Abstract
Background
This study aimed to investigate the associations of circulating long, non‐coding (lncRNA) IFNG‐AS1, lncRNA ANRIL and lncRNA ITSN1 relative expressions with disease risk, severity and inflammatory cytokines levels in coronary artery disease (CAD) patients.
Methods
One hundred and ninety‐one patients suspected of CAD who underwent coronary angiography were consecutively enrolled in this casecontrol study, and divided into CAD patients (N = 102) and controls (N = 89) according to coronary angiographic results. Blood samples of all participants were collected. Plasma lncRNA IFNG‐AS1, lncRNA ANRIL and lncRNA ITSN1 expressions were detected using quantitative polymerase chain reaction (qPCR). Serum tumor necrosis factor‐α (TNF‐α), interleukin (IL)‐1β (IL‐1β), IL‐6, IL‐8, IL‐10, and IL‐17 were assessed using enzyme‐linked immunosorbent assay (ELISA). Gensini Score was used to evaluate the disease severity of CAD patients.
Results
LncRNA IFNG‐AS1 relative expression in CAD patients was upregulated compared with that in controls (P < .001), and the receiver operating characteristic (ROC) curve showed that the area under curve (AUC) of lncRNA‐IFNG‐AS1 for predicting the risk of CAD was 0.755 (95% CI: 0.688‐0.821). lncRNA IFNG‐AS1 relative expression was remarkably associated with Gensini Score (r = .259, P = .009). Additionally, lncRNA IFNG‐AS1 relative expression was positively associated with high‐sensitivity C‐reactive protein (hs‐CRP) (r = .283, P = .004), TNF‐α (r = .269, P = .006), and IL‐6 levels (r = .425, P < .001), while it was negatively correlated with IL‐10 level (r = −.263, P = .008). lncRNA ANRIL or lncRNA ITSN1 was not correlated with CA D risk, Gensini Score, hs‐CRP, ESR, TNF‐α, IL‐1β, IL‐6, IL‐8, IL‐10, or IL‐17 levels (all P > .05).
Conclusion
Circulating lncRNA IFNG‐AS1 expression correlates with increased disease risk, higher disease severity and elevated inflammation in CAD patients.
Keywords: coronary artery disease, disease risk, Gensini Score, inflammatory cytokines, lncRNA IFNG‐AS1
1. INTRODUCTION
Coronary artery disease (CAD) is an ischemic heart disease characterized by abnormal lipid metabolism, accumulation of atherosclerotic substances in the arterial intima, resulting in narrowing of the arterial lumen, obstruction of blood flow, heart ischemia, and angina. It is the most common type of cardiovascular diseases.1 In 2015, CAD affects 110 million people and leads to 8.9 million deaths that makes up 15.9% of all deaths caused by diseases, making it the most common cause of death globally.2, 3 Hyperlipidemia, high blood pressure, smoking, diabetes, obesity, high blood cholesterol, and family history are risk factors for CAD.4, 5, 6, 7 Many methods are used to assist in the diagnosis of CAD, including electrocardiogram, cardiac stress testing, coronary computed tomographic angiography and coronary angiogram, among which angiography is the gold standard in the diagnosis of CAD, while the invasive technique is not suitable for all patients.8, 9 Therefore, the exploration of biomarkers for diagnosis and disease monitoring of CAD is of great importance to patients.
Long non‐coding RNAs (lncRNA) are a class of RNAs that are more than 200 nucleotides in length, while they do not encode protein functions.10 In recent years, studies illustrate that lncRNAs function in various aspects of cell biology, whose potential role in various diseases etiologies including cardiovascular diseases is of great concern.11, 12, 13, 14 lncRNA IFNG‐AS1 is reported to be an enhancer of inflammation in ulcerative colitis (UC)15; induction of lncRNA ANRIL prevents coronary atherosclerosis by regulating apoptosis in vascular endothelial cells and inflammatory factor expression16; and lncRNA ITSN1 is found to be elevated in rheumatoid arthritis patients and correlated with C‐reactive protein level.17 These 3 lncRNAs are disclosed to be pro‐inflammatory genes contributing to inflammation and immune response, which might be involved in the pathogenesis of CAD. However, their roles in CAD development and progression have not yet been explored. Thus, this study aimed to investigate the associations of circulating lncRNA Ifng antisense RNA 1 (IFNG‐AS1), lncRNA CDKN2B antisense RNA 1 (ANRIL), and lncRNA intersectin 1 (ITSN1) relative expressions with disease risk, severity and inflammatory cytokines levels in CAD patients.
2. MATERIALS AND METHODS
2.1. Patients
A total of 191 patients who underwent diagnostic coronary angiography due to symptoms of suspected CAD between April 2015 and January 2017 at Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University, were consecutively enrolled in this case‐control study. Patients were excluded if there was evidence of hemodynamically significant valvular heart disease or a history of coronary artery bypass graft surgery, vasospastic angina, cardiomyopathy, pulmonary embolism, malignant tumors, serious infection, systematic hematological disease, systematic inflammatory disease, heart failure, or other important organs failure. Meanwhile, patients who were younger than 18 years, pregnant women, or lactating women were also excluded. After undergoing the diagnostic coronary angiography, 102 patients were diagnosed with CAD and categorized into case group, while other 89 patients were classified into controls. CAD was defined as at least 1 coronary arteries had a stenosis ≥50% by coronary angiography.
This study was performed according to the Declaration of Helsinki. The protocol of the study was approved by the Ethics Committee of Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University. Participants in the study were voluntary and gave their written informed consents before they participated in the study.
2.2. Data collection
After enrollment, all participants’ demographic information, clinical characteristics and laboratory indexes were collected including age, gender, body mass index (BMI), hypertension, diabetes, smoke, family history of CAD, triglyceride (TG), total cholesterol (TC), fasting high‐density lipoprotein cholesterol (HDL‐C), fasting low‐density lipoprotein cholesterol (LDL‐C), high‐sensitivity C‐reactive protein (hs‐CRP) and erythrocyte sedimentation rate (ESR).
2.3. Disease severity assessment
Gensini Score was used to assess the disease severity of CAD in this study. It was calculated based on the narrowing of the lumen diameter and the position of the lesion. Scoring standards were as follows: 1 point for ≤25%, 2 points for 26%‐50%, 4 points for 51%‐75%, 8 points for 76%‐90%, 16 points for 91%‐99% and 32 points for complete occlusion. Each lesion score was multiplied by a factor that took into account the importance of the lesion's position in the coronary circulation. More specifically, the left main was assigned the significant multiplier ×5; the proximal segment of the left anterior descending (LAD) was given ×2.5; the proximal segment of the circumflex artery was weighted by a factor of ×2.5; the mid segment of the LAD was assigned a factor of ×1.5; the right coronary artery, the distal segment of the LAD, the posterolateral artery and the obtuse marginal artery were all given ×1; and all other areas were assigned a factor of ×0.5.
2.4. Sample collection
Blood samples of all participants were collected and stored in the ethylene diamine tetra acetic acid (EDTA) 3k tubes as well as in tubes without any additives. Plasma was separated from the blood that was stored in EDTA‐containing tubes by centrifugation at 1410 g for 15 minutes. Then, the plasma was used to detect relative expressions of lncRNA IFNG‐AS1, lncRNA ANRIL, and lncRNA ITSN1. At the same time, serum was separated from blood and stored in the tubes that did not contain any additives using centrifugation at 1410 g for 5 minutes, and serum was used to detect the levels of inflammatory cytokines using enzyme‐linked immunosorbent assay (ELISA).
2.5. Reverse transcription quantitative polymerase chain reaction (RT‐qPCR) assay
LncRNA expression was evaluated using RT‐PCR. Total RNA was extracted from plasma using TRIzol solution (Invitrogen, Carlsbad, USA) following the instructions of manufacturer. Then, nanodrop ND‐1000 (Biotek, Vermont, USA) was used to detect the concentration and purity of total RNA, and the integrity of total RNA was detected using agarose gel electrophoresis. Finally, 1 μg of total RNA with A260/A280 value of 1.92.1, A260/A230 >2.0 and RNA integrity number >8 was used for cDNA synthesis using transcription kit (TOYOBO, Osaka, Japan). Subsequently, the SYBR Premix EX Taq kit (KAPA, Boston, USA) was used to evaluate lncRNA IFNG‐AS1/lncRNA ANRIL/lncRNA ITSN1 relative expressions using U6 as an internal reference. The relative quantity of lncRNAs was calculated by the 2−△△Ct formula. The cDNA was amplified in triplicate (3 technical replicates) per reference gene and biological replicate (sample). The intra‐assay coefficient of variation of PCR was 0.3%‐1.1%, and the inter‐assay coefficient of variation was 0.71%‐1.46%.
2.6. Enzyme‐linked immunosorbent assay
The expressions of serum tumor necrosis factor‐α (TNF‐α), interleukin (IL)‐1β (IL‐1β), IL‐6, IL‐8, IL‐10, and IL‐17 were assessed using commercial ELISA kit (R&D, Minnesota, USA) following the instructions of the manufacturer.
2.7. Statistics
Statistical analysis was performed using the SPSS software 21.0 (IBM, New York, NY, USA) and GraphPad prism V5.01 (IBM). Data were presented as count (%), mean ± standard deviation or median (25th‐75th value). Comparison between 2 groups was determined using t test, Chi‐square test, or Wilcoxon rank sum test. Receiver operating characteristic (ROC) curve was performed to assess the value of LncRNA IFNG‐AS1, lncRNA ANRIL, and lncRNA ITSN1 for predicting the risk of CAD. The correlations of lncRNA IFNG‐AS1, lncRNA ANRIL and lncRNA ITSN1 relative expressions with Gensini Score, hs‐CRP, ESR, TNF‐α, IL‐1β, IL‐6, IL‐8, IL‐10, and IL‐17 levels were evaluated using Spearman 2‐way test. P < .05 was considered significant.
3. RESULTS
3.1. Baseline characteristics of CAD patients and controls
As listed in Table 1, the mean age in CAD patients and controls was 60.3 ± 9.4 years and 58.2 ± 8.0 years (P = .100), respectively. There were 74 males and 28 females in CAD patients, and 64 males and 25 females in controls (P = .922). Also, the percentage of patients with hypertension (P < .001), hyperglycemia (P = .047) and family history of CAD (P = .017) were higher in CAD patients compared with that in controls. Additionally, the TC (P = .026), HDL‐C (P = .012), LDL‐C (P = .013) and Gensini Score (P < .001) in CAD patients were higher than those in the controls. The other clinical and pathological characteristics of CAD patients and controls were presented in Table 1.
Table 1.
Characteristics of patients
| Parameters | CAD patients (n = 102) | Controls (n = 89) | P Value |
|---|---|---|---|
| Age (y) | 60.3 ± 9.4 | 58.2 ± 8.0 | .100 |
| Gender (Male/Female) | 74/28 | 64/25 | .922 |
| BMI (kg/m2) | 24.8 ± 3.4 | 23.9 ± 3.7 | .082 |
| Hypertension (n/%) | 85 (83.3) | 55 (61.8) | <.001 |
| Diabetes (n/%) | 31 (30.4) | 16 (18.0) | .047 |
| Smoke (n/%) | 53 (52.0) | 42 (41.2) | .511 |
| Family history of CAD (n/%) | 47 (46.1) | 26 (29.2) | .017 |
| TG (mmol/L) | 1.81 ± 0.96 | 1.72 ± 0.71 | .468 |
| TC (mmol/L) | 4.53 ± 1.60 | 4.04 ± 1.38 | .026 |
| HDL‐C (mmol/L) | 1.01 ± 0.36 | 1.15 ± 0.40 | .012 |
| LDL‐C (mmol/L) | 2.99 ± 1.13 | 2.63 ± 0.81 | .013 |
| Gensini Score | 47.50 (24.63‐74.00) | 1.00 (1.00‐2.00) | <.001 |
BMI, body mass index; CAD, coronary artery disease; HDL‐C, fasting high‐density lipoprotein cholesterol; LDL‐C, fasting low‐density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride.
Data were presented as mean value ± standard deviation, median and 25th‐75th value or count (percentage). Significance of the comparison was determined by t test, Wilcoxon rank sum test or Chi‐square test. P Value <.05 was considered significant. Bold values indicate the statistical significance.
3.2. lncRNA IFNG‐AS1, lncRNA ANRIL, and lncRNA ITSN1 relative expression in CAD patients
The lncRNA IFNG‐AS1 relative expression in CAD patients was remarkably upregulated compared with that in controls (P < .001, Figure 1A). But the lncRNA ANRIL and lncRNA ITSN1 relative expressions have no difference between CAD patients and controls (all P > .05, Figure 1B,C). The values of lncRNA IFNG‐AS1, lncRNA ANRIL, and lncRNA ITSN1 relative expressions in CAD patients and controls were presented in Table S1.
Figure 1.
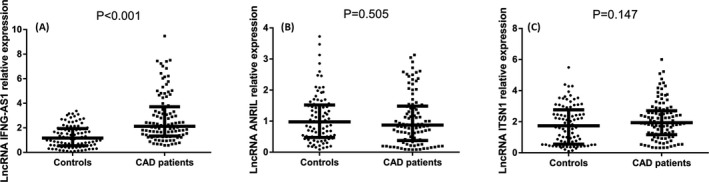
LncRNA IFNG‐AS1, lncRNA ANRIL, and lncRNA ITSN1 relative expressions in CAD patients and controls. Relative expression of lncRNA IFNG‐AS1 was increased in CAD patients compared with controls (A); however, the lncRNA ANRIL (B) and lncRNA ITSN1 (C) relative expressions were similar in both CAD patients and controls. Comparison between 2 groups was determined using Wilcoxon rank sum test. P < .05 was considered significant. CAD, coronary artery disease
3.3. The value of lncRNA IFNG‐AS1, lncRNA ANRIL, and lncRNA ITSN1 relative expressions for predicting the risk of CAD
To assess the value of lncRNA IFNG‐AS1, lncRNA ANRIL and lncRNA ITSN1 in distinguishing CAD patients from controls, ROC curves were performed. As shown in Figure 2, the area under curve (AUC) of lncRNA‐IFNG‐AS1 for predicting the risk of CAD was 0.755 (95% CI: 0.688‐0.821, Figure 2A), and the AUC of lncRNA ANRIL and lncRNA ITSN1 were 0.528 (95% CI: 0.446‐0.610) and 0.561 (95% CI: 0.478‐0.644), respectively (Figure 2B,C).
Figure 2.
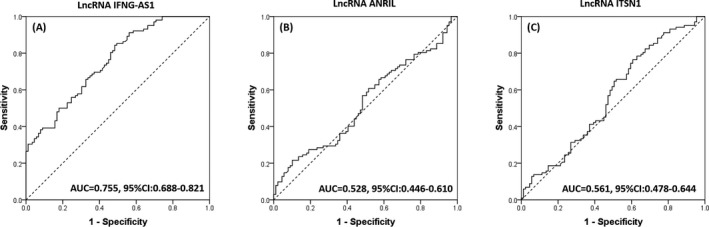
Diagnostic value of lncRNA IFNG‐AS1, lncRNA ANRIL and lncRNA ITSN1 for CAD patients. ROC curve showed that lncRNA INFG‐AS1 had a good AUC (A), but lncRNA ANRIL (B) or lncRNA ITSN1 (C) did not. Comparison between 2 groups was determined using Wilcoxon rank sum test. P < .05 was considered significant. AUC, area under curve; CAD, coronary artery disease
3.4. Correlation of lncRNA IFNG‐AS, lncRNA ANRIL and lncRNA ITSN1 relative expression with Gensini Score
The Spearman test was sued to analyze the correlations between lncRNA IFNG‐AS1/lncRNA ANRIL/lncRNA ITSN1 relative expressions and Gensini Score, which discovered that lncRNA IFNG‐AS1 relative expression was positively correlated with Gensini Score (r = .259, P = .009, Figure 3A). However, lncRNA ANRIL (r = .150, P = .133) or lncRNA ITSN1 (r = −.035, P = .724) relative expressions were not correlated with Gensini Score (Figure 3B,C).
Figure 3.
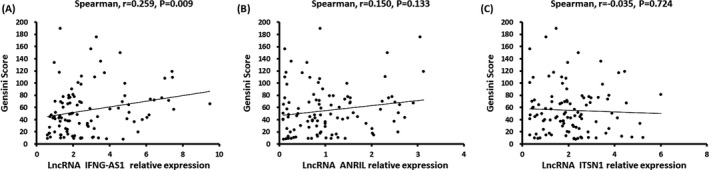
Correlation of lncRNA IFNG‐AS1, lncRNA ANRIL and lncRNA ITSN1 relative expressions with Gensini Score. The relative level of lncRNA INFG‐AS1 was positively associated with Gensini score (A), but lncRNA ANRIL (B) or lncRNA ITSN1 was not correlated with Gensini score (C). Spearman 2‐way test was used to assess the correlations of lncRNA IFNG‐AS1, lncRNA ANRIL, and lncRNA ITSN1 relative expressions with Gensini Score. P < .05 was considered significant
3.5. Correlations of lncRNA IFNG‐AS1, lncRNA ANRIL and lncRNA ITSN1 relative expressions with hs‐CRP and ESR
The relative expression of lncRNA IFNG‐AS1 was positively associated with hs‐CRP level in CAD patients as presented in Figure 4A (r = .283, P = .004), while lncRNA ANRIL (r = .129, P = .196) and lncRNA ITSN1 (r = −.044, P = .661) relative expressions were not associated with hs‐CRP level (Figure 4B,C). In addition, no correlation of lncRNA IFNG‐AS1 (r = .135, P = .177), lncRNA ANRIL (r = .163, P = .101) or lncRNA ITSN1 (r = −.076, P = .450) relative expression with ESR was found (Figure 4D,E,F). And those results suggested that lncRNA IFNG‐AS1 might act as a pro‐inflammatory factor in CAD patients.
Figure 4.
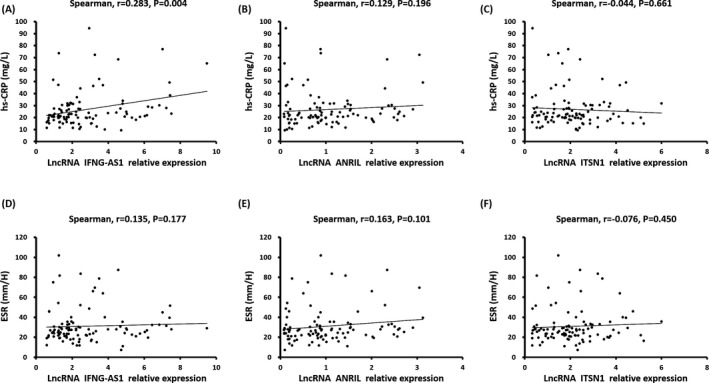
Correlations of lncRNA IFNG‐AS1, lncRNA ANRIL, and lncRNA ITSN1 relative expressions with hs‐CRP and ESR. Relative expression of lncRNA INFG‐AS1 positively correlated with hs‐CRP (A), but was not correlated with ESR (D). However, no correlation of lncRNA ANRIL expression with hs‐CRP (B) or ESR (E) was found. And lncRNA ITSN1 level was not associated with hs‐CRP (C) or ESR (F). Spearman 2‐way test was used to assess the correlation of lncRNA IFNG‐AS1, lncRNA ANRIL and lncRNA ITSN1 relative expression with hs‐CRP and ESR. P < .05 was considered significant. ESR, erythrocyte sedimentation rate; hs‐CRP, high‐sensitivity C‐reactive protein
3.6. Correlation of lncRNA IFNG‐AS, lncRNA ANRIL and lncRNA ITSN1 relative expressions with inflammatory cytokines
As displayed in Figure 5, lncRNA IFNG‐AS1 relative expression was found to be positively correlated with TNF‐α (r = .269, P = .006, Figure 5A) and IL‐6 levels, (r = .425, P < .001, Figure 5G) but negatively associated with IL‐10 level (r = −.263, P = .008, Figure 5M), while no correlation of lncRNA IFNG‐AS1 relative expression with other inflammatory cytokines expressions was found (all P > .05). Furthermore, no correlation of lncRNA ANRIL and lncRNA ITSN1 relative expression with these inflammatory cytokines was found (all P > .05). These results suggested that lncRNA IFNG‐AS1 relative expression may serve as a pro‐inflammatory gene in CAD patients.
Figure 5.
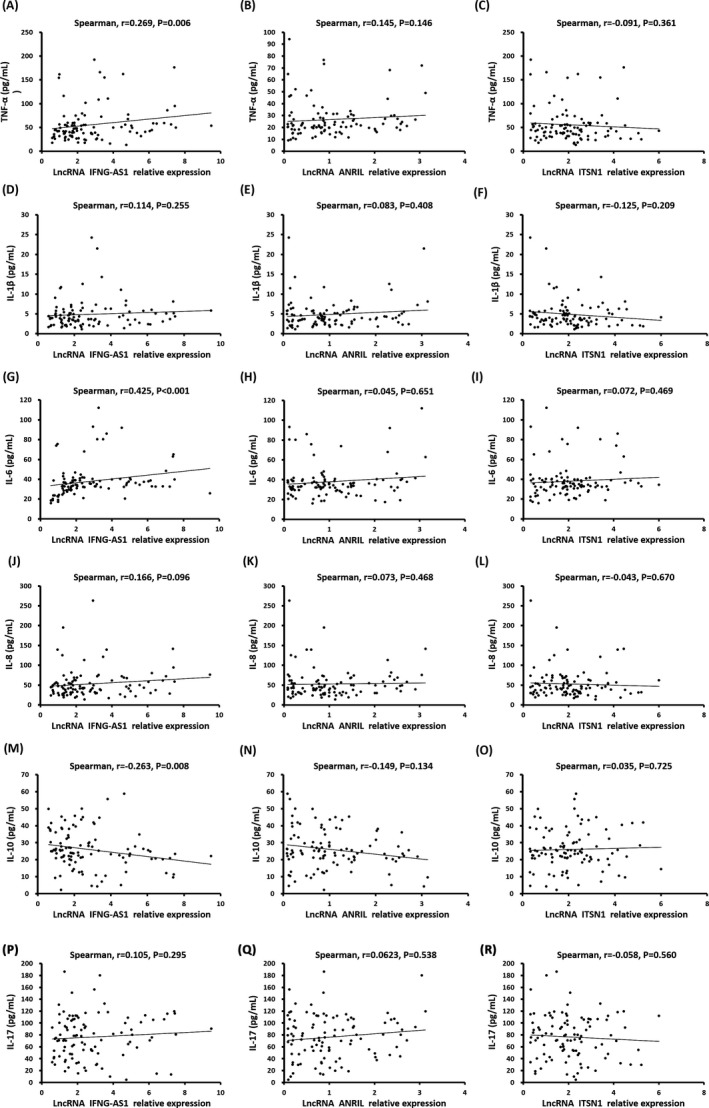
Correlation of lncRNA IFNG‐AS1, ANRIL and lncRNA ITSN1 relative expression with inflammatory cytokines. lncRNA INFG‐AS1 relative expression was positively correlated with the levels of TNF‐α (A) and IL‐6 (G); no correlation of lncRNA INFG‐AS1 relative level with IL‐1β (D), IL‐8 (J) or IL‐17 (P) was discovered; but a negative correlation of lncRNA INFG‐AS1 expression with IL‐10 was observed (M). And lncRNA ANRIL level was not correlated with TNF‐α (B), IL‐1β (E), IL‐6 (H), IL‐8 (K), IL‐10 (N), or IL‐17 (Q). In addition, no correlation of lncRNA ITSN1 level with TNF‐α (C), IL‐1β (F), IL‐6 (I), IL‐8 (L), IL‐10 (O) or IL‐17 (R) was found. Spearman 2‐way test was used to assess the correlation of lncRNA IFNG‐AS1, lncRNA ANRIL, and lncRNA ITSN1 relative expression with inflammatory cytokines. P < .05 was considered significant
4. DISCUSSION
Atherosclerosis is the main pathological mechanism of CAD, which is usually characterized by endothelial damage, inflammatory cells, and vascular smooth muscle cell (VSMC) accumulation, as well as by extracellular lipid and fibrous tissue deposition.18, 19 Studies suggest that local or systemic coronary artery inflammation plays an important role in the development of atherosclerosis.20, 21, 22 lncRNAs are regulators that mediate gene expression at epigenetic, transcription, and translation levels, coordinating and integrating multiple signaling pathways involved in cellular differentiation, proliferation to maintain cell homeostasis, and organ development.23, 24 A growing number of recent studies display that lncRNAs play pivotal roles in diverse physiological and pathological processes in the cardiovascular system. For example, increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of CAD.25 A study reports that long intergenic noncoding RNA 00305 sponges miR‐136 to regulate the hypoxia‐induced apoptosis of human umbilical vein endothelial cells.26 And 1 study exhibits that lncRNA MEG3 is regulated by dNK‐derived interferon gamma (IFN‐γ), stimulates cells migration, inhibits cells proliferation, and promotes cell apoptosis in human aorta VSMC (HA‐VSMC) cell line.27 Besides, lncRNA TUG1 sponges miR‐204‐5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification.28 These indicate that lncRNAs are involved in the pathogenesis of CAD.
lncRNA IFNG‐AS1 is located on human chromosome 12q15, which is reported to be associated with abnormalities in inflammation and cell activity.15 LncRNA IFNG‐AS1 regulates the expression of IFNG at the transcriptional and translational levels in human CD4+ T cells that contributes to Th1 cell response in Hashimoto's Thyroiditis (HT) patients, indicating lncRNA IFNG‐AS1 may promote the inflammatory responses in HT patients.29 In the autoimmune myasthenia gravis (EAMG) model, lncRNA IFNG‐AS1 proliferates T helper type 1 (Th1)/Treg cells and regulates the expression of their transcription factors by decreasing the expression of HLA‐DRB and human leukocyte antigen major histocompatibility complex, class II, DO beta (HLA‐DOB) to promote the EAMG development.30 Moreover, another prior study illuminates that lncRNA IFNA‐AS1 regulates CD4+ T cell activation in myasthenia gravis through regulating major histocompatibility complex, class II, DR beta 1 (HLA‐DRB1) as well.30 These results of the previous studies suggest lncRNA IFNG‐AS1 may be a pro‐inflammatory gene. However, the association between lncRNA IFNG‐AS1 and CAD has not been reported so far.
In our study, we found lncRNA IFNG‐AS1 was upregulated in CAD patients compared to controls, and ROC curve showed a good prediction value of lncRNA IFNG‐AS1 relative expression for CAD risk. These might result from that lncRNA IFNG‐AS1 positively regulates the systematic inflammation leading to the CAD development, which was disclosed by the positive correlation with CRP and inflammatory cytokines in our study as well. This study also elucidated that lncRNA IFNG‐AS1 relative expression was positively associated with Gensini Score, which is used to evaluate the disease severity of CAD patients: this might be explained as lncRNA IFNG‐AS1 regulating the levels of inflammatory factors transcriptions, enhancing the expression of inflammatory cytokines, and accelerating the occurrence and severity of atherosclerosis; thus, its expression positively correlates with disease severity of CAD.15, 29, 30 To demonstrate the correlation of lncRNA IFNG‐AS1 with inflammation, the levels of hs‐CRP, ESR, and 6 common inflammatory cytokines in serum were also detected in CAD patients in this study, which illuminated that lncRNA IFNG‐AS1 was positively correlated with hs‐CRP, TNF‐α and IL‐6, while negatively associated with IL‐10 in patients with CAD. These results suggested that lncRNA IFNG‐AS1 was indeed an inflammation‐related gene in CAD. However, the molecular mechanism of the effect of lncRNA IFNG‐AS1 on regulating inflammation needs to be further explored in cells or animal experiments.
lncRNA ANRIL is located on human chromosome 9p21.3, which was reported to have cancer‐promoting effects in various cancers, such as non‐small cell lung cancer (NSCLC) and cervical cancer.31, 32 More importantly, lncRNA ANRIL is reported to be involved in the inflammatory diseases such as inflammatory bowel disease and atherosclerosis.33, 34 An animal experiment reveals that compared with control rats, atherosclerotic plaques and thrombi are appeared in rats with lncRNA ANRIL high expression, and serum IL‐1, IL‐6, and CRP levels are increased in rats with high expression lncRNA ANRIL compared with low‐expression lncRNA ANRIL rat.16 These results in their study suggest that lncRNA ANRIL was not only an oncogene but also a pro‐inflammatory gene. As for lncRNA ITSN1, which is located on human chromosome 21q22.11, 1 study indicates that circulating lncRNA ITSN1 is highly expressed in rheumatoid arthritis patients, and the lncRNA ITSN1 level is positively associated with ESR and disease activity score in 28 joints (DAS28).17 These suggest that lncRNA ANRIL and lncRNA ITSN1 have pro‐inflammatory effects in multiple inflammatory diseases. In this study, there was no difference between the lncRNA ANRIL and lncRNA ITSN1 expressions in CAD patients compared with that in controls, and no association of their expressions with the Gensini score was discovered. In addition, their expressions were not correlated with hs‐CPR level, ESR level, or inflammatory cytokines levels in CAD patients. The possible explanation of those results might be that lncRNA ANRIL and lncRNA ITSN1 were not causative genes in CAD.
There were some limitations in this study: (i) the sample size in our study was relatively small. Thus, study with larger sample size should be conducted in the future. (ii) This is a single‐center study, which causes selection bias, thus multi‐center research is necessary in the future.
In conclusion, circulating lncRNA IFNG‐AS1 expression correlates with increased disease risk, higher disease severity, and elevated inflammation in CAD patients.
Supporting information
Xu Y, Shao B. Circulating lncRNA IFNG‐AS1 expression correlates with increased disease risk, higher disease severity and elevated inflammation in patients with coronary artery disease. J Clin Lab Anal. 2018;32:e22452 10.1002/jcla.22452
REFERENCES
- 1. GBD 2013 Mortality and Causes of Death Collaborators . Global, regional, and national age‐sex specific all‐cause and cause‐specific mortality for 240 causes of death, 1990‐2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117‐171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. GBD 2015 Disease and Injury Incidence and Prevalence Collaborators . Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990‐2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1545‐1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. GBD 2015 Mortality and Causes of Death Collaborators . Global, regional, and national life expectancy, all‐cause mortality, and cause‐specific mortality for 249 causes of death, 1980‐2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1459‐1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mehta PK, Wei J, Wenger NK. Ischemic heart disease in women: a focus on risk factors. Trends Cardiovasc Med. 2015;25:140‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Charlson FJ, Moran AE, Freedman G, et al. The contribution of major depression to the global burden of ischemic heart disease: a comparative risk assessment. BMC Med. 2013;11:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dai X, Wiernek S, Evans JP, Runge MS. Genetics of coronary artery disease and myocardial infarction. World J Cardiol. 2016;8:1‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaudhary R, Chauhan A, Singhal M, Bagga S. Risk factor profiling and study of atherosclerotic coronary plaque burden and morphology with coronary computed tomography angiography in coronary artery disease among young Indians. Int J Cardiol. 2017;240:452‐457. [DOI] [PubMed] [Google Scholar]
- 8. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281‐297. [DOI] [PubMed] [Google Scholar]
- 9. Athanasiadis A, Sechtem U, European Society of Cardiology . [Diagnostics and therapy of chronic stable coronary artery disease: new guidelines of the European Society of Cardiology]. Herz. 2014;39:902‐912. [DOI] [PubMed] [Google Scholar]
- 10. Quinn JJ, Chang HY. Unique features of long non‐coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47‐62. [DOI] [PubMed] [Google Scholar]
- 11. Gao W, Zhu M, Wang H, et al. Association of polymorphisms in long non‐coding RNA H19 with coronary artery disease risk in a Chinese population. Mutat Res. 2015;772:15‐22. [DOI] [PubMed] [Google Scholar]
- 12. Yang Y, Cai Y, Wu G, et al. Plasma long non‐coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin Sci (Lond). 2015;129:675‐685. [DOI] [PubMed] [Google Scholar]
- 13. Shahmoradi N, Nasiri M, Kamfiroozi H, Kheiry MA. Association of the rs555172 polymorphism in SENCR long non‐coding RNA and atherosclerotic coronary artery disease. J Cardiovasc Thorac Res. 2017;9:170‐174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li X, Zhao Z, Gao C, et al. Identification of a peripheral blood Long non‐coding RNA (Upperhand) as a potential diagnostic marker of coronary artery disease. Cardiol J. 2017. 10.5603/CJ.a2017.0133. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 15. Padua D, Mahurkar‐Joshi S, Law IK, et al. A long noncoding RNA signature for ulcerative colitis identifies IFNG‐AS1 as an enhancer of inflammation. Am J Physiol Gastrointest Liver Physiol. 2016;311:G446‐G457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song CL, Wang JP, Xue X, et al. Effect of circular ANRIL on the inflammatory response of vascular endothelial cells in a rat model of coronary atherosclerosis. Cell Physiol Biochem. 2017;42:1202‐1212. [DOI] [PubMed] [Google Scholar]
- 17. Gong X, Fan X, Zhang Z, et al. Circulating lnc‐ITSN1‐2 expression presents a high value in diagnosis of rheumatoid arthritis and correlates with disease activity. Int J Clin Exp Pathol. 2017;10:10451‐10458. [PMC free article] [PubMed] [Google Scholar]
- 18. Abrams J. Clinical practice. Chronic stable angina. N Engl J Med. 2005;352:2524‐2533. [DOI] [PubMed] [Google Scholar]
- 19. Simoons ML, Windecker S. Controversies in cardiovascular medicine: chronic stable coronary artery disease: drugs vs. revascularization. Eur Heart J. 2010;31:530‐541. [DOI] [PubMed] [Google Scholar]
- 20. Ott SJ, El Mokhtari NE, Musfeldt M, et al. Detection of diverse bacterial signatures in atherosclerotic lesions of patients with coronary heart disease. Circulation. 2006;113:929‐937. [DOI] [PubMed] [Google Scholar]
- 21. Wick G, Knoflach M, Xu Q. Autoimmune and inflammatory mechanisms in atherosclerosis. Annu Rev Immunol. 2004;22:361‐403. [DOI] [PubMed] [Google Scholar]
- 22. Warnatsch A, Ioannou M, Wang Q, Papayannopoulos V. Inflammation. Neutrophil extracellular traps license macrophages for cytokine production in atherosclerosis. Science. 2015;349:316‐320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904‐914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Z, Gao W, Long QQ, et al. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci Rep. 2017;7:7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang BY, Jin Z, Zhao Z. Long intergenic noncoding RNA 00305 sponges miR‐136 to regulate the hypoxia induced apoptosis of vascular endothelial cells. Biomed Pharmacother. 2017;94:238‐243. [DOI] [PubMed] [Google Scholar]
- 27. Liu W, Liu X, Luo M, et al. dNK derived IFN‐gamma mediates VSMC migration and apoptosis via the induction of LncRNA MEG3: a role in uterovascular transformation. Placenta. 2017;50:32‐39. [DOI] [PubMed] [Google Scholar]
- 28. Yu C, Li L, Xie F, et al. LncRNA TUG1 sponges miR‐204‐5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification. Cardiovasc Res. 2017;114:168‐179. [DOI] [PubMed] [Google Scholar]
- 29. Peng H, Liu Y, Tian J, et al. The long noncoding RNA IFNG‐AS1 promotes T helper type 1 cells response in patients with Hashimoto's thyroiditis. Sci Rep. 2015;5:17702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luo M, Liu X, Meng H, et al. IFNA‐AS1 regulates CD4(+) T cell activation in myasthenia gravis though HLA‐DRB1. Clin Immunol. 2017;183:121‐131. [DOI] [PubMed] [Google Scholar]
- 31. Nie FQ, Sun M, Yang JS, et al. Long noncoding RNA ANRIL promotes non‐small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14:268‐277. [DOI] [PubMed] [Google Scholar]
- 32. Zhang D, Sun G, Zhang H, Tian J, Li Y. Long non‐coding RNA ANRIL indicates a poor prognosis of cervical cancer and promotes carcinogenesis via PI3K/Akt pathways. Biomed Pharmacother. 2017;85:511‐516. [DOI] [PubMed] [Google Scholar]
- 33. Zacharopoulou E, Gazouli M, Tzouvala M, Vezakis A, Karamanolis G. The contribution of long non‐coding RNAs in Inflammatory Bowel Diseases. Dig Liver Dis. 2017;49:1067‐1072. [DOI] [PubMed] [Google Scholar]
- 34. Chi JS, Li JZ, Jia JJ, Zhang T, Liu XM, Yi L. Long non‐coding RNA ANRIL in gene regulation and its duality in atherosclerosis. J Huazhong Univ Sci Technolog Med Sci. 2017;37:816‐822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


