Abstract
Background
Recent studies have found circular RNAs (circRNAs) involved in the biological process of cancers. However, little is known about their functional roles in glioblastoma.
Methods
Human circRNA microarray analysis was performed to screen the expression profile of circRNAs in IDH1 wild‐type glioblastoma tissue. The expression of hsa_circ_0008344 in glioblastoma and normal brain samples was quantified by qRT‐PCR. Functional experiments were performed to investigate the biological functions of hsa_circ_0008344, including MTT assay, colony formation assay, transwell assay, and cell apoptosis assay.
Results
CircRNA microarray revealed a total of 417 abnormally expressed circRNAs (>1.5‐fold, P < .05) in glioblastoma tissue compared with the adjacent normal brain. Hsa_circ_0008344, among the top differentially expressed circRNAs, was significantly upregulated in IDH1 wild‐type glioblastoma. Further in vitro studies showed that knockdown of hsa_circ_0008344 suppressed glioblastoma cell proliferation, colony formation, migration, and invasion, but increased cell apoptotic rate.
Conclusions
Hsa_circ_0008344 is upregulated in glioblastoma and may contribute to the progression of this malignancy.
Keywords: apoptosis, glioblastoma, hsa_circ_0008344, invasion, proliferation
1. INTRODUCTION
Gliomas represent the most common primary brain tumors, with the most biologically aggressive type, glioblastoma that accounts for more than 80% of malignant glioma.1, 2, 3 Despite the improvement in multimodal treatment involving surgical resection, radiotherapy, and temozolomide (TMZ) chemotherapy, patients with glioblastoma derive little benefit from the current standard of care.4, 5 The median survival time of glioblastoma was only 12‐15 months, and the 5‐year survival rate was only 4%‐5%.6, 7 Recently, substantial efforts have been undertaken in examining molecular mechanisms involved in glioblastoma and allowed for a novel understanding of tumor development and progression.
Circular RNA (circRNA) is a novel type of RNA molecule formed by a covalently closed loop and is found to exist widely in eukaryotes.8 Emerging evidence indicates that many circRNAs have cell‐type‐specific expression and are linked to physiological development and various diseases including cancer.9 Similar to long noncoding RNA, circRNAs can function as a miRNAs sponge and play a role in the pathological process of glioma.10 For example, upregulated circ‐TTBK2 in glioma was found to act as the miR‐217 sponge and promote cell proliferation, migration, and invasion.11 Current studies showed another circRNA hsa_circ_0046701 significantly overexpressed in glioma and functioning as a sponge for miR‐142‐3p to promote carcinogenesis.12 Interestingly, circRNAs were also been found to be targeted by microRNAs that CDR1‐AS, a miR‐7 sponge, could be degraded by miR‐671‐5p in glioma.13 Moreover, endogenous circRNAs could encode functional proteins, which was confirmed in glioblastoma that circ‐FBXW7 encoded a novel 21‐kDa protein FBXW7‐185aa and inhibited tumor cell proliferation and cell cycle acceleration.14 Deep insight into the roles of circRNA in tumor biology of glioblastoma accelerates to pave the way for treating this disease.
In this study, we applied circRNA microarray to investigate the difference between glioblastoma and normal brain and observed a novel circRNA, hsa_circ_0008344, overexpressed in glioblastoma. Knockdown of hsa_circ_0008344 inhibited glioblastoma cell proliferation, migration, and invasion, and increased cell apoptosis, suggesting that hsa_circ_0008344 might play an oncogenic role in the development and progression of glioblastoma.
2. MATERIALS AND METHODS
2.1. Acquisition of tissue samples
Ten patients with primary glioblastoma, who underwent surgery at Changzheng Hospital from January 2016 to December 2016, were enrolled in the study. Pairs of glioblastoma tissue and adjacent noncancerous brain tissue were excised from patients and immediately frozen for further analysis. This study was approved by the Specialty Committee on Ethics of Biomedicine Research, Second Military Medical University of China. Informed consent was obtained from all participants.
2.2. Human circular RNA microarray
Human circRNA expression analysis was performed on 4 pairs of glioblastoma tissue and corresponding adjacent brain tissue. Total RNA was extracted, of which linear RNA was removed by RNAse R kit (Epicentre, Inc. Madison, WI, USA). Human circRNA microarray hybridization was performed according to Arraystar's standard protocols. The enriched circRNA was amplified to cDNA and transcribed into cRNA using Arraystar Super RNA Labeling Kit (Arraystar, Rockville, MD, USA). Labeled cRNAs were then hybridized using Arraystar Human circRNA Array (8 × 15K, Arraystar) and scanned by the Agilent Scanner G2505C.
2.3. RNA extraction and quantitative real‐time PCR
The total RNA was extracted from cell lines using TRIzol Reagent (Invitrogen, Karlsruhe, Germany) following the manufacturer's protocol. First‐strand cDNA was synthesized using ReverTra Ace (Toyobo, Osaka, Japan). The quantitative real‐time PCR was conducted by ABI PRISM 7900HT Sequence Detection System in the presence of SYBR Green dye (Toyobo). Each sample was tested in triplicate. GAPDH was used as an internal control. All PCR primers used are as follows: hsa_circ_0008344 forward primer, GCAGGAGGTAATGACGGAAG and reverse primer, TGGTCGAAGTCAGCAGACAC; GAPDH forward primer, GCGAGATCCCTCCAAAATCAA and reverse primer, GTTCACACCCATGACGAACAT.
2.4. Cell line and culture
The human glioblastoma cell lines (U87 and U251) were obtained from the American Type Culture Collection (ATCC). All cells were maintained in a humidified atmosphere of 5% CO2 at 37°C and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
2.5. Plasmid and lentivirus infection
To establish cell lines in which hsa_circ_0008344 expression would be stably knocked down, the 21‐nucleotide sequence (shRNA, 5′‐GCCAAGCCAGTTCCATTAAAT‐3′) was used to target hsa_circ_0008344.
Sequence (shNC, 5′‐TTCTCCGAACGTGTCACGT‐3′) served as a control. Corresponding sense and antisense oligonucleotides were synthesized from GENECHEM (Shanghai, China), and then annealed and cloned into the AgeI‐EcoRI sites of the GV493 lentiviral vector. The procedures of packaging and infecting with lentivirus were performed according to the previous study.15
2.6. MTT assay
Cells in the logarithmic phase of growth were seeded in 96‐well plates (5000 cells/well) in sextuple and allowed to grow for 1, 2, 3, and 4 days. Cell proliferation assay was analyzed by MTT kit following the manufacturer's instructions. The optical density was measured at 490‐nm wavelength using a microplate reader (EL × 800, BIO‐TEK, USA). Data were obtained from 3 independent assays performed in triplicate.
2.7. Colony formation assay
For colony formation assay, 1 × 103 cells were independently seeded onto 60‐mm culture plates in triplicate. After about 2 weeks, visible colonies were fixed with 100% methanol and then stained with crystal violet. Colony‐forming ability was evaluated by counting the number of colonies.
2.8. Transwell assay
Transwell compartments with a 24‐well 8‐μm pore size (Costar, Washington, DC, USA) were coated with or without Matrigel. Cells were resuspended in 250 μL serum‐free medium and placed in the upper chamber, and 600 μL medium with 20% FBS was added to the lower chamber. After cultured for 36 hours, the migrated and invaded cells on the lower membrane surface were removed with a cotton swab and fixed by 4% paraformaldehyde and stained with crystal violet solution (Sigma‐Aldrich, Saint Louis, MO, USA).
2.9. Cell apoptosis analysis
Cells were plated into 6‐well plates at 6 × 104 cells/well. Forty‐eight hours after transfection, the cells were harvested and washed with PBS. Cellular apoptosis was evaluated by flow cytometry using Annexin V‐FITC apoptosis detection kit (BD Biosciences, Piscataway, NJ, USA) as described by the manufacturer's instructions.
2.10. Statistical analysis
Statistical analyses were all performed by SPSS 19.0 software. Data were calculated from 3 independent experiments presented as the mean ± standard deviation (SD). The difference between 2 groups was analyzed using Student's t test. Difference (P < .05) was considered to be statistically significant.
3. RESULTS
3.1. Human circular RNA microarray identified hsa_circ_0008344 upregulated in glioblastoma tissue
To investigate the abnormal expression of circRNAs, human circular RNA microarray analysis was performed in 4 pairs of IDH1 wild‐type glioblastoma tissue and corresponding nontumor normal brain. Results revealed there a total of 417 abnormally expressed circRNAs (>1.5‐fold, P < .05), of which 136 were increased and 281 were downregulated in glioblastoma (Figure 1A). The abnormal circRNA expression profiles in the malignancy provided potential functional circRNAs related to glioblastoma progression. Among the top differentially expressed circRNAs (Table 1), hsa_circ_0008344 further detected its expression level in another group of 16 IDH1 wild‐type glioblastoma samples and 7 normal brain tissues, which confirmed the significantly upregulated hsa_circ_0008344 in glioblastoma (Figure 1B).
Figure 1.
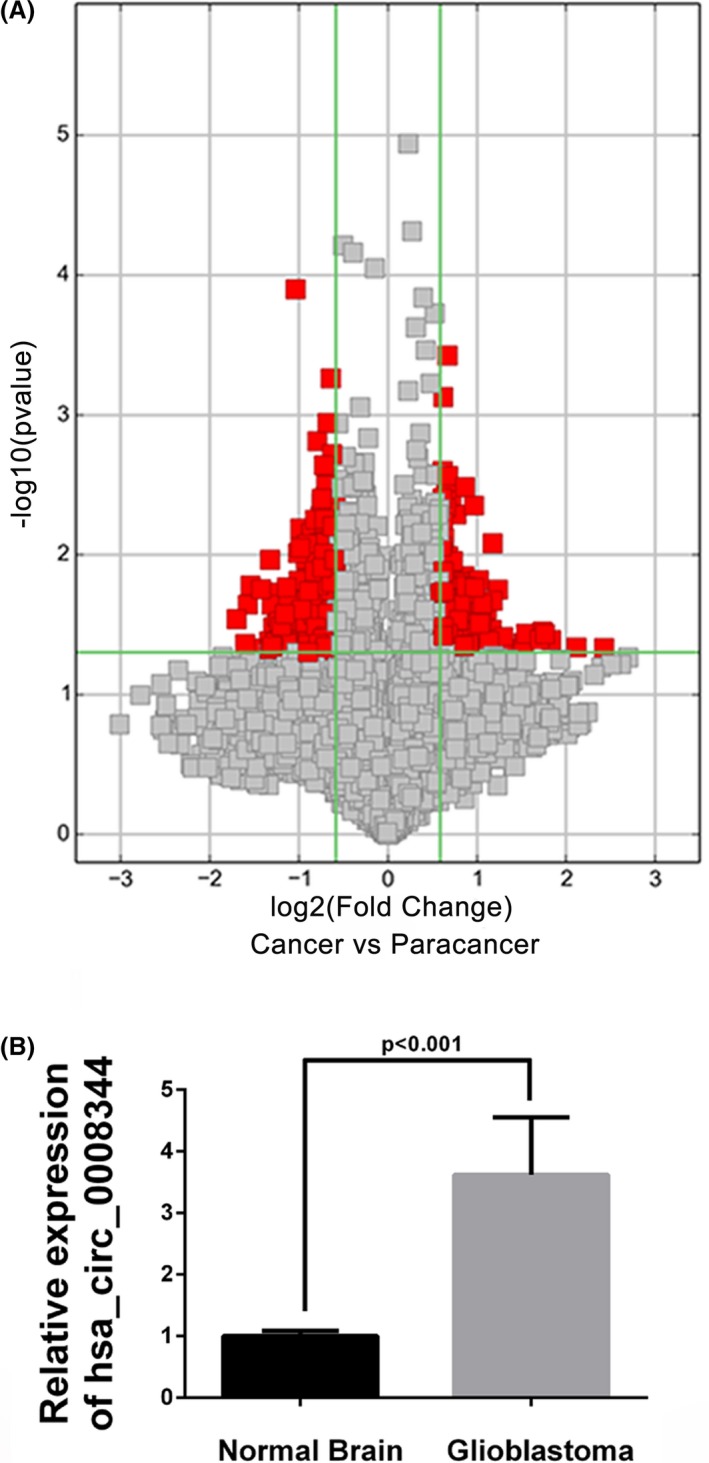
Hsa_circ_0008344 is upregulated in glioblastoma. A, Volcano plot of circRNAs. The vertical lines correspond to 1.5‐fold up and down, and the horizontal line represents a P value of .05. The red point in the plot represents the differentially expressed circRNAs with statistical significance. B, Hsa_circ_0008344 expression level is examined by qRT‐PCR and significantly higher in glioblastoma tissues than normal brain samples (P < .001)
Table 1.
The top 10 differentially expressed circRNAs in glioblastoma
| circRNA | Expression |
|---|---|
| hsa_circRNA_0008344 | Up |
| hsa_circRNA_005198 | Up |
| hsa_circRNA_001587 | Up |
| hsa_circRNA_101085 | Up |
| hsa_circRNA_101066 | Up |
| hsa_circRNA_405962 | Up |
| hsa_circRNA_018440 | Down |
| hsa_circRNA_006240 | Down |
| hsa_circRNA_403717 | Down |
| hsa_circRNA_104105 | Up |
3.2. Hsa_circ_0008344 knockdown suppressed the proliferation of glioblastoma cells
Given its high level of glioblastoma, hsa_circ_0008344 was targeted to study its possible biological roles in tumor progression. Then, specifically shRNA inhibiting hsa_circ_0008344 was transfected into U87 and U251 cells, and the expression of hsa_circ_0008344 was markedly decreased compared to control groups (Figure 2A). MTT assay was performed to determine glioblastoma cell proliferation, which showed that hsa_circ_0008344 knockdown suppressed the proliferation activity in both U87 and U251 (Figure 2B‐C). Consistently, colony formation assay also revealed fewer numbers of clones in U87 and U251 groups transfected with shRNA compared with that in shNC and blank control cells (Figure 2D‐E).
Figure 2.
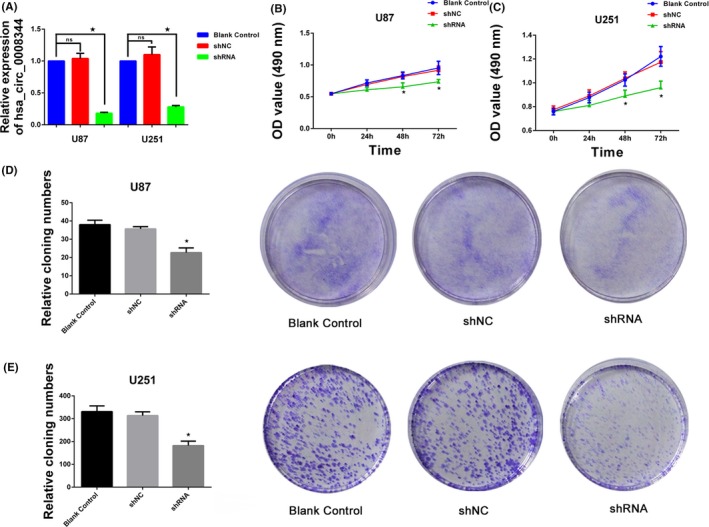
Hsa_circ_0008344 promotes glioblastoma cell growth. A, The efficiency of shRNA silencing hsa_circ_0008344 in U87 and U251 is detected by qRT‐PCR. B‐C, Decreased expression of hsa_circ_0008344 inhibits tumor cell proliferation. D‐E, Comparing with the cells carrying shNC and blank control, the depleted hsa_circ_0008344 cells display fewer colonies (*P < .05)
3.3. Suppression of hsa_circ_0008344 inhibited the glioblastoma cell migration and invasion
To explore the impact of hsa_circ_0008344 on migration and invasion of glioblastoma cells, transwell assays were conducted in the 2 selected cells. As shown in Figure 3A‐B, downregulating hsa_circ_0008344 leads to remarkably impaired migration capacity of glioblastoma cells compared with the shNC and blank control groups. The transwell assay confirmed the role of hsa_circ_0008344 on cell migration. Moreover, knockdown of hsa_circ_0008344 by shRNA also caused fewer cells traversed the membrane, which indicated a decreased invasive potential of glioblastoma cells (Figure 3C‐D).
Figure 3.
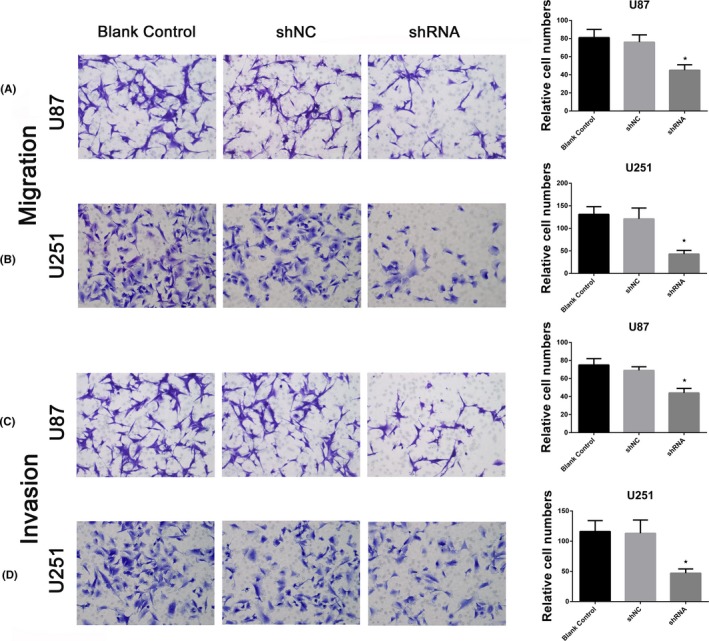
Hsa_circ_0008344 promotes glioblastoma cell migration and invasion. A‐B, Inhibition of hsa_circ_0008344 expression decreases cell migration as determined by transwell assay. C‐D, The numbers of invasive cells in the depleted hsa_circ_0008344 groups are much smaller than those in shNC and blank control groups (*P < .05)
3.4. Hsa_circ_0008344 knockdown promoted tumor cell apoptosis in glioblastoma
We then investigated other aspects of hsa_circ_0008344 functions, such as apoptosis, in glioblastoma by flow cytometry. In accordance with previous results, u87 and U251 cells in the group of depleted hsa_circ_0008344 displayed markedly increased apoptotic rate compared with the shNC and blank control groups (Figure 4). The apoptosis assay suggested that hsa_circ_0008344 might have an anti‐apoptosis character in glioblastoma cells.
Figure 4.
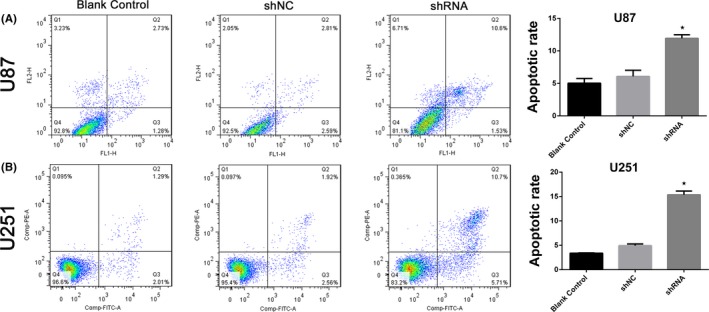
Hsa_circ_0008344 regulates glioblastoma cell apoptosis. A‐B, Downregulation of hsa_circ_0008344 increases the apoptotic rates of tumor cells, as compared to the shNC and blank control (*P < .05)
4. DISCUSSION
The rapid advances in high‐throughput sequencing for noncoding RNA have sparked new interest in circRNA, which is a special class of endogenous RNA, take a great proportion of all spliced transcripts, and wildly exist in human cells.16, 17 Recent studies have demonstrated that circRNA plays a crucial role in various physiological and pathological processes.18 Remarkably, hundreds of circRNAs have been shown to be deregulated in distinct cancers, including liver, lung, colorectal, breast, prostate, bladder, ovarian, kidney, and gastric cancer, as well as malignant glioma, and are likely to affect several of the hallmarks of cancer.19 In our study, we identified a novel circRNA, hsa_circ_0008344, upregulated in glioblastoma and underscored its roles in tumor cell proliferation, migration, invasion, and apoptosis, which implied the great potential in clinical application.
Current studies indicated that circRNAs might regulate gene expression by acting as miRNA sponges, thereby regulating linear RNA transcription and protein production in cancer.20, 21 Compared with their linear isomers, circRNAs have much higher expression levels and more miRNA binding sites, which may lead to more effective in sequestering miRNAs.21, 22, 23 As another type of noncoding RNA, miRNA can negatively modulate the translation process and target mRNA stability, thus suppress gene expression at the post‐transcriptional level.24, 25 Further studies have demonstrated that miRNA can also function as a tumor suppressor gene, oncogene, or both, depending on the cell context, and is closely correlated with cancer profiling.26, 27 Therefore, the carcinogenic mechanisms driven by circRNAs may occur through their interaction with miRNAs on regulating gene expression. Increased evidence has confirmed this idea that circular RNA‐ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR‐130a/miR‐107 and modulating PTEN, circ‐ITCH inhibited bladder cancer progression by sponging miR‐17/miR‐224 and regulating p21 and PTEN expression, and hsa_circ_0046701 promoted carcinogenesis by increasing the expression of miR‐142‐3p target ITGB8 in glioma.12, 28, 29 Furthermore, hsa_circ_0008344 may bind to several miRNAs, such as miR‐433‐3p and miR‐450b‐3p, predicted by bioinformatics, while miR‐433‐3p has been found to suppress cell growth and enhance chemosensitivity by targeting CREB in glioblastoma.30 Accordingly, it is presumed that hsa_circ_0008344 can sponge tumor suppressor miRNAs followed by disinhibition of the expression of some targeted oncogenic genes, which contributes to glioblastoma progression.
Collectively, the present study was first to investigate the expression and functional role of hsa_circ_0008344 in glioblastoma, which may expedite the appraisal of a novel therapeutic target for this malignancy.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (81572501) and Program for Academic Leaders of Shanghai (No. 043).
Zhou J, Wang H, Chu J, et al. Circular RNA hsa_circ_0008344 regulates glioblastoma cell proliferation, migration, invasion, and apoptosis. J Clin Lab Anal. 2018;32:e22454 10.1002/jcla.22454
Jinxu Zhou, Hongxiang Wang and Junsheng Chu contributed equally to this work.
Contributor Information
Juxiang Chen, Email: juxiangchen@smmu.edu.cn.
Yuhai Wang, Email: wangyuhai67@126.com.
REFERENCES
- 1. Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842‐1850. [DOI] [PubMed] [Google Scholar]
- 2. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492‐507. [DOI] [PubMed] [Google Scholar]
- 3. Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, Delattre JY. Primary brain tumours in adults. Lancet. 2012;379:1984‐1996. [DOI] [PubMed] [Google Scholar]
- 4. Wang H, Xu T, Jiang Y, et al. The challenges and the promise of molecular targeted therapy in malignant gliomas. Neoplasia. 2015;17:239‐255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alifieris C, Trafalis DT. Glioblastoma multiforme: pathogenesis and treatment. Pharmacol Ther. 2015;152:63‐82. [DOI] [PubMed] [Google Scholar]
- 6. Aldape K, Zadeh G, Mansouri S, et al. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829‐848. [DOI] [PubMed] [Google Scholar]
- 7. Batash R, Asna N, Schaffer P, et al. Glioblastoma multiforme, diagnosis and treatment. Recent Literature Review. Curr Med Chem. 2017;24:3002‐3009. [DOI] [PubMed] [Google Scholar]
- 8. Yang P, Qiu Z, Jiang Y, et al. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/beta-catenin signaling pathway. Oncotarget. 2016;7:63449‐63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen B, Huang S. Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41‐50. [DOI] [PubMed] [Google Scholar]
- 10. Panda AC, Grammatikakis I, Munk R, et al. Emerging roles and context of circular RNAs. Wiley Interdiscip Rev RNA. 2017;8:e1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng J, Liu X, Xue Y, et al. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1beta/Derlin-1 pathway. J Hematol Oncol. 2017;10:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li G, Yang H, Han K, et al. A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis by increasing the expression of miR-142-3p target ITGB8 in glioma. Biochem Biophys Res Commun. 2018;498:254‐261. [DOI] [PubMed] [Google Scholar]
- 13. Barbagallo D, Condorelli A, Ragusa M, et al. Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7:4746‐4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen Y, Meng D, Wang H, et al. VAMP8 facilitates cellular proliferation and temozolomide resistance in human glioma cells. Neuro Oncol. 2014;17:407‐418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2017;32:e22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang MS, Zhu T, Li L, et al. LncRNAs and CircRNAs from the same gene: masterpieces of RNA splicing. Cancer Lett. 2018;415:49‐57. [DOI] [PubMed] [Google Scholar]
- 18. Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992‐999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555‐565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6:563‐579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472‐480. [PMC free article] [PubMed] [Google Scholar]
- 22. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440‐441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522‐531. [DOI] [PubMed] [Google Scholar]
- 25. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848‐5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang H, Xu T, Jiang Y, et al. MicroRNAs in human glioblastoma: from bench to beside. Front Biosci (Landmark Ed). 2015;20:105‐118. [DOI] [PubMed] [Google Scholar]
- 28. Liu T, Liu S, Xu Y, et al. Circular RNA-ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR-130a/miR-107 and modulating PTEN. Cancer Res Treat. 2018. 10.4143/crt.2017.537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Yang C, Yuan W, Yang X, et al. Circular RNA circ‐ITCH inhibits bladder cancer progression by sponging miR‐17/miR‐224 and regulating p21. PTEN expression. Mol Cancer. 2018;17:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun S, Wang X, Xu X, et al. MiR‐433‐3p suppresses cell growth and enhances chemosensitivity by targeting CREB in human glioma. Oncotarget. 2017;8:5057‐5068. [DOI] [PMC free article] [PubMed] [Google Scholar]


