Abstract
Background
Detection of circulating cell‐free mRNA serves as noninvasive tools for cancer diagnosis. As an oncofetal protein, HMGA2 (high mobility group AT‐hook 2) is upregulated in colorectal cancer (CRC) tissues. However, it is not clear whether the increased levels of circulating cell‐free HMGA2 mRNA functions as potential biomarkers for improved diagnosis of CRC.
Methods
To assess its clinical significance in diagnosis and prediction, we evaluated serum levels of circulating HMGA2 mRNA in CRC patients and in healthy controls. In this study, 83 CRC patients and 11 normal controls were enrolled in this study. We used real‐time quantitative reverse transcription‐PCR to evaluate the plasma mRNA levels of HMGA2 and analyze the correlation between their expression and clinicopathologic characteristics.
Results
We found that the levels of HMGA2 mRNA were significantly higher in CRC patients compared with healthy volunteers. The patients with right‐sided CRC, colon cancer, positive nerve infiltration, positive vascular invasion, negative microsatellite instability (MSI), and increasing in serum carbohydrate antigen (CA) 199 had higher levels of plasma HMGA2 mRNA. A strong positive correlation between circulating cell‐free HMGA2 mRNA and CA199 level in serum was found in our study. Furthermore, statistical analysis revealed that levels of HMGA2 mRNA in plasma and in tumors were strictly correlated.
Conclusions
Collectively, our data suggested that cell‐free HMGA2 mRNA in plasma might function as a novel diagnostic marker for CRC.
Keywords: biomarker, circulating cell‐free mRNA, colorectal cancer, high mobility group AT‐hook 2, serum
1. INTRODUCTION
Colorectal cancer (CRC) is the third most prevalent cancer and the third leading cause of cancer‐related death worldwide.1 The carcinogenesis of CRC is a multi‐step process which is characterized by genomic alterations.2 It is more frequent in the industrialized countries adopting western lifestyle and dietary habits, than in developing countries. Despite the great advance in overall survival, prognosis of CRC is highly dependent on the stage at diagnosis. The 5‐year survival rates range from 90% for stage I to 10% for stage IV. However, approximately 30% of patients show distant metastases at diagnosis.1 Therefore, early detection and diagnosis of CRC positively correlates with substantial improvements in survival. Several clinical tests are considered as the powerful tools for reduction of cancer‐related incidence and mortality, including endoscopy, fecal occult blood tests (FOBT), and radiology tests.3 Therefore, it is crucial to identify a novel and noninvasive biomarker for the early diagnosis of CRC.
High mobility group AT‐hook 2 belongs to a family of nonhistone chromatin‐binding proteins which regulate the process of transcription by altering the structure of chromatin.4 It is absent in normal human adult tissues, but abundantly expressed in many human malignancies, such as lung, ovarian, breast, pancreatic, and colorectal cancer. As an oncoprotein, it influences a variety of biologic processes, including cell proliferation, apoptosis, differentiation, embryogenesis, and metastasis.5
Previous studies have focused on endoscopic examination and histological evaluation of tumor specimens obtained by biopsy or surgical resection for CRC diagnosis. However, the above methods are either insufficient or invasive. Although serum carcinoembryonic antigen (CEA) and CA199 are the most widely used blood biomarkers for diagnosing CRC,6, 7 lack of sufficient sensitivity and specificity limit the extensive application. To find new tools for the early diagnosis of CRC, expressions of circulating cell‐free HMGA2 mRNA were detected in the plasma of patients and healthy controls, and relationships with clinicopathologic characteristics were also analyzed.
In this study, we evaluated the plasma mRNA levels of HMGA2 in 83 CRC patients and 11 normal controls. Our analyses showed that circulating cell‐free HMGA2 mRNA was significantly elevated in the plasma of CRC patients. High levels of HMGA2 were associated with tumor location, positive nerve infiltration, positive vascular invasion, negative microsatellite instability (MSI) status, and elevated serum CA199 levels. All of these results demonstrated that HMGA2 mRNA from plasma could be used as a potential marker for diagnosis in CRC.
2. MATERIALS AND METHODS
2.1. Patients and samples
In all, 83 CRC patients who underwent surgery at the Second Affiliated Hospital of Zhejiang University and 11 healthy donors were enrolled in this study. Tissues and plasma specimens were obtained at the time of surgery. The clinicopathologic parameters of these cases are summarized in Table 1. This study was approved by the Ethical Committee of Zhejiang University.
Table 1.
Relationship between the expression of circulating cell‐free HMGA2 mRNA in peripheral blood and clinicopathologic characteristics in 83 CRC patients
| Parameters | Patients (n) | Circulating mRNA levels | P value | |
|---|---|---|---|---|
| Mean | P25, P75 | |||
| Gender | ||||
| Female | 25 | 0.109 743 561 | 0.091 742 722, 0.140 931 388 | .322 |
| Male | 58 | 0.097 293 03 | 0.065 395 301, 0.125 066 853 | |
| Age | ||||
| <60 | 26 | 0.099 724 859 | 0.0657 648 44, 0.130 175 587 | .878 |
| ≥60 | 57 | 0.101 644 534 | 0.073 990 558, 0.136 786 893 | |
| Position | ||||
| Colon | 45 | 0.116 346 148 | 0.097 555 575, 0.139 118 238 | .006a |
| Rectum | 38 | 0.082 921 266 | 0.032 337 544, 0.105 247 53 | |
| Position | ||||
| Left | 55 | 0.092 575 405 | 0.0597 328 11, 0.124 199 426 | .011a |
| Right | 28 | 0.117 676 339 | 0.096 030 901, 0.141 137 55 | |
| Differentiation | ||||
| Low | 15 | 0.097 151 658 | 0.080 429 096, 0.124 199 426 | .692 |
| Middle + High | 66 | 0.103 157 112 | 0.067 057 415, 0.137 884 661 | |
| Tumor size (cm) | ||||
| <5 | 56 | 0.102 646 83 | 0.068 168 753, 0.138 425 668 | .961 |
| ≥5 | 24 | 0.103 273 861 | 0.072 744 688, 0.121 528 341 | |
| T stage | ||||
| T1 + T2 | 8 | 0.086 534 894 | 0.039 638 599, 0.135 736 415 | .380 |
| T3 + T4 | 73 | 0.103 656 138 | 0.072 343 228, 0.136 786 893 | |
| Lymph node | ||||
| Negative | 48 | 0.0967 132 19 | 0.068 168 753, 0.124 474 066 | .285 |
| Positive | 33 | 0.109 360 099 | 0.072 343 228, 0.139 562 53 | |
| Metastasis | ||||
| Negative | 63 | 0.099 118 668 | 0.065 429 811, 0.136 543 291 | .351 |
| Positive | 19 | 0.111 820 784 | 0.072 765 432, 0.137 030 495 | |
| AJCC stage | ||||
| 1 + 2 | 39 | 0.095 395 084 | 0.065 291 769, 0.124 207 830 | .283 |
| 3 + 4 | 42 | 0.107 874 035 | 0.072 554 330, 0.137 968 207 | |
| Nerve infiltration | ||||
| Negative | 61 | 0.093 331 443 | 0.065 584 145, 0.123 324 789 | .004a |
| Positive | 18 | 0.133 5957 48 | 0.090 526 172, 0.143 513 707 | |
| Vascular invasion | ||||
| Negative | 44 | 0.091 363 932 | 0.060 442 372, 0.124 205 729 | .048a |
| Positive | 35 | 0.114 544 887 | 0.080 429 096, 0.139 582 078 | |
| MSI | ||||
| Negative | 11 | 0.156 564 993 | 0.097 856 934, 0.232 692 177 | .022a |
| Positive | 49 | 0.094 413 756 | 0.065 584 145, 0.137 384 986 | |
| Chemotherapy | ||||
| No | 66 | 0.102 621 82 | 0.071 735 88, 0.137 884 661 | .591 |
| Yes | 17 | 0.094 914 391 | 0.063 318 757, 0.110 674 8 | |
| CEA | ||||
| <5 | 45 | 0.095 849 921 | 0.065 360 79, 0.124 203 628 | .327 |
| ≥5 | 38 | 0.107 193 114 | 0.072 554 33, 0.139 787 002 | |
| CA199 | ||||
| <37 | 62 | 0.093 464 516 | 0.064 270 753, 0.124 296 575 | .022a |
| ≥37 | 21 | 0.123 418 323 | 0.077 289 948, 0.141 579 124 | |
MSI, microsatellite instability; CRC, colorectal cancer; HMGA2, high mobility group AT‐hook 2.
P < .05, statistical significance.
2.2. RNA extraction from plasma
RNA was extracted using QIAamp Circulating Nucleic Acid kit (Qiagen, Shanghai, China) according to the manufacturer's instructions. Plasma of patients and healthy controls was obtained from peripheral blood by centrifugation. Plasma samples were lysed with proteinase K, at 60°C for 30 min. Lysates were then transferred onto a QIAamp Mini column by vacuum pressure. RNA was dissolved in 30 μl of elution buffer, at 37°C for 10 min.
2.3. Real‐time quantitative reverse transcription‐PCR
cDNA was synthesized from RNA (100 ng) using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher, Shanghai, China) according to instructions provided by the manufacturer. Then, real‐time PCR assays were performed by SYBR Green PCR Master Mix (Applied Biosystems, Shanghai, China) using an ABI Prism 7900 Detection System. The primers used in this study were as follows: HMGA2 forward 5′‐ATCTACTACCAAGAACCA‐3′, HMGA2 reverse 5′‐ACACATAAGGCTCATAGA‐3′, GAPDH forward 5′‐CCCTTCATTGACCTCAACTACATG‐3′, GAPDH reverse 5′‐TGGGATTTCCATTGATGACAAGC‐3′.
2.4. Statistical analyses
Statistical analyses were conducted by SPSS version 22.0 software. The results were expressed as mean ± standard deviation and analyzed by Student's t test or one‐way ANOVA. P < .05 was considered as statistically significant. To determine the performance of plasma HMGA2 mRNA in diagnosing CRC, receiver operating characteristic (ROC) curve was plotted and the corresponding area under the curve (AUC) was analyzed.
3. RESULTS
3.1. Circulating HMGA2 mRNA served as a potential biomarker for CRC diagnosis
Circulating cell‐free HMGA2 mRNA was amplified by real‐time qRT‐PCR from the plasma of 83 CRC patients prior to surgery and the plasma of 11 healthy volunteers. The detail clinical characteristics of the recruited patients are shown in Table 1. Compared to healthy controls, statistically significant higher levels of circulating cell‐free HMGA2 mRNA were observed in CRC patients (P < .001; Figure 1A).
Figure 1.
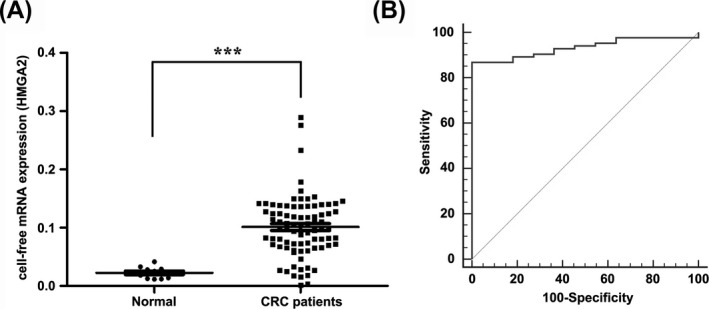
Circulating HMGA2 mRNA expression is increased in the plasma of colorectal cancer (CRC) patients. (A) Dot plot analysis showed that higher levels of circulating cell‐free HMGA2 mRNA were observed in CRC patients. Each dot represented as one case. The results were presented as mean ± SEM. ***P < .001. (B) ROC curves showed that plasma HMGA2 mRNA discriminated between CRC patients and healthy controls. The AUC of HMGA2 mRNA was 0.932. AUC, area under the curve; ROC, receiver operating characteristic
Subsequently, ROC curve analysis was performed to evaluate the diagnostic capabilities of circulating HMGA2 mRNA for CRC. As shown in Figure 1B, the AUC was 0.932 (95% CI, 0.882‐0.983). And the best cutoff value was 0.044 which provided a sensitivity of 86.75% (95% CI, 77.5%‐93.2%). More intriguingly, a strong positive correlation was found between plasma HMGA2 mRNA and CA199 levels (r = .380, P < .001; Figure 2). All these results demonstrated that circulating HMGA2 mRNA might function as a novel diagnostic marker for CRC.
Figure 2.

Correlation between HMGA2 mRNA and CA199 was shown in the dot plot. r = .380, P < .001
3.2. Plasma HMGA2 mRNA correlates with tumor location, nerve infiltration, vascular invasion, and MSI status
To further explore the role of HMGA2 in the progression of CRC, the association between plasma HMGA2 mRNA level and clinicopathologic parameters was evaluated. As shown in Table 1, circulating HMGA2 mRNA levels were significantly correlated with tumor location, nerve infiltration, vascular invasion, MSI status, and serum CA199 levels. However, there was no difference between HMGA2 and other characteristics. As for tumor location, the patients with colon cancer (P = .006; Figure 3A) and right‐sided CRC (P = .011; Figure 3B) showed higher levels of circulating cell‐free HMGA2 mRNA. In addition, positive nerve infiltration (P = .004; Figure 3C), positive vascular invasion (P = .048; Figure 3D), negative MSI status (P = .022; Figure 3E), or elevated serum CA199 levels (P = .022; Figure 3F) significantly correlated with higher levels of plasma HMGA2 mRNA, suggesting that circulating HMGA2 mRNA increased with tumor progression.
Figure 3.

Dot plot analysis showed that levels of circulating cell‐free HMGA2 mRNA were correlated with tumor location (A and B), nerve infiltration (C), vascular invasion (D), microsatellite instability (MSI) status (E), and serum CA199 levels (F). The results were presented as mean ± SEM. *P < .05; **P < .01
3.3. Circulating HMGA2 mRNA in plasma correlated with HMGA2 mRNA expression in tissues
To explore whether levels of HMGA2 mRNA in plasma were correlated with HMGA2 levels in tumors, real‐time qRT‐PCR analysis was employed for measurement of HMGA2 expressions in 54 primary colorectal cancer samples. Notably, levels of HMGA2 mRNA in plasma and HMGA2 protein in tumors were found to be strictly correlated (r = .767, P < .001, Figure 4).
Figure 4.
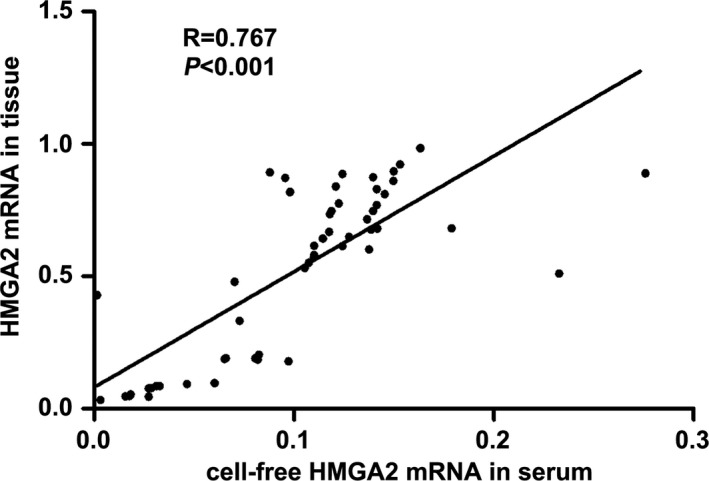
Correlation between HMGA2 mRNA in plasma and HMGA2 mRNA expression in tissues was shown in the dot plot. r = .767, P < .001
4. DISCUSSION
Accumulating studies have focused on cell‐free nucleic acids (cfNAs) as noninvasive biomarkers for diagnostic and prognostic purposes.8, 9 The field of cfNAs in cancer has grown very fast in the past 20 years. It is known that they originate from circulating tumor cells and increased levels of cfNAs (such as DNA, RNA, and miRNA) are significantly associated with tumor behaviors.10, 11, 12, 13 Analyses of circulating cell‐free nucleic acids can serve as “liquid biopsies” approaches, which can be exploited for diagnosis in the early stages of cancer, assessment of prognosis, detection of recurrence, prediction of response to treatment, and identification of different genomic alterations during the course of treatment.8, 12, 14, 15 cfNAs were first identified in human bloodstream in 1948 by Mandel et al.16 Chan and co‐workers conducted a prospective study and reported that circulating cell‐free Epstein‐Barr virus (EBV) DNA could be used for screening early asymptomatic nasopharyngeal carcinoma.17 Body fluids are the main sources for screening and detection of many noninvasive biomarkers, including sputum, saliva, and pleural effusion.18, 19 However, the causes of the increase in cfNA during cancer development are still not well understood. Besides the cfDNA, circulating mRNA was also observed in the plasma of cancer patients.20 Joosse and colleagues reported that the levels of circulating cell‐free MAGE‐A, BORIS mRNA, as well as let‐7b were significantly increased in the patients with malignant breast carcinomas than in benign breast diseases.21 Kim et al found that the urine levels of cell‐free UBE2C RNA could differentiate between bladder cancer and hematuria, and it proved to be an independent indicator of prognosis.22 In patients with breast cancer, cell‐free mammaglobin and telomerase mRNAs could serve as useful cancer markers for diagnosis.23, 24 Presence of cyclin D1 mRNA in plasma was associated with poor clinical outcome in a subset of patients with a good prognosis and it could be used as a marker of resistance to endocrine therapies.25 Miura et al found that hTERT mRNA, combined with EGFR mRNA, was significantly elevated in the plasma of lung cancer patients, and might be useful to assess clinical stage.26 In CRC, the presence of EGFR, CK19, CK20, REG4, uPA, and NPRL2 mRNA in peripheral blood could discriminate CRC from healthy controls, and furthermore, they were also correlated with cancer progression.27 Thus, the applications of cfRNA‐based assays enable the assessment of cancers through gene expression profiling. Although they have a great potential, many problems must be tackled before proceeding to the clinic.8, 14 Because of the low stability, high heterogeneity, and low occurrence frequencies of cfRNAs in plasma, one of the most important issues is the development and standardization of more powerful and reliable technologies.8 It extends to each step in the process, including the blood collection procedure to improve the stability, cfRNAs extraction and quantification methods, and the approaches for detection of molecular and genetic alterations. With the rapid progress of technology,28, 29, 30, 31 sensitivity and specificity have been greatly improved.
As an oncofetal protein, HMGA2 is highly expressed in a variety of neoplastic tissues, but low or absent in normal adult tissues.32 However, there have been few studies focusing on the detection of circulating HMGA2 mRNA in blood. In breast cancer, expression of cell‐free HMGA2 mRNA in plasma may serve as a powerful indicator for predicting worse prognosis.33, 34 In their study of circulating HMGA2 in the peripheral blood of patients with epithelial ovarian cancer, Galdiero et al showed for the first time the expression of HMGA2 in the plasma of patients, instead of healthy donors.35 In the present study, we found that patients with higher levels of circulating HMGA2 mRNA were more likely to be malignant. All these results demonstrated that circulating HMGA2 mRNA could serve as a potential biomarker for cancer diagnosis.
To our knowledge, this is the first study focusing on the detection of cell‐free HMGA2 mRNA in the plasma of CRC patients. The level of circulating HMGA2 mRNA was found to be substantially higher in patients than in healthy volunteers. Our results demonstrated that HMGA2 gave a high diagnostic performance. Its expression was found to be significantly associated with tumor location, nerve infiltration, vascular invasion, MSI status, and serum CA199 levels. Levels of HMGA2 mRNA in plasma and HMGA2 mRNA in tissues were found to be strictly correlated. In summary, our study strongly indicated that cell‐free HMGA2 mRNA in the plasma might act as a potential biomarker in the diagnostic prediction of CRC.
ACKNOWLEDGMENTS
We would like to thank the Zhejiang Provincial Natural Science Foundation of China (LY17H160034), National Natural Science Foundation of China (81772527, 81672342, 81302073, and 81302072), and Fundamental Research Funds for the Central Universities (2017QNA7004).
Sahengbieke S, Wang J, Li X, Wang Y, Lai M, Wu J. Circulating cell‐free high mobility group AT‐hook 2 mRNA as a detection marker in the serum of colorectal cancer patients. J Clin Lab Anal. 2018;32:e22332 10.1002/jcla.22332
Sana Sahengbieke and Jian Wang contributed equally to this work.
REFERENCES
- 1. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490‐1502. [DOI] [PubMed] [Google Scholar]
- 2. Peters U, Bien S, Zubair N. Genetic architecture of colorectal cancer. Gut. 2015;64:1623‐1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robertson DJ, Imperiale TF. Stool testing for colorectal cancer screening. Gastroenterology. 2015;149:1286‐1293. [DOI] [PubMed] [Google Scholar]
- 4. Hammond SM, Sharpless NE. HMGA2, microRNAs, and stem cell aging. Cell. 2008;135:1013‐1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Young AR, Narita M. Oncogenic HMGA2: short or small? Genes Dev. 2007;21:1005‐1009. [DOI] [PubMed] [Google Scholar]
- 6. Stiksma J, Grootendorst DC, van der Linden PW. CA 19‐9 as a marker in addition to CEA to monitor colorectal cancer. Clin Colorectal Cancer. 2014;13:239‐244. [DOI] [PubMed] [Google Scholar]
- 7. Basbug M, Arikanoglu Z, Bulbuller N, et al. Prognostic value of preoperative CEA and CA 19‐9 levels in patients with colorectal cancer. Hepatogastroenterology. 2011;58:400‐405. [PubMed] [Google Scholar]
- 8. Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531‐548. [DOI] [PubMed] [Google Scholar]
- 9. Suraj S, Dhar C, Srivastava S. Circulating nucleic acids: an analysis of their occurrence in malignancies. Biomed Rep. 2017;6:8‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calapre L, Warburton L, Millward M, Ziman M, Gray ES. Circulating tumour DNA (ctDNA) as a liquid biopsy for melanoma. Cancer Lett. 2017;404:62‐69. [DOI] [PubMed] [Google Scholar]
- 11. Song CX, Yin S, Ma L, et al. 5‐Hydroxymethylcytosine signatures in cell‐free DNA provide information about tumor types and stages. Cell Res. 2017. 10.1038/cr.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Armand‐Labit V, Pradines A. Circulating cell‐free microRNAs as clinical cancer biomarkers. Biomol Concepts. 2017. 10.1515/bmc-2017-0002. [DOI] [PubMed] [Google Scholar]
- 13. Zhou D, Tang W, Liu X, An HX, Zhang Y. Clinical verification of plasma messenger RNA as novel noninvasive biomarker identified through bioinformatics analysis for lung cancer. Oncotarget. 2017;8:43978‐43989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mader S, Pantel K. Liquid biopsy: current status and future perspectives. Oncol Res Treat. 2017;40:404‐408. [DOI] [PubMed] [Google Scholar]
- 15. von Bubnoff N. Liquid biopsy: approaches to dynamic genotyping in cancer. Oncol Res Treat. 2017;40:409‐416. [DOI] [PubMed] [Google Scholar]
- 16. Mandel P, Metais P. Not available. C R Seances Soc Biol Fil. 1948;142:241‐243. [PubMed] [Google Scholar]
- 17. Chan KCA, Woo JKS, King A, et al. Analysis of plasma Epstein‐Barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med. 2017;377:513‐522. [DOI] [PubMed] [Google Scholar]
- 18. Chan AK, Chiu RW, Lo YM. Cell‐free nucleic acids in plasma, serum and urine: a new tool in molecular diagnosis. Ann Clin Biochem. 2003;40(Pt 2):122‐130. [DOI] [PubMed] [Google Scholar]
- 19. Swarup V, Rajeswari MR. Circulating (cell‐free) nucleic acids–a promising, non‐invasive tool for early detection of several human diseases. FEBS Lett. 2007;581:795‐799. [DOI] [PubMed] [Google Scholar]
- 20. Schwarzenbach H, Hoon DS, Pantel K. Cell‐free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426‐437. [DOI] [PubMed] [Google Scholar]
- 21. Joosse SA, Muller V, Steinbach B, Pantel K, Schwarzenbach H. Circulating cell‐free cancer‐testis MAGE‐A RNA, BORIS RNA, let‐7b and miR‐202 in the blood of patients with breast cancer and benign breast diseases. Br J Cancer. 2014;111:909‐917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim WT, Jeong P, Yan C, et al. UBE2C cell‐free RNA in urine can discriminate between bladder cancer and hematuria. Oncotarget. 2016;7:58193‐58202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gal S, Fidler C, Lo YM, et al. Detection of mammaglobin mRNA in the plasma of breast cancer patients. Ann N Y Acad Sci. 2001;945:192‐194. [DOI] [PubMed] [Google Scholar]
- 24. Chen XQ, Bonnefoi H, Pelte MF, et al. Telomerase RNA as a detection marker in the serum of breast cancer patients. Clin Cancer Res. 2000;6:3823‐3826. [PubMed] [Google Scholar]
- 25. Garcia V, Garcia JM, Pena C, et al. Free circulating mRNA in plasma from breast cancer patients and clinical outcome. Cancer Lett. 2008;263:312‐320. [DOI] [PubMed] [Google Scholar]
- 26. Miura N, Nakamura H, Sato R, et al. Clinical usefulness of serum telomerase reverse transcriptase (hTERT) mRNA and epidermal growth factor receptor (EGFR) mRNA as a novel tumor marker for lung cancer. Cancer Sci. 2006;97:1366‐1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yeh CS, Wang JY, Wu CH, et al. Molecular detection of circulating cancer cells in the peripheral blood of patients with colorectal cancer by using membrane array with a multiple mRNA marker panel. Int J Oncol. 2006;28:411‐420. [PubMed] [Google Scholar]
- 28. Ehlert T, Tug S, Brahmer A, et al. Establishing PNB‐qPCR for quantifying minimal ctDNA concentrations during tumour resection. Sci Rep. 2017;7:8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mehrotra M, Singh RR, Chen W, et al. Study of preanalytic and analytic variables for clinical next‐generation sequencing of circulating cell‐free nucleic acid. J Mol Diagn. 2017;19:514‐524. [DOI] [PubMed] [Google Scholar]
- 30. Spornraft M, Kirchner B, Haase B, Benes V, Pfaffl MW, Riedmaier I. Optimization of extraction of circulating RNAs from plasma–enabling small RNA sequencing. PLoS One. 2014;9:e107259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wood‐Bouwens C, Lau BT, Handy CM, Lee H, Ji HP. Single‐color digital PCR provides high‐performance detection of cancer mutations from circulating DNA. J Mol Diagn. 2017;19:697‐710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899‐910. [DOI] [PubMed] [Google Scholar]
- 33. Langelotz C, Schmid P, Jakob C, et al. Expression of high‐mobility‐group‐protein HMGI‐C mRNA in the peripheral blood is an independent poor prognostic indicator for survival in metastatic breast cancer. Br J Cancer. 2003;88:1406‐1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sezer O, Langelotz C, Blohmer JU, Schmid P, Akrivakis K, Possinger K. Detection of HMGI‐C in the peripheral blood of breast cancer patients. Eur J Cancer. 2000;36:1944‐1948. [DOI] [PubMed] [Google Scholar]
- 35. Galdiero F, Romano A, Pasquinelli R, et al. Detection of high mobility group A2 specific mRNA in the plasma of patients affected by epithelial ovarian cancer. Oncotarget. 2015;6:19328‐19335. [DOI] [PMC free article] [PubMed] [Google Scholar]


