Abstract
Background
Peroxisome proliferator‐activated receptor‐γ (PPARγ) is a regulator of inflammation. This study aimed to explore associations between PPARγ gene single‐nucleotide polymorphisms (SNPs) and susceptibility to and clinical outcome of sepsis in the North China Han population.
Methods
This study included 303 patients with sepsis and 303 controls. We conducted genetic typing for 13 common PPARγ gene SNPs (improved multiplex ligation detection reaction), linkage disequilibrium mapping, and haplotype inference. Associations between SNP genotypes/haplotypes and sepsis susceptibility and outcome (septic shock, organ dysfunction, or death) were assessed using unconditional logistic regression analysis.
Results
For rs2972164, patients with genotypes CT/CT+TT had higher risk of sepsis than genotype CC (odds ratio [95% CI]: 1.74 [1.05‐2.86], P = .03 and 1.72 [1.06‐2.80], P = .026, respectively); the T allele was associated with increased sepsis risk compared with the C allele (1.64 [1.04‐2.58], P = .033). For rs1801282, genotypes CG/CG+GG had lower risk of sepsis than genotype CC (0.55 [0.33‐0.92], P = .024 and 0.57 [0.35‐0.95], P = .03, respectively); the G allele was associated with decreased sepsis risk compared with the C allele (0.62 [0.39‐1.01], P = .055). For rs4135275, genotypes AG/AG+GG had higher risk of severe organ dysfunction (multiple organ dysfunction syndrome score >8) than genotype AA (2.66 [1.16‐6.09], P = .038 and 2.21 [1.00‐4.85], P = .042, respectively). Haplotype TAT (rs2972164, rs4684846, and rs17036188) was associated with increased sepsis risk (1.66 [1.03‐2.67], P = .038).
Conclusions
No mutation was correlated with septic shock or death. PPARγ gene polymorphisms may play a role in the occurrence and progression of sepsis in the North China Han population.
Keywords: peroxisome proliferator‐activated receptor gamma, polymorphism, prognosis, risk, sepsis, single nucleotide
1. INTRODUCTION
Sepsis is a complex clinically syndrome resulting from a dysregulated host inflammatory response to the invasion of pathogenic microorganisms.1 The prevalence and overall mortality of sepsis is still high; indeed, sepsis and its complications, septic shock and multiple organ dysfunction syndrome (MODS), remain the leading noncardiac causes of death in patients in the intensive care unit (ICU).2, 3 Individuals differ greatly in their response to infection, likely due to environmental factors (such as type and virulence of the pathogenic microorganism and site of infection) and host factors (such as age, underlying disease, and resistance). Importantly, host genetic factors are now recognized to have a greater impact on the occurrence and progression of sepsis than environmental factors.4, 5, 6, 7 The identification of genetic factors influencing the occurrence and progression of sepsis will allow the early detection of high‐risk patients with sepsis, facilitating the prompt initiation of anti‐infection therapy and treatment to support organ function and thereby improving prognosis in patients who develop this syndrome.
The mass production of inflammatory mediators is an important feature of sepsis.3 Peroxisome proliferator‐activated receptor‐g (PPARγ) is a member of the nuclear receptor superfamily and a ligand‐dependent nuclear transcription factor with multiple biological effects, including modulation of lipid and glucose metabolism and maintenance of homeostasis.8 PPARγ is widely distributed in immune cells such as monocytes‐macrophages, T/B lymphocytes, and dendritic cells and therefore can act as a regulator for inflammation and immune responses. PPARγ has been reported to exert a potent anti‐inflammatory effect by modulating the activation and migration of neutrophils, enhancing macrophage phagocytosis, reducing the production of pro‐inflammatory mediators such as tumor necrosis factor‐a (TNF‐a), interleukin‐6 (IL‐6), IL‐8, IL‐1b, and active oxygen/nitrogen species, and promoting the secretion of anti‐inflammatory mediators such as IL‐10.9, 10 Numerous in vivo and in vitro studies have shown that administration of a PPARγ agonist can suppress the inflammatory responses associated with sepsis,11 acute pancreatitis,12 inflammatory bowel disease,13 mastitis14 and chronic airway inflammation,15, 16 increasing the host's ability to kill and clear pathogenic microorganisms and improving prognosis. Therefore, PPARγ and its ligands have become a new therapeutic target for the treatment of sepsis and other inflammatory diseases.17
Patients carrying a mutated PPARγ gene may be susceptible to excessive inflammatory responses, manifesting as a high susceptibility to sepsis and/or poor prognosis. However, investigations of the relation of PPARγ single‐nucleotide polymorphisms (SNPs) with sepsis are rare and each concentrated on only one or a small number of SNPs.18, 19 Hence, some SNPs of the PPARγ gene have yet to be investigated for their possible influence on the susceptibility to sepsis or the prognosis of this condition. Therefore, the aim of this study was to analyze the association between common SNPs in the PPARγ gene (a total of 13 SNPs) and the susceptibility to and clinical outcome of sepsis in a North China Han population. The identification of SNPs associated with the susceptibility to or prognosis of sepsis could potentially allow for the screening of patients at high risk for sepsis, permitting the instigation of early aggressive therapy and improving prognosis.
2. MATERIALS AND METHODS
2.1. Subjects and design
Clinical data were consecutively collected from 310 patients with sepsis admitted at the Department of Emergency, Chinese People's Liberation Army (PLA) General Hospital, between June 2011 and June 2012. The diagnostic and grading criteria for sepsis developed by the American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference in 2001 were referred to as the diagnostic criteria in the current study.1 The inclusion criteria were as follows: (i) age ≥ 18 years old; (ii) Han race, native to Beijing and its surrounding areas (Tianjin, Hebei, Shanxi, and Inner Mongolia), and not biologically related to any other study participants; (iii) definitive diagnosis for sepsis; and (iv) complete data available for medical history and clinical laboratory and auxiliary examination results. The exclusion criteria included: (i) death within 24 hours after admission; (ii) the presence of certain comorbidities, such as acquired immunodeficiency syndrome, malignant tumor, autoimmune disease, post‐organ transplantation, and end‐stage failure of the heart, liver, kidney, lung, brain, or other important organ; (iii) history of intake or application of cytokines capable of regulating the inflammatory process, such as TNF‐α antibody and interferon‐γ; (iv) treated with immunosuppressive agents or hormone therapy; and (v) pregnant women.
While enrolling the patients with sepsis, we also consecutively included sepsis‐free controls who participated in physical examinations at the Health Examination Center of the Chinese PLA General Hospital as controls. The exclusion criteria included history or present evidence of sepsis, malignancy or autoimmune diseases, current acute/chronic severe organ system disease, and biological relationship with any other study participant. After matching with the sepsis group in terms of age, sex, race, and birthplace, a total of 309 individuals were confirmed as the controls. This study was approved by the Ethics Committee of the Chinese PLA General Hospital, and signed written informed consent was obtained from all participants.
2.2. Collection of data
Socio‐demographic data (age, sex, race, birthplace, height, and weight) and previous medical history, including history of tobacco and alcohol use, were collected from the sepsis and control groups using a questionnaire. The following baseline information was collected from patients in the sepsis group: laboratory and auxiliary examination results, site of infection, basic indices for the diagnosis of sepsis (body temperature, heart rate, respiratory rate, blood pressure, white blood cell count, and microbial culture results), and acute physiology, and chronic health evaluation II (APACHE II) score within 24 hours after the diagnosis of sepsis. In addition, the following clinical outcome measures were recorded: highest MODS score during hospitalization (calculated using Marshall's criteria),20 severity of MODS (mild=MODS score of 0‐8 points, severe=MODS score of 9‐18 points), occurrence/absence of septic shock, and death/survival. Septic shock was defined as a systolic blood pressure <90 mm Hg despite adequate volume resuscitation as suggested by the 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference.1
2.3. SNP selection
After referring to the dbSNP database of the National Center for Biotechnology Information (NCBI), SNPs located at functional regions (exons and 3′‐ and 5′‐untranslated regions) of the PPARγ gene were selected. Then, a literature search for published studies on disease‐related PPARγ SNPs was conducted using the PubMed database. Finally, tag SNPs were obtained from a Beijing Han population sample in the HapMap database, with the condition r 2 > 0.8. For the SNPs obtained, only those with a minor allele frequency (MAF) > 5% in the Chinese population were considered; therefore, a total of 13 SNPs was investigated (Table 1).
Table 1.
Information of the single‐nucleotide polymorphisms of the peroxisome proliferator‐activated receptor‐γ gene examined in this study PPARγ
| SNP ID | Location | Alleles | Call rate | HWE (P value) |
|---|---|---|---|---|
| control | ||||
| rs11128601 | intron1 | C < T | 99.84 | 1.00 |
| rs1175543 | intron6 | G < A | 99.84 | .81 |
| rs17036188 | intron1 | C < T | 99.84 | .16 |
| rs17817276 | intron1 | G < A | 99.84 | .82 |
| rs1801282 | nonsynon_exon1 | G < C | 99.84 | .24 |
| rs2920502 | 5′‐flanking | G < C | 99.84 | .13 |
| rs2972164 | intron1 | T < C | 99.84 | .35 |
| rs3856806 | synon_exon7 | T < C | 99.84 | 1.00 |
| rs4135275 | intron4 | A < G | 99.84 | .35 |
| rs4684846 | intron1 | A < G | 99.84 | .91 |
| rs4684847 | intron1 | T < C | 99.84 | .24 |
| rs709156 | intron6 | A < G | 99.84 | .06 |
| rs709158 | intron6 | G < A | 99.84 | .72 |
HWE, Hardy‐Weinberg equilibrium; ID, identification; SNP, single‐nucleotide polymorphism.
2.4. Genotyping
For every patient and control enrolled, a 3‐mL sample of venous blood was obtained, anti‐coagulated by sodium citrate and preserved at −80°C until used. The QIAGEN™ DNA Extraction Kit (Qiagen, Hilden, Germany) was used to extract the DNA from the peripheral venous blood. The improved multiplex ligation detection reaction (iMLDR) technique21, 22 was adopted for genotyping. Primers were designed according to the genome sequence provided by the dbSNP database. Negative controls were set during the polymerase chain reaction (PCR). After the PCR, 20 samples each from the sepsis and control groups were randomly selected in a double‐blinded manner for tests of repeatability, and a repeatability rate of 100% was observed. Haploview software23 was used to conduct linkage disequilibrium (LD) analysis, while the haplotype block was inferred based on the D′ value. For each haplotype block, haplotype inference was performed using Phase software.24, 25
2.5. Statistical analysis
SPSS software (version 19.0; SPSS Inc., Chicago, IL, USA) was adopted for the statistical analysis. Continuous variables are expressed as mean ± standard deviation (SD), and comparisons between groups were made using Student's t test or the Mann‐Whitney U test, depending on the distribution of the data. Categorical data are presented as frequencies and comparisons between groups were made using the χ2 test or Fisher's exact test. For the analyses, data were grouped as follows: patients with sepsis vs controls; sepsis shock vs no sepsis shock; mild MODS vs severe MODS; and death vs survival. Genotype frequencies for every SNP were counted for each of these various groups and analyzed using the goodness‐of‐fit test to determine whether they were in Hardy‐Weinberg equilibrium. Unconditional logistic regression analysis was utilized to assess the associations between each SNP genotype frequency and each inferred haplotype frequency with the susceptibility to sepsis and with clinical outcome under different genetic models. The following factors were corrected for during the logistic regression analysis: age, sex, body mass index (BMI), tobacco/alcohol use, comorbidities (hypertension, coronary heart disease [CHD], Type‐2 diabetes mellitus [T2DM], hyperlipidemia, and chronic obstructive pulmonary disease [COPD]), and (in the clinical outcome model but not susceptibility to sepsis model) APACHE II score. The P values, odds ratios (ORs), and 95% confidence intervals (CIs) were calculated. All statistical tests were two‐sided, and P values < .05 were considered statistically significant.
3. RESULTS
3.1. Characteristics of the study participants
A total of 310 patients with sepsis and 309 controls were enrolled. During the genotyping process, some unreliable samples (seven from the sepsis group and six from the control group) were excluded due to low effective DNA concentrations or serious contamination; therefore, the final analysis included 303 participants each in the sepsis and control groups. Table 2 lists the sociodemographic data and clinical characteristics of the study participants. The mean ages were 59.35 ± 18.79 (range 18‐95) years in the sepsis group and 59.50 ± 18.12 (range 19‐94) years in the control group. The sepsis and control groups did not differ significantly in terms of age, gender, BMI, tobacco use, alcohol use, or incidences of hypertension, CHD, T2DM, or hyperlipidemia (Table 2), although the sepsis group had more participants with COPD than the control group (6.93% vs 3.30%, P = .043).
Table 2.
Socio‐demographic data and clinical characteristics of individuals participants in the sepsis and control groups
| Index | Sepsis group (n = 303) | Control group (n = 303) | P value |
|---|---|---|---|
| Age (y), mean ± SD | 59.35 ± 18.79 | 59.50 ± 18.12 | .693 |
| Sex, n (%) | |||
| Male | 187 (61.72%) | 169 (55.78%) | .137 |
| Female | 116 (38.28%) | 134 (44.22%) | |
| BMI (kg/m2), mean ± SD | 23.51 ± 3.97 | 23.50 ± 3.35 | .911 |
| Tobacco use, n (%) | 88 (29.04%) | 75 (24.75%) | .264 |
| Alcohol use, n (%) | 33 (10.89%) | 44 (14.52%) | .099 |
| Hypertension, n (%) | 112 (36.96%) | 121 (39.93%) | .452 |
| Coronary heart disease, n (%) | 43 (14.19%) | 44 (14.52%) | .908 |
| Type‐2 diabetes mellitus, n (%) | 68 (22.44%) | 72 (23.76%) | .773 |
| Hyperlipidemia, n (%) | 20 (6.60%) | 18 (5.94%) | .738 |
| COPD, n (%) | 21 (6.93%) | 10 (3.30%) | .043 |
BMI, body mass index; COPD, chronic obstructive pulmonary disease; SD, standard deviation.
3.2. Clinical features in patients with sepsis
Additional clinical features of the patients with sepsis are shown in Table 3. The most common cause of sepsis was pulmonary infection (n = 180, 59.41%); 25 (8.25%) patients had infections at 2 or more sites (most commonly lungs and urinary tract). Microbial culture showed no microbial growth in 102 (33.66%) patients. The most common pathogens in patients with Gram‐negative bacteria (85/303, 28.05%) were Pseudomonas aeruginosa, Acinetobacter baumannii, and Stenotrophomonas maltophilia, while Staphylococcus aureus was most common in patients with Gram‐positive bacteria (22/303, 7.26%). Combined Gram‐positive/Gram‐negative infections were found in 76 (25.08%) patients. APACHE II score within 24 hours after the diagnosis of sepsis was 16.51 ± 6.06 points. Organ dysfunction most commonly occurred in the respiratory system (172/303, 56.77%).
Table 3.
Clinical characteristics of the septic patients
| Clinical characteristic | Value |
|---|---|
| Number of patients with sepsis | 303 |
| Site of infection, n (%) | |
| Lung | 180 (59.41%) |
| Biliary system | 60 (19.80%) |
| Gastrointestinal system | 18 (5.94%) |
| Liver abscess | 5 (1.65%) |
| Intracranial | 2 (0.66%) |
| Urinary tract system | 3 (0.99%) |
| Combined infectiona | 25 (8.25%) |
| Others | 10 (3.30%) |
| Pathogenic microbe, n (%) | |
| Gram‐positive bacterium | 22 (7.26%) |
| Gram‐negative bacterium | 85 (28.05%) |
| Fungus | 18 (5.94%) |
| Mixed | 76 (25.08%) |
| None | 102 (33.66%) |
| APACHE II score, mean ± SD | 16.51 ± 6.06 |
| Organ dysfunction, n (%) | |
| Respiratory system | 172 (56.77%) |
| Circulatory system | 86 (28.38%) |
| Kidney | 71 (23.43%) |
| Liver | 58 (19.14%) |
| Central nervous system | 49 (16.17%) |
| Coagulation system | 18 (5.94%) |
| MODS score, mean ± SD | 6.11 ± 3.09 |
| Clinical outcome, n (%) | |
| Septic shock | 74 (24.42%) |
| Severe organ dysfunction | 65 (21.45%) |
| Death | 79 (26.07%) |
MODS, multiple organ dysfunction syndrome; SD, standard deviation.
Infection at 2 or more sites.
3.3. Clinical outcomes in patients with sepsis
Septic shock occurred in 74/303 (24.42%) patients (Table 4). Patients who developed septic shock were significantly older than those who did not (66.38 ± 18.21 vs 56.78 ± 18.38 years, P < .001) and had higher APACHE II score, MODS score, and mortality rate (all P < .001; Table 4). Other than hypertension incidence, there were no significant differences in other clinical characteristics between patients with septic shock and those without (Table 4). Patients with severe organ dysfunction (65/303, 21.45%) had significantly higher APACHE II score, MODS score, and mortality rate than those with mild organ dysfunction (all P < .001); other clinical features were similar (Table 4). Mortality rate was 79/303 (26.07%). Patients who died were significantly older and had higher APACHE II and MODS scores than those who survived (P < .001; Table 4).
Table 4.
Characteristics of the patients with sepsis stratified by septic shock occurrence, severity of organ dysfunction, and mortality
| Index | Without septic shock | With septic shock | Mild organ dysfunction | Severe organ dysfunction | Survival | Death |
|---|---|---|---|---|---|---|
| n = 229 | n = 74 | n = 238 | n = 65 | n = 224 | n = 79 | |
| Age (y), mean ± SD | 56.78 ± 18.38a | 66.38 ± 18.21b | 58.41 ± 18.71 | 61.96 ± 18.88 | 56.47 ± 18.61a | 66.63 ± 17.24d |
| Sex, n (%) | ||||||
| Male | 139 (60.70%) | 48 (64.86%) | 146 (61.34%) | 41 (63.08%) | 135 (60.27%) | 52 (65.82%) |
| Female | 90 (39.30%) | 26 (35.14%) | 92 (38.66%) | 24 (36.92%) | 89 (39.73%) | 27 (34.18%) |
| BMI (kg/m2), mean ± SD | 23.63 ± 3.98 | 23.24 ± 3.91 | 23.51 ± 4.00 | 23.64 ± 3.92 | 23.62 ± 3.96 | 23.32 ± 3.98 |
| Tobacco use, n (%) | 72 (31.44%) | 16 (21.62%) | 72 (30.25%) | 16 (24.62) | 68 (30.36%) | 20 (25.32%) |
| Alcohol use, n (%) | 27 (11.79%) | 6 (8.11%) | 26 (10.92%) | 7 (10.77) | 22 (9.82%) | 11 (13.92%) |
| APACHE II score, mean ± SD | 14.80 ± 5.20a | 21.62 ± 5.34b | 14.97 ± 5.22a | 22.06 ± 5.36c | 15.11 ± 5.39a | 20.38 ± 5.95d |
| MODS score, mean ± SD | 5.13 ± 2.47a | 9.03 ± 2.90b | 4.81 ± 1.80a | 10.75 ± 2.13c | 5.37 ± 2.79a | 8.13 ± 2.96d |
| Hypertension, n (%) | 76 (33.19%)a | 36 (48.65%)b | 83 (34.87%) | 29 (44.62%) | 76 (33.93%) | 36 (45.57%) |
| Coronary heart disease, n (%) | 31 (13.54%) | 12 (16.22%) | 30 (12.61%) | 13 (20.00%) | 28 (12.50%) | 15 (18.99%) |
| Type‐2 diabetes mellitus, n (%) | 47 (20.52%) | 21 (28.38%) | 52 (21.85%) | 16 (24.62%) | 46 (20.54%) | 22 (27.85%) |
| Hyperlipidemia, n (%) | 12 (5.24%) | 8 (10.81%) | 15 (6.30%) | 5 (7.69%) | 13 (5.80%) | 7 (8.86%) |
| COPD, n (%) | 16 (6.99%) | 5 (6.76%) | 15 (6.30%) | 6 (9.23%) | 16 (7.14%) | 5 (6.33%) |
| Death, n (%) | 15 (6.55%)a | 64 (86.49%)b | 55 (23.11%)a | 24 (36.92%)c | ||
APACHE II, Acute Physiology and Chronic Health Evaluation II; BMI, body mass index; COPD, chronic obstructive pulmonary disease; MODS, multiple organ dysfunction syndrome; SD, standard deviation.
P < .001.
P < .001 vs patients without septic shock.
P < .001 vs patients with mild organ dysfunction.
P < .001 vs patients who survived.
3.4. Association of PPARγ gene polymorphisms with susceptibility to and clinical outcome of sepsis
The frequency distributions of all SNPs in the control group were in Hardy‐Weinberg equilibrium. The results of the logistic regression analysis, after adjusting for age, sex, BMI, tobacco use, alcohol use, and underlying diseases, are presented in Table 5. For rs2972164, the CT/CT+TT genotypes was associated with increased risk of sepsis when compared with the CC genotype (OR [95%CI]: 1.74 [1.05‐2.86], P = .03 and 1.72 [1.06‐2.80], P = .026, respectively), while the T allele was associated with increased the risk of sepsis relative to that of the C allele (1.64 [1.04‐2.58], P = .033). For rs1801282, the CG/CG+GG genotypes was related to decreased risk of sepsis when compared with the CC genotype (0.55 [0.33‐0.92], P = .024 and 0.57 [0.35‐0.95], P = .03, respectively), while the G allele showed a trend of lower risk of sepsis as compared with the C allele (0.62 [0.39‐1.01], P = .055). On the other hand, patients with the AG/AG+GG genotypes at rs4135275 had a higher risk of severe organ dysfunction than those with the AA genotype (2.66 [1.16‐6.09], P = .038 and 2.21 [1.00‐4.85], P = .042, respectively; Table 6). No differences in SNP frequency distributions were observed between patients with septic shock or death (data not shown).
Table 5.
Association between polymorphisms of the PPARγ gene and the susceptibility to sepsis
| SNP | Genotype | Sepsis group (n = 303) | Control group (n = 303) | OR (95% CI) | P value |
|---|---|---|---|---|---|
| rs2972164 | CC | 250 (82.51) | 266 (87.79) | 1 | |
| CT | 50 (16.50) | 35 (11.55) | 1.74 (1.05‐2.86) | .03 | |
| TT | 3 (0.99) | 2 (0.66) | 1.52 (0.24‐9.45) | .65 | |
| CC | 250 (82.51) | 266 (87.79) | 1 | ||
| CT+TT | 53 (17.49) | 37 (12.21) | 1.72 (1.06‐2.80) | .026 | |
| CC+CT | 300 (99.01) | 301 (99.34) | 1 | ||
| TT | 3 (0.99) | 2 (0.66) | 1.41 (0.23‐8.73) | .71 | |
| C allele | 550 (90.8) | 567 (93.6) | 1 | ||
| T allele | 56 (9.2) | 39 (6.4) | 1.64 (1.04‐2.58) | .033 | |
| rs1801282 | CC | 272 (89.77) | 257 (84.82) | 1 | |
| CG | 30 (9.90) | 46 (15.18) | 0.55 (0.33‐0.92) | .024 | |
| GG | 1 (0.33) | 0 (0.00) | ‐ | 1 | |
| CC | 272 (89.77) | 257 (84.82) | 1 | ||
| CG+GG | 31 (10.23) | 46 (15.18) | 0.57 (0.35‐0.95) | .03 | |
| CC+CG | 302 (99.67) | 303 (100.00) | 1 | ||
| GG | 1 (0.33) | 0 (0.00) | ‐ | .23 | |
| C allele | 574 (94.72) | 558 (92.08) | 1 | ||
| G allele | 32 (5.28) | 48 (7.92) | 0.62 (0.39‐1.01) | .055 |
Data shown as n (%).
Bold values meant values which were statistically significant.
CI, confidence interval; OR, odds ratio.
Table 6.
Association between polymorphisms of the PPARγ gene and organ dysfunction of sepsis
| SNP | Genotype | Mild (n = 238) | Severe (n = 65) | OR (95% CI) | P value |
|---|---|---|---|---|---|
| rs4135275 | AA | 71 (29.83) | 15 (23.08) | 1 | |
| AG | 130 (54.62) | 42 (64.62) | 2.66 (1.16‐6.09) | .038 | |
| GG | 37 (15.55) | 8 (12.31) | 1.18 (0.37‐3.71) | .78 | |
| AA | 71 (29.83) | 15 (23.08) | 1 | ||
| AG+GG | 167 (70.17) | 50 (76.92) | 2.21 (1.00‐4.85) | .042 | |
| AA+AG | 201 (84.45) | 57 (87.69) | 1 | ||
| GG | 37 (15.55) | 8 (12.31) | 0.65 (0.24‐1.75) | .38 | |
| A allele | 272 (57.14) | 72 (55.38) | 1 | ||
| G allele | 204 (42.86) | 58 (44.62) | 1.22 (0.76‐1.96) | .41 |
Data shown as n (%).
Bold values meant values which were statistically significant.
CI, confidence interval; OR, odds ratio.
In this study, we inferred 3 haplotype blocks from the PPARγ gene (Figure 1). In haplotype block 1, haplotype TAT (rs2972164, rs4684846 and rs17036188) was found to correlate with increased risk of sepsis when compared with haplotype CAT (1.66 [1.03‐2.7], P = .038; Table 7). No mutation was observed to correlate with septic shock or death.
Figure 1.
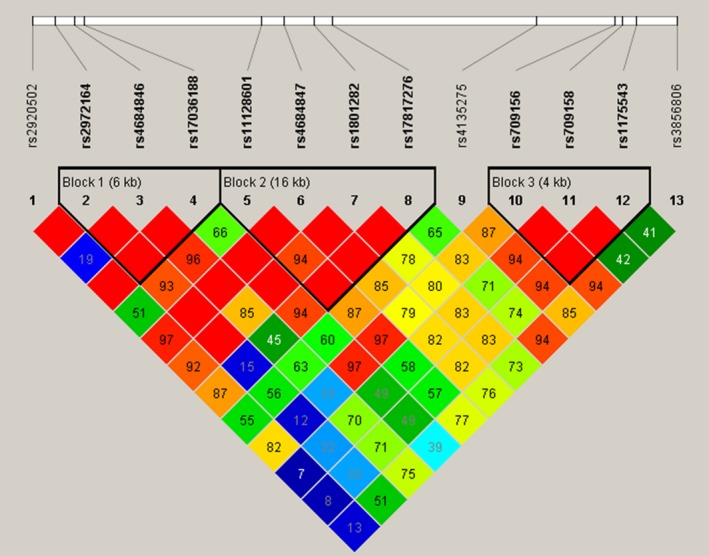
Haplotype blocks of the PPARγ gene found in the present study. Graphical representation of the SNP locations and linkage disequilibrium (LD) structure of PPARγ obtained using 13 SNPs in 303 North China Han patients with sepsis. The SNP positions are described in Table 1. Three haplotype blocks were defined using Haploview software. The SNP names are listed at the top of the figure. The numbers in the squares are D′ values (|D′| × 100). The measure of LD (D′) among all possible pairs of SNPs is shown by the shade of red where white represents very low D′ and dark red represents very high D′
Table 7.
Association between haplotype from haplotype block 1 in the PPARγ gene and sepsis
| Haplotype | Susceptibility to sepsis | Septic shock | Severity of organ dysfunction | Death | ||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| CAT | 1 | ‐ | 1 | ‐ | 1 | ‐ | 1 | ‐ |
| CGC | 1.066 (0.807‐1.409) | .653 | 1.201 (0.695‐2.075) | .511 | 1.135 (0.651‐1.980) | .656 | 1.316 (0.811‐2.136) | .267 |
| CGT | 0.949 (0.689‐1.307) | .748 | 1.523 (0.818‐2.837) | .185 | 1.159 (0.613‐2.191) | .649 | 0.952 (0.535‐1.695) | .868 |
| TAT | 1.657 (1.028‐2.671) | .038 | 0.569 (0.228‐1.416) | .225 | 0.699 (0.282‐1.735) | .44 | 0.760 (0.339‐1.703) | .505 |
Bold values meant values which were statistically significant.
CI, confidence interval.
4. DISCUSSION
This study analyzed the association between 13 common SNPs from the PPARγ gene and the occurrence and progression of sepsis in a North China Han population. The main finding was that there were 3 SNPs related to the occurrence or progression of sepsis: (i) rs1801282 (C→G) was associated with a reduced risk of sepsis; (ii) rs2972164 (C→T) was associated with an increased risk of sepsis; and (iii) rs4135275 (G→A) was associated with an increase in the severity of organ dysfunction following the occurrence of sepsis. No SNP was found to correlate with septic shock or death. In addition, the haplotype TAT (rs2972164, rs4684846, and rs17036188), containing the rs2972164 T allele, was related to increased risk of sepsis, whereas haplotype CTGA, containing the rs1801282 G allele, was related to decreased risk of sepsis (P = .09, not shown in the text), which was consistent with the individual SNP analyses. To the best of our knowledge, this is the first study to investigate the influences of such an extensive range of PPARγ gene SNPs (13 in total). These results suggest that certain PPARγ gene mutations could be correlated with the occurrence and progression of sepsis.
The PPARγ gene is located on chromosome 3p25 and contains 9 exons. The rs1801282 polymorphism, located in exon 2, is the most extensively studied SNP in the PPARγ gene and results in a CCA→GCA nonsynonymous mutation encoding a proline→alanine substitution at amino acid residue 12 (Pro12Ala). The incidence of this polymorphism varies between different populations (fluctuating between 2% and 25%) and is significantly lower in the Asian population than the Caucasian population.26 In this study, the frequency of the G allele was 7.92% in the control group, similar to that reported by Wang et al27 (7.2%), Shi et al28 (7.9%), and Pei et al29 (7.55%). Due to the low frequency and sample size, occurrence of the homozygous mutation (GG) of this SNP was zero in the control group and 1 in the sepsis group, respectively.
A notable finding of our study was that rs1801282 (C→G) appeared to associate with decreased risk of sepsis. In a previous study analyzing the correlation between rs1801282 and sepsis, Ma et al18 enrolled 308 patients with sepsis and 345 healthy controls from the Department of Emergency and ICU of the Affiliated Hospital of Guangdong Medical College and found that rs1801282 (G) was not associated with susceptibility to sepsis, in contrast to our findings. However, patients carrying the G allele had lower disease severity and a better prognosis than those with the C allele.18 The reasons underlying the apparent inconsistency between our study and that of Ma et al18 remain unknown, but may include a small sample size in both studies or differences in the ethnicities of the participants, the pathogenic microbes involved or environmental factors. Nonetheless, both our study and that of Ma et al18 implicate rs1801282 (C→G) as having a protective role against sepsis, either in terms of the risk of acquiring it or in terms of its prognosis.
Several previous studies have examined the correlation between rs1801282 and T2DM, with most suggesting that the rs1801282 variant was a metabolic protective factor. A recent meta‐analysis revealed that this mutation was related to low risk of T2DM,30 and Pei et al29 also found similar results in the Chinese population. In a Caucasian population, Barbieri et al31 found that this variant was associated with low BMI, insulin resistance, and low triglyceride level. Studies on the correlation between rs1801282 and inflammatory diseases are relatively fewer. Some studies have indicated that rs1801282 (G) correlated with low risk of inflammatory bowel disease (IBD)32 whereas others ,have suggested no effect.33 A recent meta‐analysis34 concluded that the rs1801282 (GG) variant was associated with low risk of Crohn's disease in the European Caucasian population but not the East Asian population. Penyige et al35 found that the rs1801282 (G) variant was associated with decreased risk of COPD in an Austrian population. Overall, the available studies indicate that the rs1801282 variant plays a protective role in inflammatory diseases, which would be consistent with our findings that rs1801282 (G) was related to decreased risk of sepsis and that the CTGA haplotype (rs11128601, rs4684847, rs1801282, and rs17817276) also demonstrated a trend toward decreased risk of sepsis. Functional studies have shown that the proline→alanine substitution (Pro12Ala) results in a decreased binding capacity of PPARγ to the target gene, reducing the role of PPARγ in transcriptional activation of target genes.26 The anti‐inflammatory effects of PPARγ are mainly realized through transrepression, ie, PPARγ interferes with the binding of nuclear factor‐κB (NF‐κB) and activator protein‐1 (AP‐1) to their target genes and represses their transcriptions of pro‐inflammatory genes; therefore, direct binding between PPARγ and a target gene is not required for this anti‐inflammatory function.9 Further functional validation studies are required to determine the specific mechanisms by which rs1801282 has relevance with sepsis.
In this study, the other 2 SNPs that associated with sepsis were both located in the intron: rs2972164 (C→T) was associated with an increased risk of sepsis, while rs4135275 (G→A) was associated with an increase in the severity of organ dysfunction following the occurrence of sepsis. To the best of our knowledge, no previous investigations have examined the possible relevance of rs2972164 to sepsis, and none have explored the association of rs4135275 with sepsis or indeed any other diseases. Peters et al36 reported that rs2972164 may modify the effectiveness of statins in reducing the risk of myocardial infarction, and Black et al37 demonstrated that rs2972164 was associated with a reduced sensitivity to insulin. Intron mutation can affect gene functions via impacts upon the cleavage, stability, and localization of mRNA. The region of intron 1 plays an important role in the regulation of gene transcription, and mutation within this region may affect gene expression by altering the binding site of the enhancer.38, 39, 40 It is possible that rs2972164 and rs4135275 may lower the expression of PPARγ by reducing the binding between the enhancer and transcriptional activator, thereby influencing the occurrence and progression of sepsis. However, it cannot be ruled out that these SNPs manifest an association with sepsis due to linkage with other sepsis‐related SNPs.
Gao et al19 conducted a study in 734 trauma patients in Chongqing district, China, and selected 3 SNPs (rs4684846, rs10865710, and rs1822825) from the PPARγ gene. Of the 734 patients, 300 experienced sepsis, and rs10865710 was found to associate with an increased risk of sepsis and higher MODS score. The rs10865710 and rs1822825 SNPs selected in their study are tag SNPs to rs11128601 and rs1175543 selected in our study. However, we did not find that rs11128601 or rs1175543 were associated with the risk of sepsis or its prognosis. As discussed above, possible reasons for this discrepancy include small sample size or differences in ethnicities, pathogenic microbes involved, or environmental factors.
The present study enrolled only subjects native to the North China area, minimizing the impact of population stratification on the results. Furthermore, since sepsis is a multifactorial disease influenced by many genes and mutations, we decided not to concentrate on just one or a small number of SNPs located at the coding or promoter region, as previous studies have done, but instead to select a wider range of SNPs (13 in total) through a combination of methods, so as to maximally represent the common genetic information from the PPAR gene. In addition to the individual SNP analyses, haplotype block and haplotype were also inferred in the current study. Since a haplotype usually has several SNPs and thus contains more genetic information, the statistical efficiency of haplotype analysis is higher. It is noteworthy that the results from the individual SNP analyses are consistent with those from the haplotype analysis, further validating the reliability of our data.
This study has some limitations. First, since all the patients with sepsis were enrolled from a hospital while the controls were enrolled from a population attending for physical examination during the same period, there is selection bias in the study. However, to maintain a minimum level of potential confounding factors, the cases and controls were matched in every aspect possible, including age, sex, tobacco and alcohol use, hypertension, CHD, hyperlipidemia, and T2DM. Second, due to inadequate statistical power resulting from a small sample size, the effects of some SNPs with a low penetrance could not be evaluated. Third, for similar reasons, certain associations may not have been revealed in the subgroup analysis. Finally, SNPs were selected from the databases of dbSNP and HapMap. Since these 2 databases are still improving with the constant discovery of emerging SNPs, our selection cannot represent all the variants of the PPARγ gene. Thus, it remains possible that other SNPs may also associate with sepsis. Our next step is to test the current results and conduct functional analyses in a larger and wider population consisting of various ethnicities.
In conclusion, this study found that 3 SNPs were associated with the occurrence or progression of sepsis: (i) rs1801282 (C→G) was associated with a reduced risk of sepsis; (ii) rs2972164 (C→T) was associated with an increased risk of sepsis; and (iii) rs4135275 (G→A) was associated with more severe organ dysfunction following the development of sepsis. No SNP was found to associate with septic shock or death. These results indicate that PPARγ gene mutations are relative to the development and progression of sepsis.
ACKNOWLEDGMENTS
This work was supported by the grant from the National Natural Science Foundation of China (No.81701890)
Contributor Information
Wenhui Wan, Email: wanwhnj@sina.com.
Fang Huang, Email: 88100224@qq.com.
REFERENCES
- 1. Levy MM, Fink MP, Marshall JC, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med. 2003;31:1250‐1256. [DOI] [PubMed] [Google Scholar]
- 2. Angus DC, Linde‐Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303‐1310. [DOI] [PubMed] [Google Scholar]
- 3. Andrades ME, Morina A, Spasic S, Spasojevic I. Bench‐to‐bedside review: sepsis ‐ from the redox point of view. Crit Care. 2011;15:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan VS, Chan KY, Chen Y, et al. Homozygous L‐SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat Genet. 2006;38:38‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sørensen TI, Nielsen GG, Andersen PK, Teasdale TW. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988;318:727‐732. [DOI] [PubMed] [Google Scholar]
- 6. Sutherland AM, Walley KR. Bench‐to‐bedside review: association of genetic variation with sepsis. Crit Care. 2009;13:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wong HR. Genetics and genomics in pediatric septic shock. Crit Care Med. 2012;40:1618‐1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kota BP, Huang HW, Roufogalis BD. An overview on biological mechanisms of PPARs. Pharmacol Res. 2005;51:85‐94. [DOI] [PubMed] [Google Scholar]
- 9. Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365‐376. [DOI] [PubMed] [Google Scholar]
- 10. Croasdell A, Duffney PF, Kim N, Lacy SH, Sime PJ, Phipps RP. PPARgamma and the Innate Immune System Mediate the Resolution of Inflammation. PPAR Res. 2015;2015:549691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Araujo CV, Campbell C, Goncalves‐de‐Albuquerque CF, et al. A PPARgamma agonist enhances bacterial clearance through neutrophil extracellular trap formation and improves survival in sepsis. Shock. 2016;45:393‐403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu P, Lou XL, Chen C, Yang ZW. Effects of peroxisome proliferator‐activated receptor‐gamma activation on apoptosis in rats with acute pancreatitis. Dig Dis Sci. 2013;58:3516‐3523. [DOI] [PubMed] [Google Scholar]
- 13. Annese V, Rogai F, Settesoldi A, Bagnoli S. PPARgamma in inflammatory bowel disease. PPAR Res. 2012;2012:620839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mingfeng D, Xiaodong M, Yue L, Taikui P, Lei X, Ming L. Effects of PPAR‐gamma agonist treatment on LPS‐induced mastitis in rats. Inflammation. 2014;37:1919‐1924. [DOI] [PubMed] [Google Scholar]
- 15. Lakshmi SP, Reddy AT, Zhang Y, et al. Down‐regulated peroxisome proliferator‐activated receptor gamma (PPARgamma) in lung epithelial cells promotes a PPARgamma agonist‐reversible proinflammatory phenotype in chronic obstructive pulmonary disease (COPD). J Biol Chem. 2014;289:6383‐6393. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Lea S, Plumb J, Metcalfe H, et al. The effect of peroxisome proliferator‐activated receptor‐gamma ligands on in vitro and in vivo models of COPD. Eur Respir J. 2014;43:409‐420. [DOI] [PubMed] [Google Scholar]
- 17. Zingarelli B, Cook JA. Peroxisome proliferator‐activated receptor‐gamma is a new therapeutic target in sepsis and inflammation. Shock. 2005;23:393‐399. [DOI] [PubMed] [Google Scholar]
- 18. Ma G, Wang H, Mo G, et al. The Pro12Ala Polymorphism of PPAR‐gamma Gene Is Associated with Sepsis Disease Severity and Outcome in Chinese Han Population. PPAR Res. 2014;2014:701971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao JW, Zeng L, Zhang AQ, et al. Identification of haplotype tag single‐nucleotide polymorphisms within the PPAR family genes and their clinical relevance in patients with major trauma. Int J Environ Res Public Health. 2016;13:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marshall JC, Cook DJ, Christou NV, Bernard GR, Sprung CL, Sibbald WJ. Multiple organ dysfunction score: a reliable descriptor of a complex clinical outcome. Crit Care Med. 1995;23:1638‐1652. [DOI] [PubMed] [Google Scholar]
- 21. Wang X, Zhang Z, Liu W, et al. Impacts and interactions of PDGFRB, MMP‐3, TIMP‐2, and RNF213 polymorphisms on the risk of Moyamoya disease in Han Chinese human subjects. Gene. 2013;526:437‐442. [DOI] [PubMed] [Google Scholar]
- 22. Chen ZJ, Zhao H, He L, et al. Genome‐wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43:55‐59. [DOI] [PubMed] [Google Scholar]
- 23. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263‐265. [DOI] [PubMed] [Google Scholar]
- 24. Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978‐989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162‐1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jeninga EH, Gurnell M, Kalkhoven E. Functional implications of genetic variation in human PPARgamma. Trends Endocrinol Metab. 2009;20:380‐387. [DOI] [PubMed] [Google Scholar]
- 27. Wang F, Han XY, Ren Q, et al. Effect of genetic variants in KCNJ11, ABCC8, PPARG and HNF4A loci on the susceptibility of type 2 diabetes in Chinese Han population. Chin Med J (Engl). 2009;122:2477‐2482. [PubMed] [Google Scholar]
- 28. Shi H, Yu X, Li Q, et al. Association between PPAR‐gamma and RXR‐alpha gene polymorphism and metabolic syndrome risk: a case‐control study of a Chinese Han population. Arch Med Res. 2012;43:233‐242. [DOI] [PubMed] [Google Scholar]
- 29. Pei Q, Huang Q, Yang GP, et al. PPAR‐gamma2 and PTPRD gene polymorphisms influence type 2 diabetes patients’ response to pioglitazone in China. Acta Pharmacol Sin. 2013;34:255‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gouda HN, Sagoo GS, Harding AH, Yates J, Sandhu MS, Higgins JP. The association between the peroxisome proliferator‐activated receptor‐gamma2 (PPARG2) Pro12Ala gene variant and type 2 diabetes mellitus: a HuGE review and meta‐analysis. Am J Epidemiol. 2010;171:645‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barbieri M, Rizzo MR, Papa M, et al. Role of interaction between variants in the PPARG and interleukin‐6 genes on obesity related metabolic risk factors. Exp Gerontol. 2005;40:599‐604. [DOI] [PubMed] [Google Scholar]
- 32. Poliska S, Penyige A, Lakatos PL, et al. Association of peroxisome proliferator‐activated receptor gamma polymorphisms with inflammatory bowel disease in a Hungarian cohort. Inflamm Bowel Dis. 2012;18:472‐479. [DOI] [PubMed] [Google Scholar]
- 33. Mwinyi J, Grete‐Wenger C, Eloranta JJ, Kullak‐Ublick GA. The Impact of PPARgamma Genetic Variants on IBD Susceptibility and IBD Disease Course. PPAR Res. 2012;2012:349469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang ZF, Yang N, Zhao G, Zhu L, Wang LX. Association between the Pro12Ala polymorphism of peroxisome proliferator‐activated receptor gamma 2 and inflammatory bowel disease: a meta‐analysis. PLoS ONE. 2012;7:e30551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Penyige A, Poliska S, Csanky E, et al. Analyses of association between PPAR gamma and EPHX1 polymorphisms and susceptibility to COPD in a Hungarian cohort, a case‐control study. BMC Med Genet. 2010;11:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peters BJ, Pett H, Klungel OH, et al. Genetic variability within the cholesterol lowering pathway and the effectiveness of statins in reducing the risk of MI. Atherosclerosis. 2011;217:458‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Black MH, Wu J, Takayanagi M, et al. Variation in PPARG is associated with longitudinal change in insulin resistance in Mexican Americans at risk for type 2 diabetes. J Clin Endocrinol Metab. 2015;100:1187‐1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285‐298. [DOI] [PubMed] [Google Scholar]
- 39. Abbott W, Gane E, Winship I, Munn S, Tukuitonga C. Polymorphism in intron 1 of the interferon‐gamma gene influences both serum immunoglobulin E levels and the risk for chronic hepatitis B virus infection in Polynesians. Immunogenetics. 2007;59:187‐195. [DOI] [PubMed] [Google Scholar]
- 40. Zhou L, Nian M, Gu J, Irwin DM. Intron 1 sequences are required for pancreatic expression of the human proglucagon gene. Am J Physiol Regul Integr Comp Physiol. 2006;290:R634‐R641. [DOI] [PubMed] [Google Scholar]


