Abstract
Background
The process of plate streaking has been automated to improve routine workflow of clinical microbiology laboratories. Although there were many evaluation reports about the inoculation of various body fluid samples, few evaluations have been reported for blood. In this study, we evaluated the performance of automated inoculating system, Previ Isola for various routine clinical samples including blood.
Methods
Blood culture, body fluid, and urine samples were collected. All samples were inoculated on both sheep blood agar plate (BAP) and MacConkey agar plate (MCK) using Previ Isola and manual method. We compared two methods in aspect of quality and quantity of cultures, and sample processing time. To ensure objective colony counting, an enumeration reading reference was made through a preliminary experiment.
Results
A total of 377 nonduplicate samples (102 blood culture, 203 urine, 72 body fluid) were collected and inoculated. The concordance rate of quality was 100%, 97.0%, and 98.6% in blood, urine, and other body fluids, respectively. In quantitative aspect, it was 98.0%, 97.0%, and 95.8%, respectively. The Previ Isola took a little longer to inoculate the specimen than manual method, but the hands‐on time decreased dramatically. The shortened hands‐on time using Previ Isola was about 6 minutes per 10 samples.
Conclusion
We demonstrated that the Previ Isola showed high concordance with the manual method in the inoculation of various body fluids, especially in blood culture sample. The use of Previ Isola in clinical microbiology laboratories is expected to save considerable time and human resources.
Keywords: blood, body fluid, inoculation, Previ Isola
1. INTRODUCTION
Identification of microorganisms and antimicrobial susceptibility testing are main tasks for a routine clinical microbiology laboratory.1 Among many analytical steps, culture of microorganisms is essential for correct identification and antimicrobial susceptibility test.2 To provide rapid culture results, accurate and fast inoculation of specimen should be supported. However, as the amount of specimens has increased, plate streaking for inoculation became time‐consuming and time‐limiting step in most clinical microbiology laboratories.3 Many skilled technical staff spend a long time for inoculation, but it is not enough to handle increased specimens. Therefore, many directors of clinical laboratory hope to solve this problem through automation.3, 4
Laboratory automation was already introduced many years ago in other parts of clinical laboratories such as hematology, chemistry, and molecular biology, increasing productivity and quality.4 However, the introduction of automation was delayed due to the complexity and variety of samples in clinical microbiology laboratories. Over the past several years, some automated microbial identification system and automated antimicrobial susceptibility testing equipment have been adopted in many clinical microbiology laboratories.5, 6 Recently, some automated plate streaking instruments have been introduced to clinical laboratories.
In fact, the first generation of automated plate streaking equipment was developed in the late 1970s.7 However, it was not suitable for routine use until third generation in throughput and accuracy aspect. Currently, several automated inoculation instruments, including the InoqulA (BD Kiestra), the Previ Isola (bioMérieux), the PreLUD (I2A), the Microstreak (LBT Innovations), and the WASP (Copan), are being used routinely in clinical microbiology laboratory.
Among these instruments, the Previ Isola system was developed for standardization as well as automation of inoculation and streaking of plates. With the aid of a circular applicator, a standard amount of inoculum is used each time and is pressure‐control streaked on agar plates. The device can streak 180 plates per hour, guaranteeing a high standard of plate processing. The Previ Isola system can be used for liquid specimens, and also used for swab systems with transport media to improve the diagnosis of aerobes, anaerobes, fastidious bacteria, and fungi.8 There are some evaluation reports about the Previ Isola system.9, 10, 11, 12 Although these reports used various samples such as urine, wound, stool, and broncopulmonary specimens, evaluation of blood samples is rare.
In this study, we prospectively evaluated the Previ Isola system for various routine specimens including blood samples, and compared manual method in aspect of quality and quantity of cultures, and sample processing time.
2. MATERIALS AND METHODS
2.1. Preliminary experiment
First, we conducted a preliminary experiment to establish a specific enumeration reading reference for the manual method and Previ Isola. Serial dilutions of Escherichia.coli reference strain (ATCC® 25922) were performed in 0.9% sodium chloride solution to obtain a calibrated suspension from 103 to 108 colony‐forming units per milliliter (CFU/mL). These suspensions were either streaked by the manual loop‐to‐plate method or by the Previ Isola streaker. Two manual quantitative plate inoculation patterns were performed by a skilled staff with 10 μL loops in quadrant streaking pattern, or a central single streaking throughout the plate followed by a zigzag pattern. Two types of culture media, BAP (bioMérieux, Craponne, France) made by sheep blood agar and MCK (bioMérieux, Craponne, France) made by MacConkey agar, were used. A total of six plates were made per dilution and incubated at 37°C in an atmosphere with 5% CO2. Cultures were checked and recorded after an 18‐ and 24‐hour incubation period.
2.2. Inoculation of plates
We prospectively tested in parallel the manual method and Previ Isola using various clinical samples received in the clinical microbiology laboratory from August 2015 to February 2016. Blood specimens were selected when showing positive signals on the blood culture bottle. All specimens except urine were manually inoculated with quadrant streaking pattern. Urine specimens were inoculated with a central single streaking throughout the plate followed by a zigzag pattern. All manual inoculations were carried out by a skilled staff. After manual inoculation, all specimens were automatically inoculated using Previ Isola. All specimens were inoculated on both BAP and MCK agar plates. Four plates were created for each specimen.
2.3. Colony counting and identification
After incubation at 37°C in 5% CO2, all plates were read at 18 and 24 hour. Gram‐positive strains were counted on the BAP agar plate, and gram‐negative strains were counted on the MCK agar plate. The number of colonies was determined in comparison with the enumeration reading reference established by preliminary experiment. Colonies were identified according to CLSI procedures by Vitek 2 (bioMérieux).
2.4. Comparison of automated specimen inoculation with the manual method
We compared the two methods in three aspects, in that, quality and quantity of cultures, and sample processing time. The consistency of cultured bacterial species was measured for quality comparison. If the bacteria were grown in only one method, or if different bacteria were found, they were judged to be inconsistent. For quantitative comparison, we directly compared the number of colony counts. If there was a difference of 1 log according to the reading reference, they were judged to be inconsistent. Sample processing time was composed of preparation time and streaking time. Preparation time included the steps of separating and dispensing the specimen for inoculation. The retest time was included in the streaking time.
3. RESULTS
3.1. Establishment of enumeration reading reference
The preliminary experiments led to the establishment of an enumeration reading reference for both Previ Isola and manual inoculation method (Figure 1). It showed the corresponding Previ Isola and manually streaked plates of quantified dilutions of E. coli reference strain. Six plates were produced per dilution. In BAP and MCK plates streaked with Previ Isola, microorganisms grew in a concentric shape with distinct difference by concentration (Figure 1A and B). Manually streaked plates with quadrant streaking pattern (Figure 1C and D) and with a central single streaking throughout the plate followed by a zigzag pattern (Figure 1E and F) also showed significant difference according to the concentration.
Figure 1.
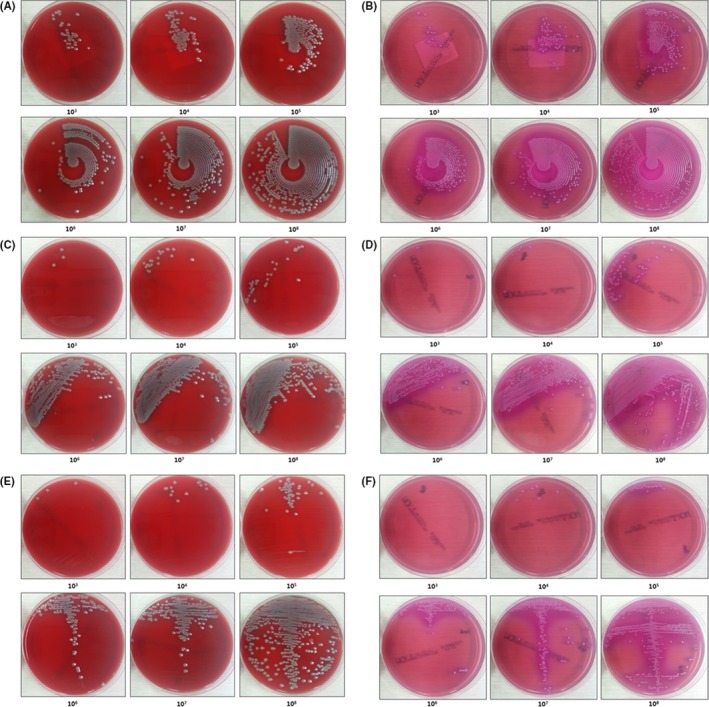
Enumeration reading reference for the automated and manual method after 24 hours of incubation. Serial dilutions of Escherichia coli ATCC 25922 were inoculated by Previ Isola on BAP (A) and MCK (B), by manual with quadrant streaking pattern on BAP (C) and MCK (D), by manual with a central single streaking throughout the plate followed by a zigzag pattern on BAP (E) and MCK (F). Units, colony‐forming units per milliliter (CFU/mL)
3.2. Clinical specimens
A total of 377 nonduplicated clinical samples were tested (Table 1). Bacteria were grown in 99 of 102 blood samples. There were 70 plates in which the gram‐positive bacteria were grown and 33 plates in which the gram‐negative bacteria were grown. Among gram‐positive bacteria, Staphylococcus epidermidis was most common, found in 25 plates, followed by Staphylococcus aureus (n = 18), Staphylococcus capitis (n = 6), and Enterococcus faecium (n = 5). Among the gram‐negative bacteria, Escherichia coli was found most frequently on 15 plates, followed by Pseudomonas aeruginosa (n = 4), Acinetobacter baumannii (n = 3), Enterobacter cloacae (n = 2). Coinfection of gram‐positive and gram‐negative bacteria was observed in four plates. E. faecium and E. coli were found together in two plates, and S. aureus and A. baumannii were found in two other plates. Among the 203 urine inoculated plates, the bacteria grew on 103 plates. There were 74 plates in which the gram‐positive bacteria were grown, and 37 plates in which the gram‐negative bacteria were grown. Among gram‐positive bacteria, Enterococcus faecalis was found most commonly, in 22 plates, followed by S. epidermidis (n = 20), Streptococcus agalactiae (n = 11), and E. faecium (n = 7). Among the gram‐negative bacteria, E. coli was found most frequently (n = 21), followed by Klebsiella pneumonia (n = 5) and A. baumannii (n = 3). Coinfection of gram‐positive and gram‐negative bacteria was observed in eight plates: S. epidemidis and E. coli, S. epidemidis and A. baumannii, S. epidermidis and C. amalonaticus, E. faecalis and P. putida, E. faecalis and A. baumannii, E. faecalis and K. pneumoniae, S. mitis and E. coli, and S. agalactiae and E. coli. Among the other specimens except blood and urine, bacteria were grown in ascitic fluid, pleural fluid, bile fluid, and closed pus. Of the 18 ascitic fluid, bacteria grew on only two plates, and were S. agalactiae and E. coli, respectively. Three of the 14 pleural fluid specimens showed bacterial growth. In one of them, S. aureus, a gram‐positive bacteria, and A. baumannii, a gram‐negative bacteria, were observed together. Seven of the 13 bile fluid specimens showed bacterial growth, and all were gram‐negative. Bacteria were grown in 9 of 10 close pus samples. K. pneumoniae, the gram‐negative bacteria, was the most common, and S. sanguis and K. pneumoniae were grown together in one plate. There was no growth on plates with cerebrospinal fluid and pancreatic fluid.
Table 1.
Characteristics of clinical specimens
| Specimen | Number (No. of growth plates) | Species | |
|---|---|---|---|
| Gram positive (No. of plates) | Gram negative (No. of plates) | ||
| Blood | 102 (99) | S. epidermidis (25), S. aureus (18), S. capitis (6), E. faecium (5), C. jeikeium (5), E. avium, 2 E. faecalis (2), S. mitis (2), C. striatum, S. haemolyticus, S. hominis, S. pyogenes, S. warneri | E. coli (15), P. aeruginosa (4), A. baumannii (3), E. cloacae (2), K. pneumonia (2), P. mirabilis (2), P. stutzeri (2), Aeromonas spp., C. freundii, S. maltophilia |
| Urine | 203 (103) | E. faecalis (22), S. epidermidis (20), S. agalactiae (11), E. faecium (7), S. haemolyticus (5), C. striatum (3), S. sanguis (2), C. jeikeium, E. aerogenes, S. anginosus, S. mitis | E. coli (21), K. pneumonia (5), A. baumannii (3), A. junii, B. cepacia, C. amalonaticus, C. freundii, C. koseri, P. aeruginosa, P. penneri, P. putida |
| Ascitic fluid | 18 (2) | S. agalactiae | E. coli |
| Pleural fluid | 14 (3) | S. aureus (2), Rhodococcus spp. | A. baumannii |
| Bile fluid | 13 (7) | E. coli (3), K. pneumoniae (2), P. stutzeri (2) | |
| Cerebrospinal fluid | 11 (0) | ||
| Closed pus | 10 (9) | S. sanguis (2), S. epidermidis, S. aureus | K. pneumoniae (4), E. coli (2) |
| Joint fluid | 5 (0) | ||
| Pancreatic fluid | 1 (0) | ||
3.3. Comparison of quality
To measure the quality of inoculation of Previ Isola and manual methods, we identified all cultured bacteria and compared them (Table 1). There was no discrepancy in blood samples. Of the 102 blood samples, three samples showed no growth with both methods. In the remaining 99 plates, the same bacteria grew. In urine samples, bacteria grew in 103 specimens with Previ Isola, while 101 specimens showed bacterial growth with manual method. Six specimens (3.0%) showed discrepancy. Three gram‐positive bacteria and one gram‐negative bacterium were grown only on the plates inoculated with Previ Isola. On the other hand, two specimens showed growth of S. epidermidis only on the plates inoculated with manual method. There was one discrepancy (1.4%) in other body fluid samples. S. aureus only grew on the plate inoculated with Previ Isola in a pleural fluid specimen.
3.4. Comparison of quantity
We determined and compared the colony count to analyze the quantitative aspect of two methods (Table 2). The colony count was determined based on the enumeration reading reference established in the preliminary experiment. There were two specimens (2.0%) showing discrepancy in blood samples. In one specimen, colony count of E. avium was 108 CFU/mL with Previ Isola, but 107 CFU/mL with manual method. In the other specimen, colony count of C. jeikeium was 108 CFU/mL with Previ Isola, but 106 CFU/mL with manual method. In urine specimens, six specimens (3.0%) showed discrepancy. In two specimens, colony count of E. faecalis was 103 and 104 CFU/mL with Previ Isola, but 105 and 102 CFU/mL with manual method, respectively. In other two specimens, colony count of E. faecium was 106 CFU/mL with Previ Isola, but 107 and 105 CFU/mL with manual method, respectively. In other specimen, colony count of E. coli was 108 CFU/mL with Previ Isola, but 107 CFU/mL with manual method. In the other specimen, colony count of A. baumannii was 105 CFU/mL with Previ Isola, but 107 CFU/mL with manual method. In other body fluid specimens, three specimens (4.2%) showed discrepant result. In two closed pus specimens, colony count of K. pneumoniae was 106 and 105 CFU/mL with manual method, but 105 and 104 CFU/mL with Previ Isola, respectively. In one pleural fluid specimen, colony count of Rodococcus spp. was 106 CFU/mL with Previ Isola, but 105 CFU/mL with manual method.
Table 2.
Comparative colony counts according to the type of sample and method of sample inoculation
| Specimen(n) | Colony count (CFU/mL) | No. of plates with Previ Isola | No. of plates with Manual | No. of discrepant specimens (%) | ||
|---|---|---|---|---|---|---|
| Gram positive | Gram negative | Gram positive | Gram negative | |||
| Blood (102) | No growth | 3 | 3 | 2 (1.96) | ||
| <103 | ||||||
| 103 | ||||||
| 104 | ||||||
| 105 | 1 | 1 | ||||
| 106 | 1 | |||||
| 107 | 5 | 3 | 6 | 3 | ||
| 108 | 65 | 29 | 63 | 29 | ||
| Urine (203) | No growth | 100 | 102 | 12 (5.91) | ||
| <103 | 9 | 9 | ||||
| 103 | 20 | 6 | 18 | 5 | ||
| 104 | 12 | 4 | 13 | 4 | ||
| 105 | 15 | 4 | 16 | 3 | ||
| 106 | 8 | 2 | 6 | 2 | ||
| 107 | 3 | 7 | 4 | 9 | ||
| 108 | 7 | 14 | 7 | 13 | ||
| Other body fluids (72) | No growth | 51 | 52 | 4 (5.56) | ||
| <103 | 1 | |||||
| 103 | 1 | 1 | ||||
| 104 | 2 | 1 | ||||
| 105 | 2 | 1 | 2 | 1 | ||
| 106 | 1 | 2 | 1 | 3 | ||
| 107 | 1 | 1 | ||||
| 108 | 3 | 9 | 3 | 9 | ||
CFU, colony‐forming units.
3.5. Comparison of sample processing time
The inoculation process was divided into preparation and streaking, and the time taken at each step for 10 samples was measured (Table 3). In blood, the time taken to transfer the sample from the blood culture bottle to the tube using a disposable sterile syringe took about 3.8 minutes in both methods. For blood sample streaking, it took 7.3 minutes with Previ Isola and 7 minutes for manual method. For the sample preparation, 1.2 minutes was required for the Previ Isola and 0.5 minutes for manual method in the case of urine specimens. The streaking time was 7.5 minutes with Previ Isola and 6.8 minutes with manual, respectively. The time required to the preparation of body fluid other than blood and urine was 1.2 minutes for manual and 2.4 minute for Previ Isola. Streaking took 8 minutes with Previ Isola and 7.6 minutes with manual method. When using Previ Isola, hands‐on time decreased from 11.1 to 3.8 minutes in blood, from 7.3 to 1.2 minutes in urine, and from 8.8 to 2.4 minutes in other body fluids, respectively.
Table 3.
Spending time for each step of inoculation process per 10 samples
| Specimen | Preparation time (min) | Streaking time (min) | Hands‐on time (min) | |||
|---|---|---|---|---|---|---|
| Previ Isola | Manual | Previ Isola | Manual | Previ Isola | Manual | |
| Blood | 3.8 | 3.8 | 7.3 | 7.0 | 3.8 | 11.1 |
| Urine | 1.2 | 0.5 | 7.5 | 6.8 | 1.2 | 7.3 |
| Other body fluids | 2.4 | 1.2 | 8.0 | 7.6 | 2.4 | 8.8 |
4. DISCUSSION
Automation in clinical microbiology laboratories continues to accelerate the detection of infectious agents. In particular, instruments for automatic sample inoculation have been claimed to improve the quality of colonization and save processing time. In 2008, Glasson et al1 reported that colony productivity per plate was increased by up to 44% using MicroStreak instrument (bioMérieux). In 2015, Croxatto et al13 reported that the automated InoqulA ((BD Kiestra) inoculation significantly saved time on laboratory workloads and laboratory costs. Using the Previ Isola system, Kim et al14 assessed the usefulness of body fluids and urine specimens, and Nebbad‐Lechani et al11 evaluated for bronchopulmonary specimens.
In this study, we conducted an evaluation of the Previ Isola for various clinical body fluid samples including blood culture sample. Blood culture is considered the standard diagnostic tool for blood stream infections. Many clinical microbiology laboratories now use automated blood culture instruments, which are equipped with a fully automated closed‐loop continuous monitoring system for microbial growth detection.15, 16, 17 Although rapid identification and antimicrobial susceptibility testing techniques have been developed for blood culture‐positive samples through molecular and biochemical technologies, agar plate inoculation is still used in many laboratories.18, 19, 20 We used 102 culture‐positive blood samples, and demonstrated that the concordance rate of quality between Previ Isola and manual method was 100%. Among 102 samples, no growth was observed in three samples with both methods. The blood culture bottle results of these specimens stated that no growth, C. sporogenes, and S. pneumoniae were observed from these samples, respectively. We inferred that one was a false positive, and the other two were due to a very small amount of bacteria.
In addition to blood and urine, we used a variety of clinical body fluid samples including ascitic fluid, pleural fluid, bile fluid, cerebrospinal fluid, closed pus, joint fluid, and pancreatic fluid. Especially, various bacteria grew in 90% of closed pus specimens, and only gram‐negative bacterium grew in bile fluid specimens. They showed 98.6% concordance rate in quality.
Colony count reading in routine clinical microbiology laboratories cannot completely exclude subjective aspects in terms of quantitative analysis. Through preliminary experiment, we were able to maintain some objectivity in further evaluation using clinical specimens. After incubation, the plates were read at 18 and 24 hours. There was almost no difference between the two. The colony counts that were obtained using the two methods were in concordance for 96.9% of all tested samples. In particular, the concordance rate between two methods in blood sample was 98.0%. In the case of two specimens showing discrepancies, the plate with Previ Isola has more colony count than that with manual method. Maybe it was because of the difference in uniformity of sample streaking, so Previ Isola was considered superior. The concordance rate between the two methods in urine samples was 97.0%. In the case of the urine sample, one method did not show superiority to the other method. The concordance rate between the two methods in other body fluids was 95.8%. In three discrepant specimens, as the same species grew within 1 log difference in quantity, if loosen the comparison standards a little more, it could be regarded as almost 100% concordant.
To inoculate blood specimens, it is necessary to transfer the specimen from the blood culture bottle to a tube using a disposable sterilizing syringe. In the case of urine or other body fluid samples, the transfer process may be very short or omitted. Because of this, the preparation time of blood took longer than that of other specimens in both methods. The Previ Isola took a little longer to streak the blood specimen to the plate than manual method. This pattern was similarly observed in other specimens. Maybe the reason for this is that we retested if an error message appears in Previ Isola, but not in the manual method. The urine sample and other body fluid specimens had a higher rate of retest than blood. However, the Previ Isola does not take longer in all situations. When the amount of specimens to be tested at one time was large, it was observed that the time of the Previ Isola took less than manual method. We have tested an average of 20 specimens at a time. If we tested more quantities at one time, we would have got different results. Not surprisingly, we found that hands‐on time was greatly reduced when using Previ Isola. Hands‐on time for inoculation refers to the time a skilled technician actually worked until the inoculated medium has been placed in the incubator from the specimen preparation. We could save about 6 minutes of hands‐on time per 10 samples. Indeed, it is expected that considerable time will be saved and resulted saving of human resources in clinical microbiology laboratories.
In summary, our study demonstrated that the Previ Isola showed high concordance with the manual method in the inoculation of various body fluid specimens. In particular, the rate of concordance in blood culture specimens, which have been rarely studied, was 100% and 98% in quality and quantity, respectively. Also, the use of the Previ Isola could save considerable time and human resources in clinical microbiology laboratories.
CONFLICTS OF INTERESTS
No potential conflicts of interests relevant to this article were reported.
Choi Q, Kim HJ, Kim JW, Kwon GC, Koo SH. Manual versus automated streaking system in clinical microbiology laboratory: Performance evaluation of Previ Isola for blood culture and body fluid samples. J Clin Lab Anal. 2018;32:e22373 10.1002/jcla.22373
REFERENCES
- 1. Glasson JH, Guthrie LH, Nielsen DJ, Bethell FA. Evaluation of an automated instrument for inoculating and spreading samples onto agar plates. J Clin Microbiol. 2008;46:1281‐1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lagier J‐C, Edouard S, Pagnier I, Mediannikov O, Drancourt M, Raoult D. Current and past strategies for bacterial culture in clinical microbiology. Clin Microbiol Rev. 2015;28:208‐236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bourbeau PP, Ledeboer NA. Automation in clinical microbiology. J Clin Microbiol. 2013;51:1658‐1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Croxatto A, Prod'hom G, Faverjon F, Rochais Y, Greub G. Laboratory automation in clinical bacteriology: what system to choose? Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2016;22:217‐235. [DOI] [PubMed] [Google Scholar]
- 5. Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G. Performance of matrix‐assisted laser desorption ionization‐time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol. 2010;48:1549‐1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson JE, Jorgensen JH, Crawford SA, Redding JS, Pruneda RC. Comparison of two automated instrument systems for rapid susceptibility testing of gram‐negative bacilli. J Clin Microbiol. 1983;18:1301‐1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tilton RC, Ryan RW. Evaluation of an automated agar plate streaker. J Clin Microbiol. 1978;7:298‐304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Van Horn KG, Audette CD, Tucker KA, Sebeck D. Comparison of 3 swab transport systems for direct release and recovery of aerobic and anaerobic bacteria. Diagn Microbiol Infect Dis. 2008;62:471‐473. [DOI] [PubMed] [Google Scholar]
- 9. Le Page S, van Belkum A, Fulchiron C, Huguet R, Raoult D, Rolain J‐M. Evaluation of the PREVI® Isola automated seeder system compared to reference manual inoculation for antibiotic susceptibility testing by the disk diffusion method. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2015;34:1859‐1869. [DOI] [PubMed] [Google Scholar]
- 10. Mischnik A, Trampe M, Zimmermann S. Evaluation of the impact of automated specimen inoculation, using Previ Isola, on the quality of and technical time for stool cultures. Ann Lab Med. 2015;35:82‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nebbad‐Lechani B, Emirian A, Maillebuau F, et al. New procedure to reduce the time and cost of broncho‐pulmonary specimen management using the Previ Isola® automated inoculation system. J Microbiol Methods. 2013;95:384‐388. [DOI] [PubMed] [Google Scholar]
- 12. Mischnik A, Mieth M, Busch CJ, Hofer S, Zimmermann S. First evaluation of automated specimen inoculation for wound swab samples by use of the Previ Isola system compared to manual inoculation in a routine laboratory: finding a cost‐effective and accurate approach. J Clin Microbiol. 2012;50:2732‐2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Croxatto A, Dijkstra K, Prod'hom G, Greub G. Comparison of inoculation with the InoqulA and WASP automated systems with manual inoculation. J Clin Microbiol. 2015;53:2298‐2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim Y, Yoon S, Sohn YS, et al. Evaluation of an automated instrument, PREVI Isola® for inoculation of body fluids and urine samples onto agar plates. Lab Med Online. 2011;1:105. [Google Scholar]
- 15. Park J, Han S, Shin S. Comparison of growth performance of the BacT/ALERT VIRTUO and BACTEC FX blood culture systems under simulated bloodstream infection conditions. Clin Lab. 2017;63:39‐46. [DOI] [PubMed] [Google Scholar]
- 16. Kim S‐C, Jeon B‐Y, Kim J‐S, et al. Performance of the BacT Alert 3D system versus solid media for recovery and drug susceptibility testing of Mycobacterium tuberculosis in a tertiary hospital in Korea. Tuberc Respir Dis. 2016;79:282‐288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nutman A, Fisher Even‐Tsur S, Shapiro G, Braun T, Schwartz D, Carmeli Y. Time to detection with BacT/Alert FA Plus compared to BacT/Alert FA blood culture media. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2016;35:1469‐1473. [DOI] [PubMed] [Google Scholar]
- 18. von Lilienfeld‐Toal M, Lehmann LE, Raadts AD, et al. Utility of a commercially available multiplex real‐time PCR assay to detect bacterial and fungal pathogens in febrile neutropenia. J Clin Microbiol. 2009;47:2405‐2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stevenson LG, Drake SK, Murray PR. Rapid identification of bacteria in positive blood culture broths by matrix‐assisted laser desorption ionization‐time of flight mass spectrometry. J Clin Microbiol. 2010;48:444‐447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Varani S, Stanzani M, Paolucci M, et al. Diagnosis of bloodstream infections in immunocompromised patients by real‐time PCR. J Infect. 2009;58:346‐351. [DOI] [PubMed] [Google Scholar]


