in each of the 96‐well plate for quality control. The primers and probe sequences utilized in the protocol are shown as follows: sense‐rs671: 5′‐ GAGCAAAGTTTAATGTCCCA‐3′, antisense‐rs671: 5′‐ACAGACCCCAATCCCCCAGC‐3′; sense‐rs4767364: 5′‐AGAGTTTCTTTTTTTTTTCT‐3′; antisense‐rs4767364: 5′‐AACTCAAAGACAAAATTATT‐3′. The primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). Amplification reaction step of PCR was conducted by ABI 7900 real‐time quantitative polymerase chain reaction (PCR) instrument (Applied Biosystem).
2.4. Follow up
Follow‐up procedures including; outpatient appointments, telephone calls and letters were employed on all 127 cases of the case group patients to keep track of their condition. The follow‐up procedure was conducted every six months from the date of our grouping to April 1st, 2015. The Survival time of each patient was calculated from the first day after the operation until the date of death or until the end of the follow‐up procedure period. The survival time was calculated by month, and the follow‐up duration was five years. In 127 patients in the case group, there were 10 lost cases, with a loss rate of 8%, among which there were two deaths in the first year and eight deaths in the second year.
2.5. Statistical analysis
Statistical package for the social sciences (SPSS) software 18.0 (SPSS Inc., Chicago, IL, USA) was applied to carry out statistical analysis for this investigation. The Measurement data were expressed as mean±standard deviation. A t test was used to statistically compare the age distribution between the two study groups, and χ2 was employed to compare the allelic and genotype frequencies between the two groups. The relative risk was calculated by an odds ratio (OR) with 95% confidence interval (95% CI). Moreover, Hardy‐Weinberg was performed to find out whether the population was well represented by our samples, and a Kaplan‐Meier estimator was used to along with Log‐Rank to estimate the survival function and thus used to draw the survival curves of patients for the two parameters that we are investigating The Cox regression analysis was applied to calculate the hazard ratios (HR) with 95% CI. All tests were two‐sided probability test, and P values of <.05 were considered as statistically significant.
3. Results
3.1. Comparisons of baseline characteristics between ESCC patients and healthy controls in the Kazak and Han populations
There was no significance in the gender, age, numbers of smokers and drinkers between the case and control groups (all P>.05) (Table 1).
Table 1.
Baseline characteristics of ESCC patients and healthy controls in the Kazak and Han populations
| Baseline characteristic | Kazak population (N=65) | Control group (N=75) | t/χ2 | P | Han population (N=62) | Control group (N=50) | t/χ2 | P |
|---|---|---|---|---|---|---|---|---|
| Mean age | 58.46±9.25 | 57.29±9.46 | 0.758 | .45 | 61.16±8.38 | 59.60±9.72 | 0.891 | .375 |
| Gender | ||||||||
| Male | 42 | 49 | 0.009 | .929 | 42 | 36 | 0.237 | .626 |
| Female | 23 | 26 | 20 | 14 | ||||
| Smoking | ||||||||
| − | 31 | 36 | 0.001 | .971 | 23 | 22 | 0.549 | .459 |
| + | 34 | 39 | 39 | 28 | ||||
| Drinking | ||||||||
| − | 29 | 44 | 2.755 | .097 | 32 | 30 | 0.788 | .375 |
| + | 36 | 31 | 30 | 20 | ||||
ESCC, esophageal squamous cell carcinoma.
3.2. Distribution of ALDH2 rs671 and c12orf30 rs4767364 genotypes in Kazak and Han populations conformed to the Hardy‐Weinberg balance
As shown in Table 2, the distribution of ALDH2 rs671 and c12orf30 rs4767364 genotypes in Kazak and Han populations conformed to Hardy‐Weinberg balance, suggesting that the frequency of ALDH2 rs671 genotypes and c12orf30 rs4767364 genotypes in the subjects was in equilibrium and showed group representation.
Table 2.
Hardy‐Weinberg equilibrium of rs671 and rs4767364 polymorphisms in Xinjiang Kazak and Han populations
| Genotype | Kazak population | Han population | ||||||
|---|---|---|---|---|---|---|---|---|
| Actual frequency (%) | Theoretical frequency (%) | χ2 | P | Actual frequency (%) | Theoretical frequency (%) | χ2 | P | |
| rs671 | ||||||||
| GG | 82 (58.57) | 79 (56.43) | 2.146 | 62 (55.36) | 63 (56.25) | 0.254 | ||
| GA | 46 (32.86) | 52 (37.14) | .143 | 44 (39.28) | 42 (37.50) | .614 | ||
| AA | 12 (8.57) | 9 (6.43) | 6 (5.36) | 7 (6.25) | ||||
| rs4767364 | ||||||||
| AA | 108 (77.14) | 106 (75.71) | 73 (65.18) | 74 (66.07) | ||||
| AG | 28 (20.00) | 31 (22.14) | 1.617 | .203 | 35 (31.25) | 34 (30.36) | 0.006 | .938 |
| GG | 4 (2.86) | 3 (2.15) | 4 (3.57) | 4 (3.57) | ||||
3.3. Genotype frequencies of ALDH2 rs671 (G>A) and C12orf30 rs4767364 (A>G) polymorphisms in ESCC patients and healthy controls in the Kazak and Han populations
Table 3 portrays the proportion of specific genotypes in the ALDH2 rs671 gene in ESCC patients and those who are in the control group. The ALDH2 rs671 genotypes GG, GA and AA were 50.39%, 40.16%, and 9.45% in the case group, and 64.00%, 31.20%, and 4.80% in the control group. The total constituent ratio of three genotypes in the two groups was not statistically significant (P>.05). On the other hand, the proportion of c12orf30 rs4767364 genotypes AA, GA and GGin the EC patients was 70.08%, 26.77%, and 3.15%, respectively, and 73.60%, 23.20%, and 3.20% in the control group. The total constituent ratio of the three genotypes also showed no significant difference between the two study groups (P>.05).
Table 3.
Relationship between 12q24 gene polymorphisms (rs671 and rs4767364) and the susceptibility to ESCC between the case and control groups (total Han and Kazak populations)
| rs671 | rs4767364 | |||||
|---|---|---|---|---|---|---|
| GG | GA | AA | AA | AG | GG | |
| Case group (%) | 64 (50.39) | 51 (40.16) | 12 (9.45) | 89 (70.08) | 34 (26.77) | 4 (3.15) |
| Control group (%) | 80 (64.00) | 39 (31.20) | 6 (4.80) | 92 (73.60) | 29 (23.20) | 4 (3.20) |
| OR (95% CI) | 1 | 0.612 (0.340‐1.040) | 0.400 (0.142‐1.125) | 1 | 0.825 (0.464‐1.466) | 0.967 (0.235‐3.988) |
| P | .069 | .075 | .512 | .963 | ||
OR, odds ratio; CI, confidence interval.
The results in Table 4 demonstrated that the frequency of ALDH2 rs671 GG, GA and AA genotypes in regard to the Kazak population in the case group 46.15%, 40.00%, 13.85%, and 68.00%, 26.67%, 5.33% in the control group. T test analysis showed that the total constituent ratio of the three genotypes between the two study groups was statistically significant (P<.05). It was also noted that GA carriers exhibited increased susceptibility to EC compared to GG carriers genotypes, and that the OR value was 2.210 (95% CI=1.057‐4.619) Additionally, AA carriers showed stronger susceptibility to EC with a higher OR value of 3.825 (95% CI=1.083‐13.500). In the Han population, the frequency of GG, GA and AA genotypes at ALDH2 rs671 was 54.84%, 40.32%, 4.84% in the case group and 58.00%, 38.00%, 4.00% in the control group, respectively. The total constituent ratio of the three genotypes between the two groups was not significantly different (P>.05). Moreover, the frequency of AA, GA and GG genotypes at the c12orf30 rs4767364 gene in the Kazak case group of was 75.38%, 21.54%, 3.08% and 78.67%, 18.67%, 2.67% in the control group. There was no significant difference in the total constituent ratio of the three genotypes (P>.05). In the Han population, the frequency of AA, GA and GG genotypes at c12orf30 rs4767364 in was 64.52%, 32.26%, 3.23%, and was 66.00%, 30.00%, 4.00% in the control group.
Table 4.
Relationship between 12q24 gene polymorphisms (rs671 and rs4767364) and the susceptibility to ESCC in Han and Kazak population (Han and Kazak population, respectively) [n (%)]
| Han | Kazak | |||||||
|---|---|---|---|---|---|---|---|---|
| Case group | Control group | OR (95% CI) | P | Case group | Control group | OR (95% CI) | P | |
| rs671 | ||||||||
| GG | 34 (54.84) | 29 (58.00) | Ref. | 30 (46.15) | 51 (68.00) | Ref. | ||
| GA | 25 (40.32) | 19 (38.00) | 0.865 (0.410‐1.935) | .771 | 26 (40.00) | 20 (26.67) | 2.210 (1.057‐4.619) | .034 |
| AA | 3 (4.84) | 2 (4.00) | 0.782 (0.122‐5.006) | .794 | 9 (13.85) | 4 (5.33) | 3.825 (1.083‐13.500) | .029 |
| rs4767364 | ||||||||
| AA | 40 (64.52) | 33 (66.00) | Ref. | 49 (75.38) | 59 (78.67) | Ref. | ||
| AG | 20 (32.26) | 15 (30.00) | 0.909 (0.403‐2.050) | .818 | 14 (21.54) | 14 (18.67) | 0.831 (0.361‐1.909) | .662 |
| GG | 2 (3.23) | 2 (4.00) | 1.212 (0.162‐9.082) | .851 | 2 (3.08) | 2 (2.67) | 0.831 (0.113‐6.117) | .855 |
OR, odds ratio; CI, confidence interval; Ref, reference.
3.4. Correlations of ALDH2 rs671 (G>A) and C12orf30 rs4767364 (A>G) polymorphisms with the prognosis of patients with ESCC in the Kazak and Han population
In the 65 Kazak patients, Cox regression analysis indicated that the ALDH2 rs671 (G>A) polymorphism was related to poor prognosis of EC (P=.009). The mutant genotype (GA+AA) carriers demonstrated higher death risks (HR=4.524; 95% CI=1.441‐14.210) in comparison homozygous carriers of GG genotype. Furthermore, the c12orf30 rs4767364 (A>G) polymorphism was also associated with poor prognosis of EC (P=.001), and the risk of death was significantly higher in AG+GG carriers (HR=9.538; 95% CI=2.302‐39.530) than the AA carriers (Table 5). According to the Kaplan‐Meier method, the 1‐, 3‐ and 5‐year survival rates of Kazak patients were 94.41%, 74.75% and 57.31%, respectively. The Kaplan‐Meier survival curves shows that rs671 and rs4767364 polymorphisms were positively correlated with poor prognosis in the Kazak EC patients (rs671: Log rank P=.002 and rs4767364: Log rank P=.001) (Figure 1A,B).
Table 5.
Relationship between 12q24 gene polymorphisms (rs671 and rs4767364) and the prognosis of ESCC in Kazak and Han populations in Xinjiang
| Gene | SNP | Genotype | Kazak | Han | ||||
|---|---|---|---|---|---|---|---|---|
| Death/Survival | HR (95% CI) | P # | Death/Survival | HR (95% CI) | P # | |||
| ALDH2 | rs671 | GG | 6/20 | 1 | 5/26 | 1 | ||
| GA+AA | 19/14 | 4.524 (1.441‐14.210) | .009 | 12/15 | 4.16 (1.226‐14.120) | .023 | ||
| c12orf30 | rs4767364 | AA | 13/31 | 1 | 11/26 | 1 | ||
| AG+GG | 12/3 | 9.538 (2.302‐39.530) | .001 | 8/13 | 0.689 (0.222‐2.125) | .569 | ||
SNP, single nucleotide polymorphisms; HR, hazard ratios; CI, confidence interval; #, correction for gender and age.
Figure 1.
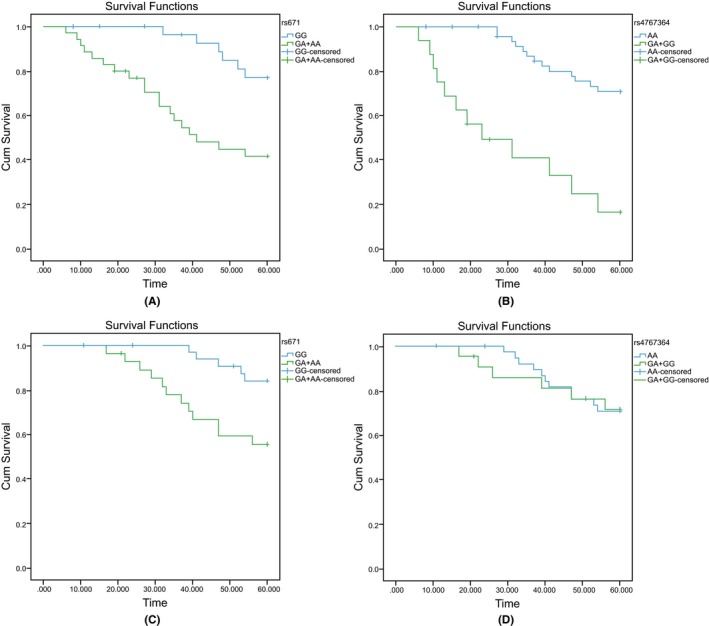
Survival curves for ESCC patients carrying different genotypes at ALDH2 rs671 and c12orf30 rs4767364. (A) The survival curve for Kazak ESCC patients carrying rs671; (B) the survival curve for Kazak ESCC patients carrying rs4767364; (C) the survival curve for Han EC patients carrying rs671; (D) the survival curve for Han EC patients carrying rs4767364
In the 62 Han patients, Cox regression analysis exhibited that the prognosis of EC was affected by ALDH2 rs671 (G>A) polymorphisms (P=.049), and that GA+AA carriers showed higher death risk (HR=3.467, 95% CI=1.083‐11.090) compared to GG carriers. However, the c12orf30 rs4767364 (A>G) polymorphism were be related to the prognosis of ESCC in Han patients, (P>.05) (Table 5). The Kaplan‐Meier method, calculated the 1‐, 3‐ and 5‐year survival rates of Han patients to be 100%, 89.76% and 70.90%, respectively. The Kaplan‐Meier survival curves revealed that the rs671 polymorphism was associated with the prognosis of ESCC in Han patients, and the survival time of GG carriers and GA+AA carriers were significantly different (Log rank P=.010) (Figure 1C). Meanwhile, the rs4767364 position failed to demonstrate an association with poor prognosis of ESCC, as well as the survival time of AA carriers and AG+GG carriers, which were not significantly different (Log rank P=.372) (Figure 1D).
4. Discussion
Over the recent years, increased gene polymorphisms have been discovered to be involved in the risk and prognosis of ESCC as more and more gene variants are identified to be potential prognostic markers in ESCC patients.14, 15 Our study aims to elucidate the effects of ALDH2 and c12orf30 gene polymorphisms on the chromosome 12q24 region in determining the risk and prognosis of ESCC. Our findings demonstrated that the ALDH2 rs671 (G>A) polymorphism is associated with increased susceptibility to ESCC in Kazak and Han population. The other gene polymorphism and c12orf30 rs4767364 (A>G) is also linked with ESCC and allows us to determine patient prognosis in the Kazak population.
We showed that the frequency of GA and AA genotypes (40.00% and 13.85%) at ALDH2 rs671 in the Kazak ESCC patients was significantly higher than those in the control group (26.67% and 5.33%). The susceptibility toward ESCC is also higher in the AA and GA carriers compared to the GG carriers. However, the frequencies of GG, GA, and AA at ALDH2 rs671 showed no statistical significance in Han patients. To the best of our knowledge, the frequency of A allele (GA+AA) carriers at the rs671 is remarkably higher in coronary artery disease (CAD) patients than the healthy controls, implying that the ALDH2 rs671 polymorphism is related to an increased risk of CAD in Han Chinese.16 Wu et al. reported that the A allele at rs671 is identified have an increase in susceptibility toward ESCC among moderate to heavy drinkers in the Chinese population, suggesting the high levels of acetaldehyde may contribute to the development of ESCC.9 Additionally, Li et al.17 also demonstrated that the ALDH2 A allele carriers who are also heavy drinkers are prominently associated with the increased risk of developing ESCC. Interestingly, a study by Ding et al.18 shows that the frequency of ALDH2 GA and AA genotypes in the ESCC patients were also higher than the controls, and they also demonstrate that the ALDH2 A/A carriers show increased susceptibility to ESCC than G/A carriers in Southeastern Chinese males, which is largely consistent with our results. Moreover, previous study has also shown that regions around Taihang Mountain, such as Henan, Hebei, Shanxi, and north Xinjiang, which is in the north and west of China, there were significantly elevated mortality rates of ESCC.19 In addition, Cox regression analysis in our study shows that ALDH2 rs671 (G>A) is a prognostic marker of ESCC in Han and Kazak patients, and that the GA+AA genotype positively correlated with the increase risk of death when compared with GG genotype. The diagnosis of lymph node metastasis and the metastatic lymph node number and ratio are extremely significant prognostic factors in ESCC20, 21 ALDH1 positivity was found significantly related to lymph node metastasis in patients with gastric cancer.22 More importantly, Li et al.23 showed that ALDH2 expression is higher in breast cancer patients with lymph node metastasis compared with those without, suggesting ALDH2 may be a potent indicator of tumor invasion and metastasis. On the basis of our findings, we can say that the ALDH2 rs671 polymorphism may affect ALDH2 expression and thus have positive correlation with the poor prognosis of ESCC.
Our study also concluded that the frequency of AA, AG, and GG genotypes at c12orf30 rs4767364 sites in both Han and Kazak ESCC patients was not significantly different in comparison with to control group. According to a article published by Gao et al., c12orf30 rs4767364 is associated with the susceptibility to ESCC in Europeans, but such associations with ESCC risk have yet to be discovered in the Chinese population.24 Moreover, our results showed that c12orf30 rs4767364 (A>G) has a strong correlation with poor prognosis of ESCC in Kazak patients but has no influence on ESCC prognosis in Han patients, and that the AG+GG carriers exhibit a higher death risk in comparison with the AA carriers. As McKay et al.12 reports, the recessive allele of rs4767364 is observed with the increased risk of upper aerodigestive tract (UADT) cancer in a smaller series of African Americans, suggesting that this correlation is also observed in other populations. This result might support our findings that c12orf30 rs4767364 (A>G) is related to poor prognosis of ESCC in Kazak patients.
In conclusion, the ALDH2 rs671 (G>A) polymorphism at the chromosome 12q24 region is positively related to the susceptibility in developing ESCC in the Kazak and Han population The c12orf30 rs4767364 (A>G) gene polymorphism is also found to be associated with poor prognosis of ESCC in Kazak population. These findings provide precise information about the role of ALDH2 rs671 and c12orf30 rs4767364 polymorphisms in individual predisposition to ESCC in Kazak and Han populations. Although we managed to obtain 65 Kazak and 62 Han individuals to participate in our study, we still needed a larger sample size. But we hope that despite our relatively small sample size, our findings helps elucidate some of our key findings that we have presented. Further studies equipped with functional evaluations on alcohol drinking and their susceptibility to ESCC, as well as effects of the studied polymorphisms on the activation of target proteins are necessary in order to shed more light on the mechanisms of the ALDH2 rs671 and c12orf30 rs4767364 polymorphisms in the prognosis of ESCC in Kazak and Han populations. This paves a huge way for potential exploration in therapeutic intervention for ESCC patients, especially for individuals with heavy alcohol consumption.
Acknowledgments
This work was supported by Natural Science Foundation of Xinjiang Uygur Autonomous Region (No. 2015211C079) and Natural Science Foundation of the First Affiliated Hospital of Xinjiang Medical University (No. 2014ZRQN02).
Liu P, Zhao H‐R, Li F, et al. Correlations of ALDH2 rs671 and C12orf30 rs4767364 polymorphisms with increased risk and prognosis of esophageal squamous cell carcinoma in the Kazak and Han populations in Xinjiang province. J Clin Lab Anal. 2018;32:e22248 10.1002/jcla.22248
References
- 1. Cui X, Chen Y, Liu L, et al. Heterozygote of PLCE1 rs2274223 increases susceptibility to human papillomavirus infection in patients with esophageal carcinoma among the Kazakh populations. J Med Virol. 2014;86:608‐617. [DOI] [PubMed] [Google Scholar]
- 2. Rustgi AK, El‐Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371:2499‐2509. [DOI] [PubMed] [Google Scholar]
- 3. Zheng S, Vuitton L, Sheyhidin I, et al. Northwestern China: a place to learn more on oesophageal cancer. Part one: behavioural and environmental risk factors. Eur J Gastroenterol Hepatol. 2010;22:917‐925. [DOI] [PubMed] [Google Scholar]
- 4. Hu N, Wang C, Hu Y, et al. Genome‐wide association study in esophageal cancer using GeneChip mapping 10K array. Cancer Res. 2005;65:2542‐2546. [DOI] [PubMed] [Google Scholar]
- 5. Warnecke‐Eberz U, Vallbohmer D, Alakus H, et al. ERCC1 and XRCC1 gene polymorphisms predict response to neoadjuvant radiochemotherapy in esophageal cancer. J Gastrointest Surg. 2009;13:1411‐1421. [DOI] [PubMed] [Google Scholar]
- 6. Cescon DW, Bradbury PA, Asomaning K, et al. p53 Arg72Pro and MDM2 T309G polymorphisms, histology, and esophageal cancer prognosis. Clin Cancer Res. 2009;15:3103‐3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Higuchi S, Matsushita S, Masaki T, et al. Influence of genetic variations of ethanol‐metabolizing enzymes on phenotypes of alcohol‐related disorders. Ann N Y Acad Sci. 2004;1025:472‐480. [DOI] [PubMed] [Google Scholar]
- 8. Gu H, Gong D, Ding G, et al. A variant allele of ADH1B and ALDH2, is associated with the risk of esophageal cancer. Exp Ther Med. 2012;4:135‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu M, Chang SC, Kampman E, et al. Single nucleotide polymorphisms of ADH1B, ADH1C and ALDH2 genes and esophageal cancer: a population‐based case‐control study in China. Int J Cancer. 2013;132:1868‐1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Douroudis K, Kisand K, Nemvalts V, Rajasalu T, Uibo R. Allelic variants in the PHTF1‐PTPN22, C12orf30 and CD226 regions as candidate susceptibility factors for the type 1 diabetes in the Estonian population. BMC Med Genet. 2010;11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prahalad S, Hansen S, Whiting A, et al. Variants in TNFAIP3, STAT4, and C12orf30 loci associated with multiple autoimmune diseases are also associated with juvenile idiopathic arthritis. Arthritis Rheum. 2009;60:2124‐2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McKay JD, Truong T, Gaborieau V, et al. A genome‐wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet. 2011;7:e1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ajani J, D'Amico TA, Hayman JA, et al. Esophageal cancer. Clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2003;1:14‐27. [DOI] [PubMed] [Google Scholar]
- 14. Ott K, Rachakonda PS, Panzram B, et al. DNA repair gene and MTHFR gene polymorphisms as prognostic markers in locally advanced adenocarcinoma of the esophagus or stomach treated with cisplatin and 5‐fluorouracil‐based neoadjuvant chemotherapy. Ann Surg Oncol. 2011;18:2688‐2698. [DOI] [PubMed] [Google Scholar]
- 15. Bradbury PA, Zhai R, Hopkins J, et al. Matrix metalloproteinase 1, 3 and 12 polymorphisms and esophageal adenocarcinoma risk and prognosis. Carcinogenesis. 2009;30:793‐798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo YJ, Chen L, Bai YP, et al. The ALDH2 Glu504Lys polymorphism is associated with coronary artery disease in Han Chinese: relation with endothelial ADMA levels. Atherosclerosis. 2010;211:545‐550. [DOI] [PubMed] [Google Scholar]
- 17. Li QD, Li H, Wang MS, et al. Multi‐susceptibility genes associated with the risk of the development stages of esophageal squamous cell cancer in Feicheng County. BMC Gastroenterol. 2011;11:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ding JH, Li SP, Cao HX, et al. Polymorphisms of alcohol dehydrogenase‐2 and aldehyde dehydrogenase‐2 and esophageal cancer risk in Southeast Chinese males. World J Gastroenterol. 2009;15:2395‐2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Z, Feng JG, Tuersun A, et al. Proteomic identification of differentially‐expressed proteins in esophageal cancer in three ethnic groups in Xinjiang. Mol Biol Rep. 2011;38:3261‐3269. [DOI] [PubMed] [Google Scholar]
- 20. Hsu WH, Hsu PK, Hsieh CC, Huang CS, Wu YC. The metastatic lymph node number and ratio are independent prognostic factors in esophageal cancer. J Gastrointest Surg. 2009;13:1913‐1920. [DOI] [PubMed] [Google Scholar]
- 21. Tanaka T, Ishiguro H, Kuwabara Y, et al. Vascular endothelial growth factor C (VEGF‐C) in esophageal cancer correlates with lymph node metastasis and poor patient prognosis. J Exp Clin Cancer Res. 2010;29:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wakamatsu Y, Sakamoto N, Oo HZ, et al. Expression of cancer stem cell markers ALDH1, CD44 and CD133 in primary tumor and lymph node metastasis of gastric cancer. Pathol Int. 2012;62:112‐119. [DOI] [PubMed] [Google Scholar]
- 23. Li R, Zhao Z, Sun M, Luo J, Xiao Y. ALDH2 gene polymorphism in different types of cancers and its clinical significance. Life Sci. 2016;147:59‐66. [DOI] [PubMed] [Google Scholar]
- 24. Gao Y, He Y, Xu J, et al. Genetic variants at 4q21, 4q23 and 12q24 are associated with esophageal squamous cell carcinoma risk in a Chinese population. Hum Genet. 2013;132:649‐656. [DOI] [PubMed] [Google Scholar]


