Abstract
Background
To discover how NLRP3 and TNFRSF1A polymorphisms affect the efficacy of traditional medicine and etanercept for ankylosing spondylitis (AS) patients.
Methods
Single nucleotide polymorphism (SNP) and haplotype analyses were conducted based on determined NLRP3 and TNFRSF1A among AS patients. We subsequently analyzed the relationship between relevant clinical indexes and polymorphisms of NLRP3 and TNFRSF1A.
Results
The 4 SNP loci on NLRP3 and 3 SNP loci on TNFRSF1A showed significant linkage disequilibrium, respectively. The T allele of NLRP3 rs4612666 and the T allele of TFRSF1A rs4149570 are both associated with AS (P<.05). The T‐A‐C‐T haplotype of NLRP3 as well as the G‐C‐C, T‐C‐C, T‐C‐T, and T‐T‐T haplotypes of TFRSF1A are associated with AS (P<.05). The morning stiffness time, BASDAI scoring, and ESR of patients receiving etanercept were significantly higher than those receiving traditional medicine. T allele of NLRP3 rs4612666 had a significantly greater negative impact on the ASAS20 improvement than C allele. Whereas the A allele of NLRP3 rs3806268 had a significantly greater positive impact on the ASAS20 improvement than G allele. There is no significant association between SNP and efficacy of traditional medicine in the treatment of AS.
Conclusion
NLRP3 and TFRSF1A (rs4149570) are associated with AS susceptibility. There is a significant association between NLRP3 polymorphisms and treatment of etanercept.
Keywords: ankylosing spondylitis, etanercept, NLRP3, single nucleotide polymorphism, TNFRSF1A
1. Introduction
Ankylosing spondylitis (AS) is a chronic inflammatory arthritic condition that mainly affects the axial skeleton, peripheral joints and extra‐articular manifestations (e.g., uveitis, psoriasis, inflammatory bowel disease, and cardiomyopathy).1, 2 The disease usually begins in the third decade of life and the ratio of male to female prevalence is 2:1.3 In light of population surveys, the overall estimated AS prevalence rate is about 0.24% in Europe, 0.17% in Asia, 0.32% in North America, 0.10% in Latin America, and 0.07% in Africa.4 The pathogenesis of AS is not completely understood. Evidence suggests that genetic factors, such as HLA B27, have a strong correlation to AS.3 Although many studies have proved that the histocompatibility antigen, human leukocyte antigen B27 (HLA‐B27), is associated with the incidence of AS, HLA‐B27 only comprised one‐third of the genetic components.5
Over the last decade, improvements in single nucleotide polymorphisms (SNPs) high‐throughput genotyping and dissection of the true polygenic nature of AS have been very rapid. To date, more than 40 genetic variants and over 36 genetic loci have been identified to be associated with AS.6 It is widely recognized that AS is a chronic inflammatory disease and that inflammasomes of NLR family pyrin domain containing 3 (NLRP3) play a crucial role in inflammatory diseases.7 Recently, Zhang et al.8 demonstrated that NLRP3 polymorphism is associated with increased susceptibility to rheumatoid arthritis in humans. Furthermore, Sode et al.9 reported that patients with NLRP3 (rs10754558) variant allele carriers and rheumatoid arthritis are more likely to have a negative response to TNF inhibitor treatment. The TNFRSF1A encodes tumor necrosis factor receptor 1 (TNF‐R1) and mutations in the gene can cause autoinflammatory disorders.10 It has been published that the TNFRSF1A variant is associated with AS risk (with an odds ratio of 1.1) and that TNF inhibitors are a highly effective method of treatment.11
Non‐steroidal anti‐inflammatory drugs (NSAIDs) and non‐biological disease‐modifying antirheumatic drugs (e.g., sulfasalazine) are mainstream pharmacologic therapies for AS.12, 13 However, many patients with AS have ongoing symptoms and they tend to develop deformities despite the use of NSAID or DMARDs. Since 2000, the use of TNF inhibitors (e.g., etanercept, infliximab, golimumab, and adalimumab) has shown rapid and sustained reductions in all clinical and laboratory measures of disease activity.14, 15 The use of TNF inhibitors is strongly recommended for patients whose clinical symptoms are not controlled by NSAIDs or DMARDs therapy, or for those whom cannot accept the adverse effects of NSAIDs or DMARDs. These agents remarkably improve both objective and subjective indicators of disease activities and functions. This includes spinal mobility, spinal stiffness, partial remission (defined as a value of two or less on a scale from 0 to 10 in each of the four domains of the ASAS 20), the bath Ankylosing Spondylitis Disease Activity Index (BASDAI), the bath ankylosing spondylitis function index (BASFI), X‐rays of the spine and the level of erythrocyte sedimentation rate (ESR) and C‐reactive protein (CRP).16
In this study, we chose etanercept as the TNF inhibitor therapy for its generally recognized efficiency in AS. Few studies have investigated the association of NLRP3 or TNFRSF1A with AS risk, or effects of NLRP3 or TNFRSF1A on the treatment efficiency of etanercept or sulfasalazine. Therefore, there are three primary aims of this study: (1) to determine the correlation between AS and the polymorphisms or haplotypes of NLRP3 and TNFRSF1A; (2) to compare the efficiency and safety of etanercept and sulfasalazine used in patients with AS; and (3) to investigate whether the polymorphisms or haplotypes of NLRP3 and TNFRSF1A affect the efficacy of etanercept or sulfasalazine. All in all, we aim to further clarify the function of genetic factors in AS and to promote the development of effective diagnosis or therapeutic strategies for AS.
2. Material and Methods
2.1. Patients and study design
A total of 200 patients with AS (155 males and 45 females) were recruited from the Department of Rheumatism and Immunology in our hospital from Jan 2014 to Jan 2017. According to the random number table, patients were divided into an etanercept treatment group (78 males and 22 females) averagely aged 46.3±6.8 years old (mean course of disease 23.9±6.8 years), and a traditional drug treatment group (77 males and 23 females) averagely aged 45.4±6.7 years old (mean course of disease 23.0±6.7 years) (Table S1). There was no significant difference in gender, age and course of disease between the experimental group and the control group (P>.05). Another group of 200 normal healthy individuals was collected over the same time period to be the healthy control group. This group will also participate in physical examinations and was made up of 104 males and 96 females with an average age of 45.7±8.3 years old.
2.2. Inclusion and exclusion criteria
Inclusion criteria: (1) Diagnosed with AS according to the revised standard of New York in 1984; (2) Not being treated with corticosteroids or other immunosuppressive drugs; (3) Have normal hepatic and renal function; (4) Participants are not allergic to antibiotics such as sulfanilamide etc.; (5) Signed informed consent.
Exclusion criteria: (1) Have cardiac insufficiency, tuberculosis infection, active HBV or other acute infectious diseases; (2) Have peptic ulcer, chronic nephritis, diabetes, chronic obstructive pneumonia, or other chronic diseases.
2.3. Treatment
All the patients accepted the same non‐steroidal anti‐inflammatory drugs (NSAIDs) as a basic treatment. They were allowed to eat 200 mg celecoxib capsules or 7.5 mg meloxicam tablets twice a day. Patients in the traditional group underwent the basic treatment and pyridine nitrogen treatment (0.75 g, three times a day, oral). The NSAIDs were discontinued after 4 weeks of disease control while the pyridine nitrogen tablets remained in use for a total of 3 months. The etanercept group was treated with the basic and etanercept treatment (25 mg, twice a week, subcutaneous injection in the upper arm). The NSAIDs were discontinued after 4 weeks of disease control while etanercept was continued and used for a total of 3 months.
2.4. Genotyping
DNA samples from the control and AS group were isolated using the Oragene™ DNA Self‐Collection kit (DNA Genotek Inc., Ottawa, Canada). Real‐time fluorescent quantitative PCR (FQ‐PCR) was used to detect the SNP polymorphisms of NLRP3 and TNFRSF1A. About 5 mL of blood was collected from each person and mixed with 0.4 mL 15 g/L ethylenediaminetetraacetic acid‐Na2 (EDTA‐Na2) for anticoagulation. Samples were digested using proteinase K, maintained at −80°C and DNA was extracted in accordance to the QIAamp DNAKit (Qiagen, Hilden, Germany). In the NCBI SNP database, the base sequences of NLRP3 (rs4612666, rs3806268, rs10925019, rs3806265) and TNFRSF1A (rs4149570, rs767455, rs4149621, rs4149569) were found and intercepted for the DNA sequence containing SNP. We used Primer 3.0 and PrimerExpress1.5 to design primers. The length of the primer was approximately 40 bp and the ratio of GC in the primer was 50–55%. The T m value was roughly 60°C. PCR primer information is shown in Table 1. PCR primers and probes technology was used by Genomics for genotyping. Specifically, real‐time quantitative polymerase chain reaction (RT‐PCR) was conducted to determine genotypes. The reaction system contained 1–5 ng of dried genomic DNA, 2.5 μL Taqman universal PCR master mix (2×), 0.25 μL 40× SNP genotyping assay, 1.25 μL ddH2O and 1 μL DNA template. All the amplification process was performed on ABI 7900 real‐time PCR amplification instrument. The reaction condition was mainly summarized as 40 cycles of 95°C for 10 minutes, 92°C for 15 seconds, and 60°C for 1 minute. Finally, the results were determined strictly according to instructions of TaqMan® Universal PCR Master Mix kit.17
Table 1.
Primer sequences of NLRP3 and TNFRSF1A
| Gene | SNP | Primer |
|---|---|---|
| NLRP3 | rs4612666 | F: 5′‐TGCTTAAGGCCATTAATTGTG‐3′ |
| R: 5′‐CTCCACCATGGACAAGGAAG‐3′ | ||
| rs3806268 | F: 5′‐GGATTGGGAAAACAATCCTGGC‐3′ | |
| R: 5′‐CTGTCTTGGTAGAGTGTCCCC‐3′ | ||
| rs10925019 | F: 5′‐GGAGACTGGTTGTTTGGGACA‐3′ | |
| R: 5′‐TGGCAGTGGGGAGAGAATTT‐3′ | ||
| rs3806265 | F: 5′‐GGACAGTGGGAACACATGCT‐3′ | |
| R: 5′‐GGGAGCATTTCTGCACTCCTA‐3′ | ||
| TNFRSF1A | rs4149570 | F: 5′‐TCTCAGACACATAACTGAAACTGT‐3′ |
| R: 5′‐CCAGGAGACAGGTTATCTCCAC‐3′ | ||
| rs767455 | F: 5′‐TAGCTGTCTGGCATGGGCCTCT‐3′ | |
| R: 5′‐CCTACTCCAAAAGGCGGATGAA‐3′ | ||
| rs4149569 | F: 5′‐TCTCTCATAGCCAAAGGGGC‐3′ | |
| R: 5′‐TCCAGAAACCCAATGTGCCA‐3′ | ||
| rs4149621 | F: 5′‐TTTTAGCTAAGAATGTGTCTTGGAC‐3′ | |
| R: 5′‐TTGGAAAACAGATCCAGACAGT‐3′ |
F, Forward primer; R, Reverse primer; SNP, single nucleotide polymorphism.
2.5. Evaluating indicators
In order to analyze the different effects of etanercept and traditional medicine in the treatment of AS, a comprehensive evaluation of the clinical indicators was measured before and after treatment. The clinical indicators included were: morning waist numb time, visual analog scale (VAS), BASDAI, BASFI, ESR, and CRP. In order to analyze different genotypes in patients with different efficacies of etanercept and traditional therapy, the rate of ASAS20 improvement was calculated after 12 weeks of treatment. The ASAS20 improvement criteria18 were as follows: (1) a >20% improvement and absolute increase of ≥1 unit of VAS, BASFI, or BASDAI scores in at least three of the four indicators overall evaluation of patient; and (2) no aggravation in the indicator that did not achieve 20% improvement.
2.6. Statistical analysis
Statistical analysis of the linkage disequilibrium between different loci of NLRP3 and TNFR1A genotypes was performed using HaploView. SPSS21.0 was used to perform the corresponding data analysis. Measurement data were displayed as mean±standard deviation (SD). The comparison between the two groups was completed using t test and the paired t test was used for data with a normal distribution. If the data were not distributed normally, the rank sum test (Mann‐Whitney assay) was applied. Single factor analysis of variance (ANOVA One‐way) or the non‐parametric Kruskal‐Wallis test was used to compare the measurement data between groups. Comparison of counting data was performed by using the Chi‐square test. Logistic regression analysis was used to analyze the related factors that influence occurrence of the disease. P<.05 was considered statistically significance.
3. Results
3.1. Linkage disequilibrium analysis
The r 2, SNP of NLRP3, and TNFRSF1A were analyzed using HaploView software. The results of the linkage disequilibrium analysis are indicated in the r 2 black‐white graph. Black blocks represent high linkage disequilibrium (r 2=.8‐1) and white blocks represent low linkage disequilibrium. As shown in Figure 1A, there is a linkage disequilibrium among the 4 SNPs of NLRP3 (rs4612666 ‐ rs3806265 ‐ rs3806268 ‐ rs10925019). Figure 1B also shows a linkage disequilibrium among the 3 SNPs of TNFRSF1A (rs4149570 ‐ rs767455 ‐ rs4149569). However, no linkage disequilibrium is seen between TNFRSF1A (rs4149621) and any other SNP (Figure 1B).
Figure 1.
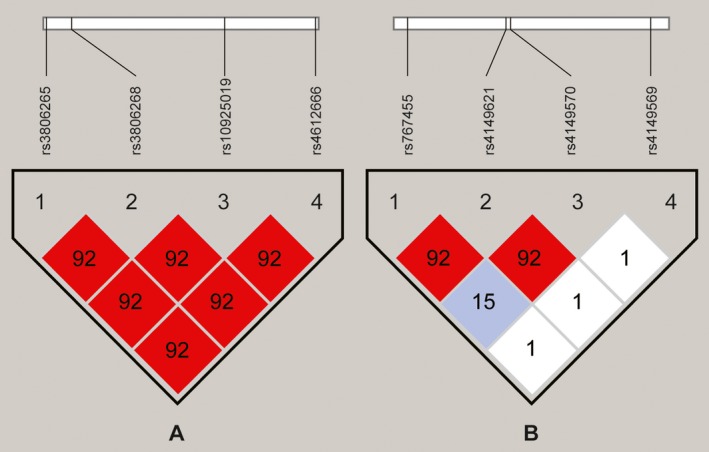
(A) Linkage disequilibrium analysis of NLRP3 (rs4612666 ‐ rs3806265 ‐ rs3806268 ‐ rs10925019). (B) Linkage disequilibrium analysis of TNFRSF1A (rs4149570 ‐ rs767455 ‐ rs4149569 ‐ rs4149621)
3.2. Association of NLRP3 and TFRSF1A polymorphisms with AS susceptibility
The loci of NLRP3 and TFRSF1A SNPs are the independent variables and the contraction of AS is the dependent variable. Logistic regression analysis was performed and the results are shown in Table 2. The T allele of NLRP3 (rs4612666) is associated with AS (P<.05) and T allele of TFRSF1A (rs4149570) is associated with AS (P<.05).
Table 2.
Association of gene polymorphisms of NLRP3 and TFRSF1A with susceptibility to AS
| SNP | Mutant | Population | Genotype (n) | HWE | Dominant model | Allele frequency | Allelic model | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 22 | P value | OR (95% CI) | P value | 1 | 2 | MAF | OR (95% CI) | P value | |||
| NLRP3 | |||||||||||||
| rs4612666 | C>T | Case (n=100) | 54 | 100 | 46 | .982 | 1.622 (1.062‐2.478) | .025 | 208 | 192 | 0.48 | 1.459 (1.102‐1.933) | .008a |
| Control (n=100) | 75 | 95 | 30 | .993 | Ref. | 245 | 155 | 0.63 | Ref. | ||||
| rs3806268 | A>G | Case (n=100) | 100 | 83 | 17 | .970 | 0.961 (0.649‐1.422) | .842 | 283 | 117 | 0.29 | 0.965 (0.712‐1.307) | .816 |
| Control (n=100) | 98 | 84 | 18 | .867 | Ref. | 280 | 120 | 0.30 | Ref. | ||||
| rs10925019 | C>T | Case (n=100) | 118 | 70 | 12 | .707 | 1.087 (0.728‐1.622) | .683 | 306 | 94 | 0.24 | 1.105 (0.793‐1.540) | .554 |
| Control (n=100) | 122 | 69 | 9 | .848 | Ref. | 313 | 87 | 0.22 | Ref. | ||||
| rs3806265 | C>T | Case (n=100) | 86 | 91 | 23 | .885 | 0.960 (0.646‐1.427) | .840 | 263 | 137 | 0.34 | 0.989 (0.739‐1.324) | .941 |
| Control (n=100) | 84 | 94 | 22 | .572 | Ref. | 262 | 138 | 0.35 | Ref. | ||||
| TNFRSF1A | |||||||||||||
| rs4149570 | G>T | Case (n=100) | 58 | 99 | 43 | .950 | 1.632 (1.076‐2.475) | .021 | 215 | 185 | 0.46 | 1.481 (1.116‐1.965) | .006a |
| Control (n=100) | 80 | 93 | 27 | .997 | Ref. | 253 | 147 | 0.37 | Ref. | ||||
| rs767455 | C>T | Case (n=100) | 85 | 91 | 24 | .962 | 0.960 (0.645‐1.428) | .839 | 261 | 139 | 0.35 | 0.963 (0.720‐1.289) | .800 |
| Control (n=100) | 83 | 89 | 26 | .781 | Ref. | 255 | 141 | 0.35 | Ref.1413 | ||||
| rs4149569 | C>T | Case (n=100) | 53 | 101 | 46 | .874 | 1.217 (0.788‐1.881) | .376 | 207 | 193 | 0.48 | 1.140 (0.863‐1.505) | .360 |
| Control (n=100) | 61 | 98 | 41 | .912 | Ref. | 220 | 180 | 0.45 | Ref. | ||||
| rs4149621 | A>G | Case (n=100) | 62 | 99 | 39 | .963 | 1.000 (0.655‐1.528) | 1.000 | 223 | 177 | 0.44 | 1.010 (0.764‐1.336) | .943 |
| Control (n=100) | 62 | 100 | 38 | .836 | Ref. | 224 | 176 | 0.44 | Ref. | ||||
SNP, single nucleotide polymorphism; 11, wild homozygote; 12, heterozygote; 22, mutant homozygote; HWE, Hardy‐Weinberg equilibrium; Dominant model: (22+12)/11; Allelic model: 2/1; OR, odds ratio; MAF, minor allele frequency; CI, confidence interval; P value.
Significant differences.
3.3. Association of NLRP3 haplotypes with AS susceptibility
The NLRP3 haplotypes are the independent variables and the contraction of AS is the dependent variable. Logistic regression analysis was performed. The results are shown in Table 3. The T‐A‐C‐T haplotype of NLRP3 (rs4612666, rs3806268, rs10925019, rs3806265) is associated with AS (P<.05). Other haplotypes did not show significant differences.
Table 3.
Association of NLRP3 haplotypes with susceptibility to AS
| Haplotypes | AS (n=400) | Control (n=386) | AOR (95% CI) | P value |
|---|---|---|---|---|
| C‐A‐C‐C | 101 | 119 | Ref | ‐ |
| C‐A‐C‐T | 30 | 38 | 1.081 (0.562‐2.079) | .816 |
| C‐A‐T‐C | 25 | 21 | 1.653 (0.795‐3.434) | .178 |
| C‐A‐T‐T | 9 | 7 | 1.795 (0.522‐6.170) | .353 |
| C‐G‐C‐C | 28 | 27 | 1.308 (0.622‐2.751) | .478 |
| C‐G‐C‐T | 10 | 10 | 0.884 (0.250‐3.131) | .849 |
| C‐G‐T‐C | 3 | 5 | 0.272 (0.041‐1.794) | .176 |
| C‐G‐T‐T | 4 | 8 | 2.461 (0.912‐6.641) | .075 |
| T‐A‐C‐C | 46 | 35 | 0.802 (0.400‐1.607) | .534 |
| T‐A‐C‐T | 38 | 25 | 2.156 (1.125‐4.129) | .021a |
| T‐A‐T‐C | 20 | 14 | 1.877 (0.782‐4.509) | .159 |
| T‐A‐T‐T | 14 | 15 | 1.232 (0.476‐3.189) | .667 |
| T‐G‐C‐C | 28 | 24 | 1.779 (0.876‐3.615) | .111 |
| T‐G‐C‐T | 25 | 22 | 1.548 (0.728‐3.290) | .256 |
| T‐G‐T‐C | 12 | 9 | 1.001 (0.330‐3.036) | .999 |
| T‐G‐T‐T | 7 | 7 | 1.023 (0.257‐4.062) | .975 |
AS, ankylosing spondylitis; Ref, Reference level; AOR, adjusted odds ratio after age and gender correction; CI, confidence interval; P value.
Significant differences.
3.4. Association of TFRSF1Ahaplotypes with AS susceptibility
The TFRSF1A haplotypes are the independent variables and the contraction of AS is the dependent variable. Logistic regression analysis was performed. The results are shown in Table 4. The G‐C‐C, T‐C‐C, T‐C‐T, and T‐T‐T haplotypes of TFRSF1A (rs4149570, rs767455, rs4149569) are associated with AS (P<.05). Other haplotypes did not show significant differences.
Table 4.
Association of TFRSF1A haplotypes with susceptibility to AS
| Haplotypes | AS (n=400) | Control (n=396) | AOR (95% CI) | P value |
|---|---|---|---|---|
| G‐C‐C | 99 | 138 | Ref | ‐ |
| G‐C‐T | 65 | 45 | 2.284 (1.319‐3.957) | .003a |
| G‐T‐C | 30 | 34 | 1.334 (0.665‐2.677) | .417 |
| G‐T‐T | 21 | 33 | 1.106 (0.540‐2.267) | .782 |
| T‐C‐C | 43 | 32 | 2.287 (1.225‐4.271) | .009a |
| T‐C‐T | 54 | 40 | 2.243 (1.258‐3.999) | .006a |
| T‐T‐C | 35 | 31 | 1.878 (0.957‐3.683) | .067 |
| T‐T‐T | 53 | 43 | 1.918 (1.075‐3.425) | .028a |
AS, ankylosing spondylitis; Ref, Reference level; AOR, adjusted odds ratio after age and gender correction; CI, confidence interval; P value.
Significant differences.
3.5. Comparison of etanercept and traditional medicine
As shown in Figure 2 and Table 5, the clinical indicators (morning stiffness time, VAS, BASFI, BASDAI, ESR, and CRP) of both the etanercept and traditional medicine groups changed after 12 weeks of treatment (P<.05). Furthermore, changes of morning stiffness time, BASDAI and ESR in the etanercept group were substantially better than that of the traditional medicine group (Table 5) (P<.05). In regards to VAS, BASFI and CRP, there is no statistical significance (P>.05) between two groups.
Figure 2.
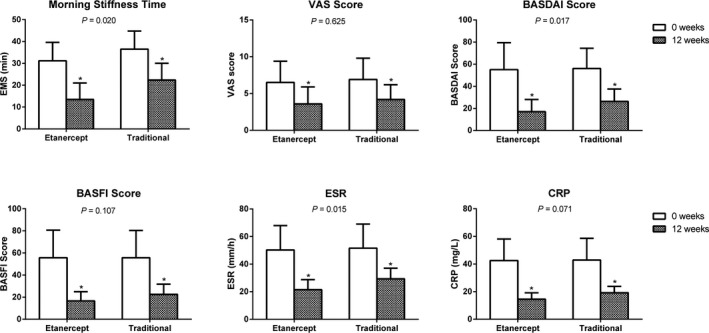
A comparison of the six clinical parameters used in the etanercept and traditional medicine groups. *Compared with previous treatment, the clinical indexes after treatments were statistically significant. P<.05 indicates the greater improvements in the etanercept group over the traditional medicine group are statistically significant
Table 5.
The situation of six clinical parameters of etanercept group and traditional medicine group
| Group (n=100) | Time | Morning stiffness time (minutes) | VAS | BASDAI | BASFI | ESR (mm/h) | CRP (mg/L) |
|---|---|---|---|---|---|---|---|
| Etanercept | 0 weeks | 31.2±8.4 | 6.5±2.9 | 55.08±24.4 | 55.7±24.9 | 50.2±17.8 | 42.5±15.6 |
| 12 weeks | 13.5±7.5a | 3.6±2.3a | 17.0±11.0a | 16.5±8.4a | 21.4±7.3a | 14.5±4.6a | |
| Difference | 17.7±10.7b | 2.9±3.6 | 38.1±26.5b | 39.1±26.3 | 28.8±18.8b | 28.0±17.1 | |
| Traditional Medicine | 0 week | 36.5±8.3 | 6.9±2.9 | 56±18.44 | 55.6±24.7 | 51.5±17.5 | 42.9±15.8 |
| 12 weeks | 22.4±7.6a | 4.2±2.0a | 26.3±11.2a | 22.6±9.2a | 29.4±7.7a | 19.2±4.6a | |
| Difference | 14.1±11.8b | 2.7±3.4 | 29.7±22.6b | 33.1±26.3 | 22.1±19.5b | 23.7±16.6 |
VAS, visual analog scale; BASDAI, bath ankylosing spondylitis disease activity index; BASFI, bath ankylosing spondylitis function index; ESR, erythrocyte sedimentation rate; CRP, C‐reactive protein.
Comparison between pre‐ and pro‐ treatmentshowed significant difference (P<.05).
Comparison between etanercept group and traditional medicine group showed significant difference (P<.05).
3.6. Association of NLRP3 and TFRSF1A polymorphisms with the efficacy of etanercept or tradition treatment for AS
After 12 weeks of etanercept or traditional treatments, ASAS20 was performed to detect the association between the SNPs and both treatments. Results are shown in Table 6. The T allele of NLRP3 (rs4612666) had a greater negative impact on the rate of ASAS20 improvement than C allele (P<.05). The G allele of NLRP3 (rs3806268) had a positive impact on the rate of ASAS20 improvement (P<.05) (Table 6). As shown in Table 7, there is no statistically significant association between SNPs and traditional drug therapy for AS (P<.05).
Table 6.
Association of gene polymorphisms of NLRP3 and TFRSF1A with the efficacy of etanercept treatment for AS
| Gene | SNP | Sample size (n) | ASAS20 effects | AOR (95% CI) | P value | ||
|---|---|---|---|---|---|---|---|
| Yes (n) | No (n) | ||||||
| NLRP3 | rs4612666 | ||||||
| Genotype | CC | 26 | 22 | 4 | Ref. | ||
| CT | 47 | 33 | 14 | 0.429 (0.125‐1.474) | .172 | ||
| TT | 27 | 14 | 13 | 0.196 (0.053‐0.723) | .011 | ||
| Allele | C | 99 | 77 | 22 | Ref. | ||
| T | 101 | 61 | 40 | 0.436 (0.235‐0.810) | .008 | ||
| rs3806268 | |||||||
| Genotype | AA | 61 | 39 | 22 | Ref. | ||
| AG | 23 | 16 | 7 | 1.289 (0.460‐3.614) | .628 | ||
| GG | 16 | 14 | 2 | 3.949 (0.820‐19.010) | .070 | ||
| Allele | A | 145 | 94 | 51 | Ref. | ||
| G | 55 | 44 | 11 | 2.170 (1.032‐4.565) | .038 | ||
| rs10925019 | |||||||
| Genotype | CC | 62 | 40 | 22 | Ref. | ||
| CT | 28 | 21 | 7 | 0.606 (0.223‐1.650) | .325 | ||
| TT | 10 | 8 | 2 | 0.455 (0.089‐2.331) | .335 | ||
| Allele | C | 152 | 101 | 51 | Ref. | ||
| T | 48 | 37 | 11 | 1.698 (0.800‐3.606) | .165 | ||
| rs3806265 | |||||||
| Genotype | CC | 54 | 36 | 18 | Ref. | ||
| CT | 27 | 20 | 7 | 1.429 (0.510‐4.003) | .496 | ||
| TT | 19 | 13 | 6 | 1.083 (0.353‐3.323) | .889 | ||
| Allele | C | 135 | 92 | 43 | Ref. | ||
| T | 65 | 46 | 19 | 1.132 (0.593‐2.158) | .707 | ||
| TNFRSF1A | rs4149570 | ||||||
| Genotype | GG | 34 | 27 | 7 | Ref. | ||
| GT | 43 | 28 | 15 | 0.484 (0.171‐1.371) | .168 | ||
| TT | 23 | 14 | 9 | 0.403 (0.124‐1.313) | .126 | ||
| Allele | G | 111 | 82 | 29 | Ref. | ||
| T | 89 | 56 | 33 | 0.600 (0.328‐1.098) | .096 | ||
| rs767455 | |||||||
| Genotype | CC | 50 | 36 | 14 | Ref. | ||
| CT | 35 | 25 | 10 | 0.972 (0.373‐2.536) | .954 | ||
| TT | 15 | 8 | 7 | 0.444 (0.136‐1.458) | .175 | ||
| Allele | C | 135 | 97 | 38 | Ref. | ||
| T | 65 | 41 | 24 | 0.669 (0.357‐1.254) | .209 | ||
| rs4149569 | |||||||
| Genotype | CC | 32 | 23 | 9 | Ref. | ||
| CT | 45 | 28 | 17 | 0.645 (0.242‐1.715) | .377 | ||
| TT | 23 | 18 | 5 | 1.409 (0.401‐4.944) | .592 | ||
| Allele | C | 109 | 74 | 35 | Ref. | ||
| T | 91 | 64 | 27 | 1.121 (0.613‐2.050) | .710 | ||
| rs4149621 | |||||||
| Genotype | AA | 34 | 26 | 8 | Ref. | ||
| AG | 43 | 26 | 17 | 0.471 (0.173‐1.281) | .136 | ||
| GG | 23 | 17 | 6 | 0.872 (0.257‐2.961) | .826 | ||
| Allele | A | 111 | 78 | 33 | Ref. | ||
| G | 89 | 60 | 29 | 0.875 (0.480‐1.598) | .664 | ||
AS, ankylosing spondylitis; SNP, single nucleotide polymorphism; AOR, adjusted odds ratio; CI, confidence interval; ASAS20, ankylosing spondylitis assessment study 20; Ref., reference.
Table 7.
Association of gene polymorphisms of NLRP3 and TFRSF1A with the efficacy of traditional treatment for AS
| Gene | SNP | Sample size (n) | ASAS20 effects | AOR (95% CI) | P value | ||
|---|---|---|---|---|---|---|---|
| Yes (n) | No (n) | ||||||
| NLRP3 | rs4612666 | ||||||
| Genotype | CC | 27 | 18 | 9 | Ref. | ||
| CT | 51 | 33 | 18 | 0.917 (0.342‐2.455) | .863 | ||
| TT | 22 | 14 | 8 | 0.875 (0.269‐2.851) | .825 | ||
| Allele | C | 105 | 69 | 36 | Ref. | ||
| T | 95 | 61 | 34 | 0.936 (0.523‐1.675) | .824 | ||
| rs3806268 | |||||||
| Genotype | AA | 52 | 34 | 18 | Ref. | ||
| AG | 34 | 21 | 13 | 0.855 (0.349‐2.098) | .733 | ||
| GG | 14 | 10 | 4 | 1.324 (0.363‐4.822) | .670 | ||
| Allele | A | 138 | 89 | 49 | Ref. | ||
| G | 62 | 41 | 21 | 1.075 (0.572‐2.021) | .823 | ||
| rs10925019 | |||||||
| Genotype | CC | 66 | 45 | 21 | Ref. | ||
| CT | 22 | 14 | 8 | 0.817 (0.297‐2.246) | .695 | ||
| TT | 12 | 6 | 6 | 0.467 (0.134‐1.620) | .223 | ||
| Allele | C | 154 | 104 | 50 | Ref. | ||
| T | 46 | 26 | 20 | 0.625 (0.319‐1.226) | .170 | ||
| rs3806265 | |||||||
| Genotype | CC | 45 | 25 | 20 | Ref. | ||
| CT | 38 | 30 | 8 | 3.000 (1.129‐7.969) | .025 | ||
| TT | 17 | 10 | 7 | 1.143 (0.369‐3.542) | .817 | ||
| Allele | C | 128 | 80 | 48 | Ref. | ||
| T | 72 | 50 | 22 | 1.364 (0.736‐2.525) | .323 | ||
| TNFRSF1A | rs4149570 | ||||||
| Genotype | GG | 29 | 20 | 9 | Ref. | ||
| GT | 46 | 29 | 17 | 0.768 (0.286‐2.064) | .600 | ||
| TT | 25 | 16 | 9 | 0.800 (0.257‐2.487) | .700 | ||
| Allele | G | 104 | 69 | 35 | Ref. | ||
| T | 96 | 61 | 35 | 0.884 (0.494‐1.582) | .678 | ||
| rs767455 | |||||||
| Genotype | CC | 42 | 28 | 14 | Ref. | ||
| CT | 42 | 26 | 16 | 0.813 (0.332‐1.987) | .649 | ||
| TT | 16 | 11 | 5 | 1.100 (0.319‐3.789) | .880 | ||
| Allele | C | 124 | 82 | 42 | Ref. | ||
| T | 76 | 48 | 28 | 0.878 (0.484‐1.594) | .669 | ||
| rs4149569 | |||||||
| Genotype | CC | 28 | 20 | 8 | Ref. | ||
| CT | 42 | 27 | 15 | 0.720 (0.256‐2.027) | .533 | ||
| TT | 30 | 18 | 12 | 0.600 (0.200‐1.800) | .360 | ||
| Allele | C | 98 | 67 | 31 | Ref. | ||
| T | 102 | 63 | 39 | 0.747 (0.417‐1.340) | .328 | ||
| rs4149621 | |||||||
| Genotype | AA | 35 | 24 | 11 | Ref. | ||
| AG | 42 | 29 | 13 | 1.022 (0.388‐2.693) | .964 | ||
| GG | 23 | 12 | 11 | 0.500 (0.169‐1.481) | .208 | ||
| Allele | A | 112 | 77 | 35 | Ref. | ||
| G | 88 | 53 | 35 | 0.688 (0.384‐1.235) | .210 | ||
AS, ankylosing spondylitis; SNP, single nucleotide polymorphism; AOR, adjusted odds ratio; CI, confidence interval; ASAS20, ankylosing spondylitis assessment study 20; Ref., reference.
3.7. Association of NLRP3 and TFRSF1A haplotypes with efficacy of etanercept or traditional drug treatment for AS
As shown in Table 8, there is no statistically significant association between NLRP3 haplotypes and etanercept treatment (P>.05). Table 9 shows that there is also no statistically significant association between TFRSF1A haplotypes and etanercept treatment for AS (P>.05). Tables 10 and 11 indicate that there is no statistically significant association between NLRP3 or TFRSF1A haplotypes and traditional treatment for AS (P>.05).
Table 8.
Association between NLRP3 haplotypes and etanercept treatment for AS
| Haplotype | ASAS20 effects/total | AOR (95% CI) | P value |
|---|---|---|---|
| C‐A‐C‐C | 35/50 | Ref | ‐ |
| C‐A‐C‐T | 9/12. | 1.29 (0.301‐5.525) | .731 |
| C‐A‐T‐C | 9/11 | 2.198 (0.417‐11.596) | .353 |
| C‐A‐T‐T | 6/6/ | ‐ | .999 |
| C‐G‐C‐C | 14/16 | 2.667 (0.53‐13.42) | .234 |
| C‐G‐C‐T | 3/4 | 1.011 (0.093‐10.991) | .993 |
| C‐G‐T‐C | 1/2 | 0.279 (0.016‐4.963) | .385 |
| C‐G‐T‐T | 2/2 | ‐ | .999 |
| T‐A‐C‐C | 14/27 | 0.396 (0.145‐1.079) | .070 |
| T‐A‐C‐T | 10/22 | 0.373 (0.129‐1.08) | .069 |
| T‐A‐T‐C | 5/10 | 0.391 (0.096‐1.599) | .191 |
| T‐A‐T‐T | 6/7 | 2.536 (0.274‐23.512) | .413 |
| T‐G‐C‐C | 9/12 | 1.206 (0.281‐5.175) | .801 |
| T‐G‐C‐T | 7/9 | 1.647 (0.3‐9.056) | .566 |
| T‐G‐T‐C | 5/7 | 0.947 (0.16‐5.596) | .952 |
| T‐G‐T‐T | 3/3 | ‐ | .999 |
AS, ankylosing spondylitis; Ref, Reference level; AOR, adjusted odds ratio after age and gender correction; CI, confidence interval; P value.
Table 9.
Association between TFRSF1A haplotypes and etanercept treatment for AS
| Haplotype | ASAS20effects/total | AOR (95% CI) | P value |
|---|---|---|---|
| G‐C‐C | 38/52 | Ref | ‐ |
| G‐C‐T | 23/29 | 1.469 (0.482‐4.483) | .499 |
| G‐T‐C | 12/19 | 0.656 (0.211‐2.036) | .465 |
| G‐T‐T | 9/11 | 1.634 (0.310‐8.625) | .563 |
| T‐C‐C | 15/24 | 0.681 (0.238‐1.943) | .472 |
| T‐C‐T | 21/30 | 0.930 (0.339‐2.545) | .887 |
| T‐T‐C | 9/14 | 0.587 (0.164‐2.110) | .415 |
| T‐T‐T | 11/21 | 0.466 (0.160‐1.361) | .163 |
AS, ankylosing spondylitis; Ref, Reference level; AOR, adjusted odds ratio after age and gender correction; CI, confidence interval; P value.
Table 10.
Association between NLRP3 haplotypes and tradition treatment for AS
| Haplotype | ASAS20effects/total | AOR (95% CI) | P value |
|---|---|---|---|
| C‐A‐C‐C | 34/51 | Ref | ‐ |
| C‐A‐C‐T | 12/18 | 0.911 (0.287‐2.890) | .874 |
| C‐A‐T‐C | 8/14 | 0.601 (0.176‐2.047) | .415 |
| C‐A‐T‐T | 1/3 | 0.215 (0.018‐2.632) | .229 |
| C‐G‐C‐C | 9/12/ | 1.394 (0.330‐5.896) | .652 |
| C‐G‐C‐T | 5/6 | 2.445 (0.259‐23.056) | .435 |
| C‐G‐T‐C | 1/1 | ‐ | 1 |
| C‐G‐T‐T | 1/2 | 0.521 (0.030‐8.993) | .653 |
| T‐A‐C‐C | 10/19 | 0.537 (0.182‐1.585) | .26 |
| T‐A‐C‐T | 12/16 | 1.437 (0.400‐5.161) | .578 |
| T‐A‐T‐C | 7/10 | 1.109 (0.252‐4.874) | .891 |
| T‐A‐T‐T | 5/7 | 1.054 (0.179‐6.213) | .953 |
| T‐G‐C‐C | 10/16 | 0.819 (0.253‐2.649) | .739 |
| T‐G‐C‐T | 12/16 | 1.485 (0.414‐5.325) | .544 |
| T‐G‐T‐C | 1/5 | 0.103 (0.010‐1.034) | .053 |
| T‐G‐T‐T | 2/4 | 0.523 (0.067‐4.069) | .536 |
AS, ankylosing spondylitis; Ref, Reference level; AOR, adjusted odds ratio after age and gender correction; CI, confidence interval; P value.
Table 11.
Association between TFRSF1A haplotypes and tradition treatment for AS
| Haplotype | ASAS20 effective/total | AOR (95% CI) | P value |
|---|---|---|---|
| G‐C‐C | 33/47 | Ref | ‐ |
| G‐C‐T | 24/36 | 0.868 (0.341‐2.214) | .768 |
| G‐T‐C | 5/11 | 0.320 (0.081‐1.263) | .104 |
| G‐T‐T | 7/10 | 0.994 (0.217‐4.55) | .994 |
| T‐C‐C | 12/19 | 0.680 (0.219‐2.111) | .504 |
| T‐C‐T | 13/24 | 0.468 (0.167‐1.311) | .149 |
| T‐T‐C | 17/21 | 1.670 (0.470‐5.932) | .428 |
| T‐T‐T | 19/32 | 0.611 (0.237‐1.574) | .307 |
Ref, Reference level; AOR, adjusted odds ratio after age and gender correction; CI, confidence interval; P value.
4. Discussion
AS is a common chronic inflammatory disease which primarily affects the spinal and sacroiliac joints. Common symptoms include soreness, rigidity, and advanced deterioration of involved joints.19, 20 Genes, such as HLA‐B27, NLRP3, and TFRSF1A, have been reported to be involved in the etiology of AS. Neeraj et al. discovered that the HLA‐B27 positive phenotype is correlated with AS. Another study conducted on a Swedish population declared there is no association between NLRP3 SNPs and AS susceptibility. However, there were still several identified SNPs predicting treatment response to the first anti‐TNF‐α agent in AS.21, 22, 23 In this study, we consistently demonstrate that there are significant correlations between NLRP3 polymorphisms, NLRP3 haplotypes, TFRSF1A rs4149570, and AS susceptibility.
The NLR family comprises three proteins of NLRP1, NLRP3, and NLRC4.24 These proteins interact with various adaptor proteins to form a macromolecular complex called inflammasome. Inflammasome induces the activation of caspase‐1 and the secretion of interleukin‐1β (IL‐1βy.24 NLRP3 is also known as CIASI, NALP3, PYPAF1, and cryopyrin. It can bind to adaptor proteins such as TUCAN (CARD8) and ASC to form inflammasome.25, 26 From there, it culminates to convert procaspase 1 into caspase 1, and also processes pro‐interleukin‐18 (proIL‐18) and pro‐interleukin‐1b (proIL‐1b), resulting in the production of active IL‐18 and IL‐1b.24 IL‐18 and IL‐1b are both pro‐inflammatory cytokines known as puissant mediators of inflammation and are associated with autoimmune disorders such as Behcet's syndrome, AIDS, Crohn's disease, and celiac disease.24, 25, 27, 28, 29 NLRP3 polymorphisms at positions rs10754558 and rs4612666 have been shown to be related to HIV infection, type 1 diabetes, food‐induced anaphylaxis, and rheumatoid arthritis. Functional analysis has previously revealed that the risk alleles of rs10754558 and rs4612666 can result in increased stability of NLRP3 mRNA and enhance NLRP3 expression.24, 26, 29, 30, 31 In the present study, we discovered that the rs4612666 (C/T) in patients could increase NLRP3 mRNA stability and enhance NLRP3 activity. This subsequently led to a series of inflammatory reactions which was consistent with the findings of Hitomi et al.31 Our study also illustrated that the NLRP3 rs3806265 in AS patients was overexpressed. We thus hypothesized that it could affect NLRP3 synthesis and culminated to the overproduction of IL‐1b (a cytokine involved in the development of AS).32 The previous studies reported that the NLRP3 rs10925019 was associated with Crohn's and ulcerative colitis disease. We found a close relationship between rs10925019 and AS occurrence. However, pathomechanism needed further study.33, 34
Tumor necrosis factor alpha (TNF‐α) is a pro‐inflammatory cytokine which acts as the ligand for TNF‐R1 and TNF‐R2. It plays a vital part in the pathomechanism of rheumatoid arthritis (RA) spondyloarthritis (SpA), psoriatic arthritis (PsA), and AS.35, 36 The expression of adhesion molecules and the increase of neutrophil activation can be stimulated by TNF‐α.37 Furthermore, at a cellular level, when ligands bind to receptors it can induce apoptosis through the exterior pathway bearing cytoplasmic death domains (death receptors) such as TNF‐R1 and TRAIL‐R1.37 The TNF‐α contains several SNPs, which have been found to be relevant to susceptibility and polymorphisms.37
The results of our study indicate that there is linkage disequilibrium in TNFRSF1A rs4149570, rs767455 and rs4149569 of AS patients. One study reported a significant positive association between carriers of allele G and treatment response. However, our experiment illustrated that allele T in rs4149570 has a significant correlation with the incidence of AS. AS and RA are different diseases, have different mechanisms and our results may offer a crucial supplement for AS genetic diagnosis.38 TNF blockers have been proven to be highly effective in reducing Spondyloarthritis (SpA) and inflammatory bowel diseases (IBDs).39, 40 Currently, there are a few TNF‐α inhibitors available for clinical treatments. These inhibitors act to block the binding of TNF‐α to its receptors and therefore, interfering with TNF‐α signaling transduction pathways, such as anti‐TNF‐α mAbs (golimumab, adalimumab, infliximab, and certolizumab pegol) and etanercept (a fusion protein which acts as a “decoy receptor” for TNF‐α).41, 42, 43 Etanercept is composed of two p75 TNF receptors fused with the Fc portion of human IgG1. It is generally administered subcutaneously 25 mg/d twice a week or 50 mg once a week.44 Etanercept binds to TNF (primarily to its soluble form) with a high affinity and blocks the effects of TNF.42, 45
Our studies have demonstrated that there are obvious differences in the sedimentation rate (ESR) level, waist duration of morning stiffness and BASFI between traditional medicine and etanercept treatment patients. We also observed no significant differences in VAS, CRP between the two groups. We conclude that the baseline factors which are significant predictors of ASAS 20 response in etanercept‐treated patients are ESR, waist duration of morning stiffness, and the BASFI score. This conclusion is partly contradictory to the findings of Davis et al.46 When patients are treated with etanercept, we discovered that the allele T has a greater negative influence on the achievement of ASAS20 than C of rs4612666 in NLRP3. Furthermore, the allele G of rs3806268 showed a better achievement of ASAS20 than A. This indicates that the two genes may be detection factors and predictors of AS.
The major limitation of this study would be the small sample size. We intended to conduct in‐depth research on a larger population for the next stage. In conclusion, we have studied the association of NLRP3 and TNFRSF1A polymorphisms and haplotypes with AS susceptibility. The correlation of NLRP3 and TNFRSF1A polymorphisms and haplotypes with the curative effect and adverse reactions of AS patients treated with traditional medicines and etanercept. Our results imply that NLRP3 SNPs and haplotypes play a critical role in the development of ankylosing spondylitis. Further research on NLRP3 inflammasome will contribute to the development of diagnostic and therapeutic methods for AS.
Supporting information
Zhao S, Chen H, Wu G, and Zhao C. The association of NLRP3 and TNFRSF1A polymorphisms with risk of ankylosing spondylitis and treatment efficacy of etanercept. J Clin Lab Anal. 2017;31:e22138 10.1002/jcla.22138
References
- 1. Elewaut D, Matucci‐Cerinic M. Treatment of ankylosing spondylitis and extra‐articular manifestations in everyday rheumatology practice. Rheumatology (Oxford). 2009;48:1029–1035. [DOI] [PubMed] [Google Scholar]
- 2. Lui NL, Thumboo J, Inman R. Cardiomyopathy in ankylosing spondylitis. Arthritis Care Res (Hoboken). 2011;63:564–569. [DOI] [PubMed] [Google Scholar]
- 3. Braun J, Sieper J. Ankylosing spondylitis. Lancet. 2007;369:1379–1390. [DOI] [PubMed] [Google Scholar]
- 4. Dean LE, Jones GT, MacDonald AG, Downham C, Sturrock RD, Macfarlane GJ. Global prevalence of ankylosing spondylitis. Rheumatology (Oxford). 2014;53:650–657. [DOI] [PubMed] [Google Scholar]
- 5. Bowness P. Hla‐B27. Annu Rev Immunol. 2015;33:29–48. [DOI] [PubMed] [Google Scholar]
- 6. Brown MA, Kenna T, Wordsworth BP. Genetics of ankylosing spondylitis–insights into pathogenesis. Nat Rev Rheumatol. 2016;12:81–91. [DOI] [PubMed] [Google Scholar]
- 7. Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. [DOI] [PubMed] [Google Scholar]
- 8. Zhang Q, Fan HW, Zhang JZ, Wang YM, Xing HJ. NLRP3 rs35829419 polymorphism is associated with increased susceptibility to multiple diseases in humans. Genet Mol Res. 2015;14:13968–13980. [DOI] [PubMed] [Google Scholar]
- 9. Sode J, Vogel U, Bank S, et al. Genetic variations in pattern recognition receptor loci are associated with anti‐TNF response in patients with rheumatoid arthritis. PLoS ONE. 2015;10:e0139781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Greco E, Aita A, Galozzi P, et al. The novel S59P mutation in the TNFRSF1A gene identified in an adult onset TNF receptor associated periodic syndrome (TRAPS) constitutively activates NF‐kappaB pathway. Arthritis Res Ther. 2015;17:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evans DM, Spencer CC, Pointon JJ, et al. Interaction between ERAP1 and HLA‐B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA‐B27 in disease susceptibility. Nat Genet. 2011;43:761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Braun J, van den Berg R, Baraliakos X, et al. 2010 update of the ASAS/EULAR recommendations for the management of ankylosing spondylitis. Ann Rheum Dis. 2011;70:896–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen J, Lin S, Liu C. Sulfasalazine for ankylosing spondylitis. Cochrane Database Syst Rev. 2014;:CD004800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barkham N, Coates LC, Keen H, et al. Double‐blind placebo‐controlled trial of etanercept in the prevention of work disability in ankylosing spondylitis. Ann Rheum Dis. 2010;69:1926–1928. [DOI] [PubMed] [Google Scholar]
- 15. Dougados M, Braun J, Szanto S, et al. Efficacy of etanercept on rheumatic signs and pulmonary function tests in advanced ankylosing spondylitis: results of a randomised double‐blind placebo‐controlled study (SPINE). Ann Rheum Dis. 2011;70:799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Escalas C, Trijau S, Dougados M. Evaluation of the treatment effect of NSAIDs/TNF blockers according to different domains in ankylosing spondylitis: results of a meta‐analysis. Rheumatology (Oxford). 2010;49:1317–1325. [DOI] [PubMed] [Google Scholar]
- 17. Stark K, Reinhard W, Neureuther K, et al. Association of common polymorphisms in GLUT9 gene with gout but not with coronary artery disease in a large case‐control study. PLoS ONE. 2008;3:e1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van der Linden S, Valkenburg HA, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum. 1984;27:361–368. [DOI] [PubMed] [Google Scholar]
- 19. Brown MA, Wordsworth BP, Reveille JD. Genetics of ankylosing spondylitis. Clin Exp Rheumatol. 2002;20(6 Suppl 28):S43–S49. [PubMed] [Google Scholar]
- 20. Brown MA. Progress in the genetics of ankylosing spondylitis. Brief Funct Genomics. 2011;10:249–257. [DOI] [PubMed] [Google Scholar]
- 21. Diaz‐Pena R, Vidal‐Castineira JR, Lopez‐Vazquez A, Lopez‐Larrea C. HLA‐B*40:01 is associated with ankylosing spondylitis in HLA‐B27‐positive populations. J Rheumatol. 2016;43:1255–1256. [DOI] [PubMed] [Google Scholar]
- 22. Kastbom A, Klingberg E, Verma D, et al. Genetic variants in CARD8 but not in NLRP3 are associated with ankylosing spondylitis. Scand J Rheumatol. 2013;42:465–468. [DOI] [PubMed] [Google Scholar]
- 23. Schiotis R, Sanchez A, Escudero A, et al. Candidate's single‐nucleotide polymorphism predictors of treatment nonresponse to the first anti‐TNF inhibitor in ankylosing spondylitis. Rheumatol Int. 2014;34:793–801. [DOI] [PubMed] [Google Scholar]
- 24. Pontillo A, Oshiro TM, Girardelli M, Kamada AJ, Crovella S, Duarte AJ. Polymorphisms in inflammasome’ genes and susceptibility to HIV‐1 infection. J Acquir Immune Defic Syndr. 2012;59:121–125. [DOI] [PubMed] [Google Scholar]
- 25. Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL‐beta. Mol Cell. 2002;10:417–426. [DOI] [PubMed] [Google Scholar]
- 26. Kastbom A, Verma D, Eriksson P, Skogh T, Wingren G, Soderkvist P. Genetic variation in proteins of the cryopyrin inflammasome influences susceptibility and severity of rheumatoid arthritis (the Swedish TIRA project). Rheumatology (Oxford). 2008;47:415–417. [DOI] [PubMed] [Google Scholar]
- 27. Kone‐Paut I, Sanchez E, Le Quellec A, Manna R, Touitou I. Autoinflammatory gene mutations in Behcet's disease. Ann Rheum Dis. 2007;66:832–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Villani AC, Lemire M, Fortin G, et al. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat Genet. 2009;41:71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pontillo A, Brandao L, Guimaraes R, Segat L, Araujo J, Crovella S. Two SNPs in NLRP3 gene are involved in the predisposition to type‐1 diabetes and celiac disease in a pediatric population from northeast Brazil. Autoimmunity. 2010;43:583–589. [DOI] [PubMed] [Google Scholar]
- 30. Pontillo A, Brandao LA, Guimaraes RL, Segat L, Athanasakis E, Crovella S. A 3′UTR SNP in NLRP3 gene is associated with susceptibility to HIV‐1 infection. J Acquir Immune Defic Syndr. 2010;54:236–240. [DOI] [PubMed] [Google Scholar]
- 31. Hitomi Y, Ebisawa M, Tomikawa M, et al. Associations of functional NLRP3 polymorphisms with susceptibility to food‐induced anaphylaxis and aspirin‐induced asthma. J Allergy Clin Immunol. 2009;124:779–785 e776. [DOI] [PubMed] [Google Scholar]
- 32. Bidoki AZ, Harsini S, Sadr M, et al. NLRP3 gene polymorphisms in Iranian patients with recurrent aphthous stomatitis. J Oral Pathol Med. 2016;45:136–140. [DOI] [PubMed] [Google Scholar]
- 33. Cummings JR, Cooney RM, Clarke G, et al. The genetics of NOD‐like receptors in Crohn's disease. Tissue Antigens. 2010;76:48–56. [DOI] [PubMed] [Google Scholar]
- 34. Zhang HX, Wang ZT, Lu XX, Wang YG, Zhong J, Liu J. NLRP3 gene is associated with ulcerative colitis (UC), but not Crohn's disease (CD), in Chinese Han population. Inflamm Res. 2014;63:979–985. [DOI] [PubMed] [Google Scholar]
- 35. Ritchlin CT. Therapeutic considerations in spondyloarthritis patients who fail tumour necrosis factor antagonists. Best Pract Res Clin Rheumatol. 2010;24:683–692. [DOI] [PubMed] [Google Scholar]
- 36. Hyrich KL, Watson KD, Silman AJ, Symmons DP. Predictors of response to anti‐TNF‐alpha therapy among patients with rheumatoid arthritis: results from the British Society for Rheumatology Biologics Register. Rheumatology (Oxford). 2006;45:1558–1565. [DOI] [PubMed] [Google Scholar]
- 37. Pundt N, Peters MA, Wunrau C, et al. Susceptibility of rheumatoid arthritis synovial fibroblasts to FasL‐ and TRAIL‐induced apoptosis is cell cycle‐dependent. Arthritis Res Ther. 2009;11:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bank S, Skytt Andersen P, Burisch J, et al. Polymorphisms in the inflammatory pathway genes TLR2, TLR4, TLR9, LY96, NFKBIA, NFKB1, TNFA, TNFRSF1A, IL6R, IL10, IL23R, PTPN22, and PPARG are associated with susceptibility of inflammatory bowel disease in a Danish cohort. PLoS ONE. 2014;9:e98815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thiebault H, Boyard‐Lasselin P, Guignant C, et al. Paradoxical articular manifestations in patients with inflammatory bowel diseases treated with infliximab. Eur J Gastroenterol Hepatol. 2016;28:876–881. [DOI] [PubMed] [Google Scholar]
- 40. Kivolowitz C, Abdulhamid A, Victorio D, et al. O‐006 expansion of immunologically‐relevant E. coli in the intestinal microbiota of patients with IBD‐associated spondyloarthritis promotes mucosal RORgt‐dependent immunity. Inflamm Bowel Dis. 2016;22(Suppl 1):S3. [Google Scholar]
- 41. Murdaca G, Colombo BM, Puppo F. Anti‐TNF‐alpha inhibitors: a new therapeutic approach for inflammatory immune‐mediated diseases: an update upon efficacy and adverse events. Int J Immunopathol Pharmacol. 2009;22:557–565. [DOI] [PubMed] [Google Scholar]
- 42. Murdaca G, Spano F, Contatore M, Guastalla A, Magnani O, Puppo F. Efficacy and safety of etanercept in chronic immune‐mediated disease. Expert Opin Drug Saf. 2014;13:649–661. [DOI] [PubMed] [Google Scholar]
- 43. Hochberg MC, Lebwohl MG, Plevy SE, Hobbs KF, Yocum DE. The benefit/risk profile of TNF‐blocking agents: findings of a consensus panel. Semin Arthritis Rheum. 2005;34:819–836. [DOI] [PubMed] [Google Scholar]
- 44. Furst DE, Breedveld FC, Kalden JR, et al. Updated consensus statement on biological agents, specifically tumour necrosis factor {alpha} (TNF{alpha}) blocking agents and interleukin‐1 receptor antagonist (IL‐1ra), for the treatment of rheumatic diseases, 2005. Ann Rheum Dis. 2005;64(Suppl 4):iv2–iv14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sprott H, Glatzel M, Michel BA. Treatment of myositis with etanercept (Enbrel), a recombinant human soluble fusion protein of TNF‐alpha type II receptor and IgG1. Rheumatology (Oxford). 2004;43:524–526. [DOI] [PubMed] [Google Scholar]
- 46. Davis JC Jr, Van der Heijde DM, Dougados M, et al. Baseline factors that influence ASAS 20 response in patients with ankylosing spondylitis treated with etanercept. J Rheumatol. 2005;32:1751–1754. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


