Abstract
Aims
To investigate the impact of 4 single nucleotide polymorphisms (SNPs) within ABO gene and their gene‐gene interactions on ischemic stroke (IS) susceptibility in Chinese Han population.
Methods
A total of 1993 participants (1375 males, 618 females) were selected, including 991 IS patients and 1002 normal controls. The SNPstats (http://bioinfo.iconcologia.net/SNPstats) was used for Hardy‐Weinberg equilibrium (HWE) test. Generalized multifactor dimensionality reduction (GMDR) was used to screen the best interaction combination among 4 SNPs within ABO gene. Logistic regression was performed to calculate the ORs (95%CI) for interaction between SNPs.
Results
Both rs579459 and rs505922 within ABO gene were associated with IS risk in additive and dominant models. IS risks were higher in those with minor alleles of rs579459 and rs505922 than those with wild‐type homozygotes, OR (95%CI) were 1.62 (1.19‐2.10) and 1.69 (1.23‐2.18), respectively. We did not find any relation of rs651007 and rs529565 with IS risk in both additive and dominant models. GMDR model indicated a significant two‐locus model (P = .0010) involving rs505922 and rs579459, indicating a potential interaction between rs505922 and rs579459, the cross‐validation consistency of the two‐locus models was 9/10, and the testing accuracy was 60.72%. We also found that participants with rs505922‐ TC/CC and rs579459‐ TC/CC genotype have the highest IS risk, compared to participants with rs505922‐ TT and rs579459‐ TT genotype, OR (95%CI) was 2.94 (1.28‐4.66).
Conclusions
We found that rs579459 and rs505922 within ABO gene and their interaction were both associated with increased IS risk in Chinese population.
Keywords: ABO gene, interaction, ischemic stroke, single nucleotide polymorphism
1. INTRODUCTION
Stroke is one of the main causes of death and adult disability around the world.1, 2 In China, there were 2.5 million new stroke cases each year and 7.5 million stroke survivors.2 In the subtypes of the stroke, the ischemic stroke (IS) is the most type, and about 43%‐79% of all strokes are ischemic in China.2, 3 Increasing evidence indicated that IS was a complex clinical syndrome resulting from environmental and genetic factors.4, 5 Many environmental risk factors for IS have been reported, including age, hypertension, type 2 diabetes mellitus (T2DM), obesity, smoking. However, these conventional risk factors could not completely explain all IS risk, family and twin‐based studies demonstrated that genetic factors also play a key role in the development of IS.6, 7
The ABO blood group system is encoded by ABO gene, which located around 9q34.2, encodes glycosyltransferases, catalyze the transfer to different carbohydrate groups onto the H antigen, thus forming A and B antigens of the ABO system.8, 9 In recent years, several single nucleotide polymorphisms (SNPs) have been reported, and the genetic variants of ABO gene were associated with several diseases, including coronary artery disease (CAD),10 venous thromboembolism,11 however, just few studies focused on the association between ABO gene SNPs and IS were reported and the results of these studies were inconsistent.12, 13, 14 In addition, no study focused on the impact of interaction among several SNPs within ABO gene on IS risk, particularly in Chinese population. So the aim of this study was to investigate the impact of ABO gene SNPs, and their gene‐gene interactions on IS risk based on Chinese population with a relatively larger sample size.
2. MATERIALS AND METHODS
2.1. Subjects
Our study sample recruited 1993 participants (1375 males, 618 females) between July 2011 and March 2015 from the first Affiliated Hospital of Jinan University and the People's Hospital of Maoming, including 991 IS patients and 1002 normal controls. The mean age of all participants was 67.9 ± 15.2 years. The diagnosis of IS the subtype of IS were determined in accordance with World Health Organization criteria15, 16 and the original TOAST20 (Trial of ORG 10172 in Acute Stroke Treatment) criteria. All patients were diagnosed by computed tomography (CT) or magnetic resonance imaging (MRI) within 48 hours after the admission. Blood vessels were evaluated with neck vascular ultrasound and brain color Doppler as well as CT angiography or magnetic resonance angiography. Controls were matched by approximate 1:1 matched to patients on the basis of age (±2 years) and sex. Normal controls with family history of IS were excluded. Written informed consent was obtained from all participants. Both doctors and study subjects provided consent to participate in this study. The protocol of this study was approved by the Ethics Committee of Jinan University.
2.2. Definition
Current cigarette smokers were those who self‐reported smoking cigarettes at least once a day for 1 year or more. Alcohol consumption was expressed as the sum of milliliters of alcohol per week from wine, beer, and spirits. Hypertension was defined as systolic blood pressure (SBP) ≥140 mm Hg and/or diastolic blood pressure (DBP) ≥90 mm Hg and/or use of antihypertensive medication. The criteria for the diagnosis of type 2 diabetes mellitus (T2DM) included a fasting glucose ≥126 mg/dL (7.0 mmol/L), or a 2 hours postprandial blood glucose ≥200 mg/dL (11.0 mmol/L), or if hypoglycemic therapy (oral agents or insulin) had been started in the interim.
2.3. Genomic DNA extraction and genotyping
SNPs within ABO gene were evaluated and selected using the HapMap database (https://hapmap.ncbi.nlm.nih.gov/) according to the following criterion: (1) a minor allele frequency (MAF) >5%; (2) which have been reported associations with IS or IS related risk factors in previous studies. At last, a total of 4 SNPs within ABO gene were selected for genotyping: rs505922, rs579459, rs651007, and rs529565. Genomic DNA from participants was extracted from EDTA‐treated whole blood, using the DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions and stored at −20°C until use. Genotyping for 4 SNPs were tested using Sequenom MassARRAY platform (San Diego, U.S) at CapitalBio Corporation (Beijing, China). Genomic DNA was isolated from human peripheral blood samples of each individual through Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA). DNA concentration was determined by DNA spectrophotometer (ND‐1000, NanoDrop, Wilmington, NC, USA). Specific assays including a locus‐specific PCR reaction based on a locus‐specific primer extension reaction were designed using the MassARRAY Assay Design software package (v3.1 Sequenom Inc., San Diego, CA, USA). Mass determination was carried out with the MALDI‐TOF mass spectrometer and Mass ARRAY Type 4.0 software was used for data acquisition.
Genotyping results were confirmed by randomly assaying 8% of the original specimens for replication to exclude genotyping errors. There were no discrepancies between genotypes determined in duplicate.
2.4. Statistical analysis
SPSS 22.0 software package (SPSS Inc, Chicago, IL, USA) for Windows 7 (Microsoft Corp, Redmond, WA, USA) was used for statistical analyses in this study. The means and SDs were calculated for normally distributed continuous variables and compared using Student's t test, percentages were calculated for categorical variables and analyzed using χ2 test. The SNPstats (http://bioinfo.iconcologia.net/SNPstats) was used to assess the Hardy‐Weinberg equilibrium for genotype frequencies and association between SNPs and IS. Generalized multifactor dimensionality reduction (GMDR)17 was used to screen the best interaction combination among 4 SNPs within ABO gene. Interaction between SNPs within ABO gene in relation to the risk of IS was estimated by multiple logistic regressions, ORs (95%CI) for interaction between SNPs were calculate. All reported P‐values were two‐tailed, and to correct for multiple testing we defined a Bonferroni corrected‐threshold in different tables.
3. RESULTS
Table 1 shows the general and clinical characteristics of all study participants in case and control group. A total of 1993 participants (1375 males, 618 females) were selected, including 991 IS patients and 1002 normal controls. The mean age of all participants was 67.9 ± 15.2 years. The rate of smoking, alcohol drinking, hypertension, T2DM and the means of body mass index (BMI), fasting plasma glucose (FPG), triglyceride (TG) and total cholesterol (TC) are significantly higher in cases than that in controls, the mean of high‐density lipoprotein (HDL) was lower in cases than that in controls. The mean age and rate of males were not different between cases and controls.
Table 1.
General characteristics of 1993 study participants in case and control group
| Variables | Case group (n = 991) | Control group (n = 1002) | P‐values |
|---|---|---|---|
| Age (y) | 68.3 ± 15.9 | 67.6 ± 16.3 | .332 |
| Males, N (%) | 680 (68.6) | 695 (69.4) | .720 |
| BMI(kg/m2) | 24.6 ± 9.3 | 23.7 ± 9.6 | .034 |
| FPG (mmol/L) | 7.1 ± 3.5 | 6.1 ± 3.7 | <.001 |
| TG (mmol/L) | 1.5 ± 0.7 | 1.3 ± 0.8 | <.001 |
| TC (mmol/L) | 4.8 ± 1.2 | 4.4 ± 1.3 | <.001 |
| HDL (mmol/L) | 1.26 ± 0.8 | 1.31 ± 0.9 | <.001 |
| Smoking, N (%) | 473 (47.7) | 368 (36.7) | <.001 |
| Alcohol drinking, N (%) | 512 (51.7) | 428 (42.7) | <.001 |
| T2DM, N (%) | 329 (33.2) | 192 (19.2) | <.001 |
| Hypertension, N (%) | 548 (55.3) | 327 (32.6) | <.001 |
BMI, body mass index; FPG, fast plasma glucose; HDL, high‐density lipoprotein; TC, total cholesterol; TG, triglyceride.
Means ± standard deviation for age, BMI, FPG, TC, TG, HDL; Number and percentages for males, smokers, drinkers.
Table 2 shows the frequencies of genotypes and alleles for 4 SNPs and results for analysis on association between SNPs within ABO gene and IS risk. Logistic regression analysis showed that rs579459 and rs505922 within ABO gene were both associated with IS risk in additive and dominant models, after adjustment for gender, age, smoking, drinking, TC, TG, HDL. The carriers of homozygous and heterozygous mutant of rs579459 and rs505922 are associated with increased IS risk than those with wild‐type homozygotes, OR (95%CI) were 1.62 (1.19‐ 2.10) and 1.69 (1.23‐2.18), respectively. We did not find any relation of rs651007 and rs529565 with IS risk in both additive and dominant models.
Table 2.
Description for allele and genotype frequencies and analysis on association between 4 SNPs and IS risk
| SNP | Genotypes and alleles | Frequencies N (%) | OR (95%CI)a | P‐values | HWE test for controls | |
|---|---|---|---|---|---|---|
| Cases (n = 991) | Controls (n = 1002) | |||||
| rs579459 | Additive | |||||
| TT | 494 (49.8) | 639 (63.8) | 1.00 | 0.257 | ||
| TC | 396 (40.0) | 315 (31.4) | 1.59 (1.21‐2.06) | <.001 | ||
| CC | 101 (10.2) | 48 (4.8) | 1.73 (1.15‐2.33) | <.001 | ||
| Dominant | ||||||
| TT | 494 (49.8) | 639 (63.8) | 1.00 | |||
| TC+CC | 497 (50.2) | 363 (36.2) | 1.62 (1.19‐2.10) | <.001 | ||
| Allele, C (%) | 598 (30.2) | 411 (20.5) | ||||
| rs651007 | Additive | |||||
| CC | 565 (57.0) | 597 (59.6) | 1.00 | 0.422 | ||
| CT | 354 (35.7) | 347 (34.6) | 1.08 (0.72‐1.51) | .431 | ||
| TT | 72 (7.3) | 58 (5.8) | 1.23 (0.66‐1.92) | .723 | ||
| Dominant | ||||||
| CC | 565 (57.0) | 597 (59.6) | 1.00 | |||
| CT+TT | 426 (43.0) | 405 (40.4) | 1.12 (0.70‐1.61) | .665 | ||
| Allele, T (%) | 498 (25.1) | 463 (23.1) | ||||
| rs505922 | Additive | |||||
| TT | 511 (51.6) | 657 (65.6) | 1.00 | 0.652 | ||
| TC | 406 (41.0) | 306 (30.5) | 1.65 (1.26‐2.12) | <.001 | ||
| CC | 74 (7.5) | 39 (3.9) | 1.83 (1.14‐2.64) | <.001 | ||
| Dominant | ||||||
| TT | 511 (51.6) | 657 (65.6) | 1.00 | |||
| TC+CC | 480 (48.4) | 345 (34.4) | 1.69 (1.23‐2.18) | <.001 | ||
| Allele, C (%) | 554 (28.0) | 384 (19.2) | ||||
| rs529565 | Additive | |||||
| TT | 521 (52.6) | 578 (57.7) | 1.00 | 0.195 | ||
| TC | 383 (38.6) | 356 (35.5) | 1.33 (0.81‐1.94) | .526 | ||
| CC | 87 (8.8) | 68 (6.8) | 1.51 (0.70‐2.41) | .742 | ||
| Dominant | ||||||
| TT | 521 (52.6) | 578 (57.7) | 1.00 | |||
| TC+CC | 470 (47.4) | 424 (42.3) | 1.40 (0.77‐2.00) | .705 | ||
| Allele, C (%) | 557 (28.1) | 492 (24.6) | ||||
Adjusted for gender, age, smoking, drinking, TC, TG, HDL. Bonferroni correction threshold: P < .00417.
Generalized multifactor dimensionality reduction model was used to screen the best interaction combination among 4 SNPs within ABO gene. Table 3 summarized the results obtained from GMDR analysis, which indicated a significant two‐locus model (P = .0010) involving rs505922 and rs579459, indicating a potential interaction between rs505922 and rs579459, the cross‐validation consistency of the two‐locus models was 9/10, and the testing accuracy was 60.72%. To obtain the odds ratios and 95% CI for the joint effects, we conducted an interaction analysis using logistic regression (Figure 1). We found that participants with rs505922‐ TC/CC and rs579459‐ TC/CC genotype have the highest IS risk, compared to participants with rs505922‐ TT and rs579459‐ TT genotype, OR (95%CI) was 2.94 (1.28‐4.66).
Table 3.
Generalized multifactor dimensionality reduction analysis on the best gene‐gene interaction combinations
| Locus no. | Best combination | Cross‐validation consistency | Testing accuracy | P‐valuesa |
|---|---|---|---|---|
| 2 | rs505922 rs579459 | 9/10 | 0.6072 | .0010 |
| 3 | rs505922 rs579459 rs651007 | 7/10 | 0.5399 | .3770 |
| 4 | rs505922 rs579459 rs651007 rs529565 | 6/10 | 0.4958 | .4258 |
Adjusted for gender, age, smoking, drinking, TC, TG, HDL.
Figure 1.
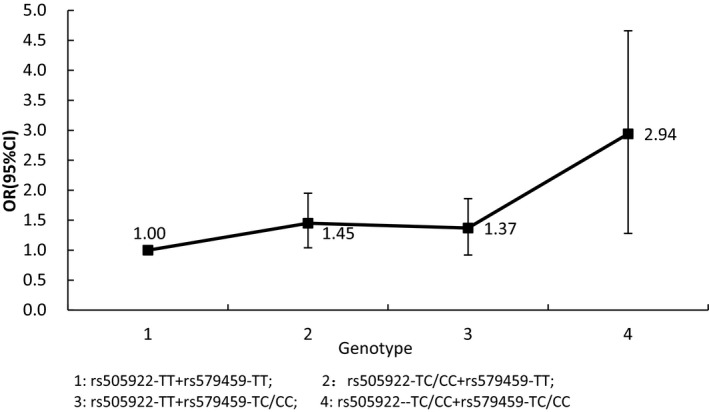
Logistic regression for interaction between rs505922 and rs579459
4. DISCUSSION
In this study, we found that both rs505922 and rs579459 within ABO gene were associated with increased IS risk in additive and dominant models. We did not find any relation of rs651007 and rs529565 in ABO gene with IS risk in two models. Some studies have focused on the association between ABO SNPs and some others diseases, but to date, few study11 has yet reported the association between ABO gene polymorphisms and the risk of IS in the Chinese population, and they concluded negative results, which maybe resulted by relatively small sample, and in addition, they did not consider the impact of interaction among several SNPs within ABO gene on IS risk. Hanson et al14 provided evidence that ABO genotype does not have a major impact in the pathophysiology of IS or any of the subtypes. But Ling et al18 concluded different results in another Chinese study, they indicated that genetic variations of ABO gene may contribute to susceptibility of large‐artery atherosclerosis (LAA) but not IS and small‐vessel diseases (SVD) in the Chinese population, but their preliminary results should be further validated in prospective independent studies with expanded sample size. Abino et al19 suggested that a relationship of non‐O blood groups in pathogenesis of thrombosis events and a possible protective effect of O blood group, mainly in young‐adults patients with diagnosis of IS. In addition to these studies, several studies have focused on the association between ABO gene and others diseases, such as coronary heart disease (CHD), MI and CAD. Williams et al13 suggest that ABO gene variants are associated with large‐vessel and cardioembolic stroke but not small‐vessel disease in the EuroCLOT Study. Previous studies found that the non‐O phenotypes were more frequent in IS patients than controls.20, 21, 22 Wu et al23 confirmed the historical impression of linkage between some vascular disorders and non‐O blood group status, although the odds ratios are similar to those predicted by the effect of ABO (H) on von Willebrand factor levels. Some subtypes of IS and CAD shared many common risk factors, for example, atherosclerosis plaque were observed in both LAA and CAD as a common pathophysiologic mechanism. Consequently, it was speculated that genetic variants of ABO gene associated with CAD,10 may be also associated with LAA.
The complex diseases, such as IS, was a result of many gene polymorphism and gene‐gene interactions, hence, it was necessary to investigate the impact of gene‐gene interactions on susceptibility to IS. In considered of the multidimensional issue, GMDR model was used to screen the best interaction combination among 4 SNPs within ABO gene, we found a potential interaction between rs505922 and rs579459 on IS risk, participants with rs505922‐TC/CC and rs579459‐TC/CC genotype have the highest IS risk, compared to participants with s505922‐TT and rs579459‐ TT genotype. To our knowledge, this study was the first study focused on the impact of ABO gene‐ gene interaction on IS risk. The results of this study suggest that IS risk may be modified by the two SNPs and they influenced with each other in susceptibility to IS. The potential mechanism for this interaction was not very clearly, it maybe that both SNPs were associated with some IS or related‐risk factors, such as T2DM, obesity, and hypertension and so on, and this combined or crossover effect could lead to the interaction between the two SNPs on IS risk.
This study also has several limitations. First, limited number of SNPs within ABO gene was included in this study, these SNPs merely represented limited genetic variability in ABO gene. Second, the sample this study was relatively small, limited size of the cohort might reduce the power to detect association, so prospective independent studies with a comparatively larger sample size are required in the future. Third, interaction between this gene and others gene or environmental risk factors should be investigated in the future studies.
In conclusion, we found that rs579459 and rs505922 within ABO gene and their interaction were both associated with increased IS risk in Chinese population.
ACKNOWLEDGMENTS
The writing of this article was supported by the first Affiliated Hospital of Jinan University and the People's Hospital of Maoming. We thank all the partners and staffs who help us in the process of this study.
Li H, Cai Y, Xu A‐D. Association study of polymorphisms in the ABO gene and their gene‐gene interactions with ischemic stroke in Chinese population. J Clin Lab Anal. 2018;32:e22329 10.1002/jcla.22329
REFERENCES
- 1. Feigin VL, Lawes CM, Bennett DA, Barker‐Collo SL, Parag V. Worldwide stroke incidence and early case fatality reported in 56 population‐based studies: a systematic review. Lancet Neurol. 2009;8:355‐369. [DOI] [PubMed] [Google Scholar]
- 2. Liu L, Wang D, Wong KS, Wang Y. Stroke and stroke care in China: huge burden, significant workload, and a national priority. Stroke. 2011;42:3651‐3654. [DOI] [PubMed] [Google Scholar]
- 3. Thrift AG, Dewey HM, Macdonell RA, McNeil JJ, Donnan GA. Incidence of the major stroke subtypes: initial findings from the North East Melbourne stroke incidence study (NEMESIS). Stroke. 2001;32:1732‐1738. [DOI] [PubMed] [Google Scholar]
- 4. Hassan A, Markus HS. Genetics and ischaemic stroke. Brain. 2000;123(Pt 9):1784‐1812. [DOI] [PubMed] [Google Scholar]
- 5. O'Donnell MJ, Xavier D, Liu L, et al. Risk factors for ischaemic and intracerebral haemorrhagic stroke in 22 countries (the INTERSTROKE study): a case‐control study. Lancet. 2010;376:112‐123. [DOI] [PubMed] [Google Scholar]
- 6. Sharma P, Yadav S, Meschia JF. Genetics of ischaemic stroke. J Neurol Neurosurg Psychiatry. 2013;84:1302‐1308. [DOI] [PubMed] [Google Scholar]
- 7. Bevan S, Traylor M, Adib‐Samii P, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43:3161‐3167. [DOI] [PubMed] [Google Scholar]
- 8. Daniels G. The molecular genetics of blood group polymorphism. Hum Genet. 2009;126:729‐742. [DOI] [PubMed] [Google Scholar]
- 9. Yamamoto F, McNeill PD, Hakomori S. Genomic organization of human histo‐blood group ABO genes. Glycobiology. 1995;5:51‐58. [DOI] [PubMed] [Google Scholar]
- 10. Mehta NN. Large‐scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Circ Cardiovasc Genet. 2011;4:327‐329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bruzelius M, Strawbridge RJ, Trégouët DA, et al. Influence of coronary artery disease‐associated genetic variants on risk of venous thromboembolism. Thromb Res. 2014;134:426‐432. [DOI] [PubMed] [Google Scholar]
- 12. Dichgans M, Malik R, König IR, et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome‐wide analysis of common variants. Stroke. 2014;45:24‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Williams FM, Carter AM, Hysi PG, et al. Ischemic stroke is associated with the ABO locus: the EuroCLOT study. Ann Neurol. 2013;73:16‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanson E, Karlsson S, Jood K, Nilsson S, Blomstrand C, Jern C. No evidence for an association between ABO blood group and overall ischemic stroke or any of the major etiologic subtypes. Thromb Res. 2012;130:339‐342. [DOI] [PubMed] [Google Scholar]
- 15. The World Health Organization . MONICA Project (monitoring trends and determinants in cardiovascular disease): a major international collaboration. WHO MONICA Project Principal Investigators. J Clin Epidemiol. 1988;41:105‐114. [DOI] [PubMed] [Google Scholar]
- 16. Adams HP Jr, Bendixen BH, Kappelle LJ, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of org 10172 in acute stroke treatment. Stroke. 1993;24:35‐41. [DOI] [PubMed] [Google Scholar]
- 17. Lou XY, Chen GB, Yan L, et al. A generalized combinatorial approach for detecting gene‐by gene and gene‐by‐ environment interactions with application to nicotine dependence. Am J Hum Genet. 2007;80:1125‐1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ling X, Zheng Y, Tao J, Zheng Z, Chen L. Association study of polymorphisms in the ABO gene with ischemic stroke in the Chinese population. BMC Neurol. 2016;16:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abino Ade P, Ribeiro DD, Domingheti CP, et al. ABO blood group polymorphisms and risk for ischemic stroke and peripheral arterial disease. Mol Biol Rep. 2014;41:1771‐1777. [DOI] [PubMed] [Google Scholar]
- 20. Tanis B, Algra A, van der Graaf Y, Helmerhorst F, Rosendaal F. Procoagulant factors and the risk of myocardial infarction in young women. Eur J Haematol. 2006;77:67‐73. [DOI] [PubMed] [Google Scholar]
- 21. Clark P, Meiklejohn DJ, O'Sullivan A, Vickers MA, Greaves M. The relationships of ABO, Lewis and Secretor blood groups with cerebral ischaemia of arterial origin. J Thromb Haemost. 2005;3:2105‐2108. [DOI] [PubMed] [Google Scholar]
- 22. Carpeggiani C, Coceani M, Landi P, Michelassi C, L'abbate A. ABO blood group alleles: a risk factor for coronary artery disease. An angiographic study. Atherosclerosis 2010;211:461‐466. [DOI] [PubMed] [Google Scholar]
- 23. Wu O, Bayoumi N, Vickers MA, Clark P. ABO(H) blood groups and vascular disease: a systematic review and meta‐analysis. J Thromb Haemost. 2008;6:62‐69. [DOI] [PubMed] [Google Scholar]


