Abstract
Background
Circular RNA (circRNA) is a new type of noncoding RNA that can serve as ideal biomarkers. Evidence has showed that circRNAs play an important role in carcinogenesis and cancer development. However, little is known about the diagnostic value of circRNAs in papillary thyroid carcinoma (PTC) as well as their associations with clinicopathologic characteristics of patients with PTC.
Methods
The expression levels of hsa_circ_0137287 were detected in 120 PTC and 60 adjacent noncancerous thyroid tissues by quantitative real‐time polymerase chain reaction. The relationships between the expression of hsa_circ_0137287 in PTC and the clinicopathologic factors were analyzed. Finally, receiver operating characteristic (ROC) curves were generated to assess the diagnostic value of hsa_circ_0137287 as a biomarker for PTC.
Results
The expression of hsa_circ_0137287 was significantly downregulated in PTC tissues compared with adjacent noncancerous tissues (P < .0001). Downregulation of hsa_circ_0137287 correlated with aggressive clinicopathologic characteristics of PTC such as extrathyroidal extension (P < .001), lymph node metastasis (P = .022), advanced T stage (P < .001) and larger tumor size (P < .001). The ROC curves revealed that hsa_circ_0137287 had a potential diagnostic value in predicting malignancy, extrathyroidal extension and lymph node metastasis. The area under curves were 0.8973 (95% CI = 0.8452‐0.9494, P < .0001), 0.6885 (95%CI = 0.5908‐0.7862, P = .0009), and 0.6691(95%CI = 0.5641‐0.7742, P = .0034), respectively.
Conclusions
Our findings suggest that hsa_circ_0137287 may serve as a novel biomarker for PTC.
Keywords: biomarker, circular RNA, extrathyroidal extension, lymph node metastasis, papillary thyroid carcinoma
1. INTRODUCTION
Thyroid cancer, the most common endocrine malignancy, has increased rapidly in incidence over the last several decades.1, 2 Papillary thyroid carcinoma (PTC) is the predominant histopathologic type of thyroid cancer, accounting for more than 80% of all cases.3 Although the majority of PTC is indolent, with a 10‐year survival of >90%, some aggressive characteristics of PTC are associated with poor prognosis, such as older age, larger tumor size, extrathyroidal extension, lymph node and distant metastases, advanced TNM stage, and certain subtypes.4 These aggressive PTCs usually require aggressive treatment such as bilateral thyroidectomy and extensive lymph node dissection, as well as high dose of postoperative thyroid hormone therapy or radioactive iodine therapy. Aggressive treatment also results in more complications and poor life quality, let along the high economic burden on patients with PTC.4 Therefore, finding novel biomarkers to improve diagnosis of PTC and to identify those with high risk of aggressiveness at an early stage is of great significance.
Circular RNAs (circRNAs) are a new type of noncoding RNAs that are widely expressed across species.5 Characterized by covalently closed continuous loops with neither 5′ to 3′ polarity nor a polyadenylated tail, circRNAs are resistant to exonucleases and appear to be stably expressed in cell‐type‐specific and developmental stage–specific manners.6, 7, 8, 9 These features make them the ideal biomarkers for disease diagnosis. Recent studies have also revealed that circRNAs are involved in a variety of cancers, functioning as microRNA (miRNA) sponges, RNA‐binding protein (RBP) sequestering agents or transcription and splicing regulators.10, 11, 12, 13, 14, 15 These pieces of evidence indicate that certain circRNAs can be used as promising biomarkers for cancer diagnosis or prognosis monitoring. However, to date, the role of circRNAs in PTC remains largely unknown.
Hsa_circ_0137287 is a 284 bp circRNA that was primarily identified in mammalian brain.16 The gene of hsa_circ_0137287 is located at chr8:92301363‐92307931 + , and its host gene symbol is SLC26A7.17 In this study, we attempt to investigate the expression of hsa_circ_0137287 and its clinical significance in PTC. The reason why we chose hsa_circ_0137287 as the candidate circRNA of this study is that it was found to be downregulated in PTC tissues in our previous RNA sequencing screening. By expanding the sample size, we found that hsa_circ_0137287 expression levels were associated with aggressive clinicopathologic characteristics of patients with PTC. Therefore, hsa_circ_0137287 might become a novel biomarker for the diagnosis of PTC.
2. MATERIALS AND METHODS
2.1. Patient samples
The study was approved by the ethics committee of Zhejiang Cancer Hospital, Hangzhou, China. Written informed consent was obtained from all patients. One hundred and twenty PTC samples and 60 adjacent normal thyroid tissues were collected from patients at the Department of Head and Neck Surgery of Zhejiang Cancer Hospital between September 2016 and October 2017. The diagnosis of PTC was pathologically confirmed by two pathologists independently. All tissue samples were immediately snap‐frozen and stored in liquid nitrogen until further use.
2.2. RNA extraction
Total RNA was extracted from tissue samples using TRIzol reagent (Life Technologies, CA, USA) following the manufacturer’s instructions. The quality of the RNA samples was determined by OD260/280 assessment using a NanoDrop ND‐2000 instrument (Thermo Fisher Scientific, MA, USA).
2.3. Quantitative real‐time polymerase chain reaction (qRT‐PCR)
Qualified RNA was reverse‐transcribed to synthesize cDNA using PrimeScript RT Master Mix (Takara, Dalian, China) and then subjected to qRT‐PCR analysis with SYBR Premix Ex Taq II (Takara) on a LightCycler 480 system (Roche). The amplification conditions were 95°C for 5 second, 55°C for 30 second, and 72°C for 30 second with a total of 40 cycles. Glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) was used as the internal control. Divergent primers specific for the back‐splicing junction of hsa_circ_0137287 were designed, and melt curves were analyzed to keep the specificity of the PCR products. The primers used for qRT‐PCR analysis were as follows: hsa_circ_0137287 (F 5′‐ ACAAGCGTGCTGGGCTTATC‐3′, R 5′‐ GGCCAATCCTGAATCACACCT‐3′) and GAPDH (F 5′‐ GGCCAATCCTGAATCACACCT‐3′, R 5′‐ GAAGGCTGGGGCTCATTT‐3′). The relative expression level of hsa_circ_0137287 was calculated using the method.
2.4. Statistical analysis
All statistical analyses were performed using GraphPad Prism 6 (GraphPad Software, CA, USA) and SPSS 19.0 (IBM, Chicago, IL, USA). Data were expressed as mean ± standard deviation (SD). Significant differences were determined by Student’s t test and Spearman correlation test. Receiver operating characteristic (ROC) curves were established to evaluate the diagnostic value of hsa_circ_0137287 for PTC. P < .05 was considered statistically significant.
3. RESULTS
3.1. Clinicopathologic characteristics of PTC patients
The medical records of the 120 PTC patients were reviewed. There were 82 females and 38 males, aged from 16 to 66 (median 44). Of the 120 cases of PTC, 39 were with extrathyroidal extension, 84 with lymph node metastases, and only one had distant metastasis. Other clinicopathologic information of the patients is summarized in Table 1.
Table 1.
Correlations between hsa_circ_0137287 expression and clinicopathologic characteristics of patients with papillary thyroid carcinoma
| Characteristics | No. of patients (%) | Mean ± SD | P value |
|---|---|---|---|
| Gender | |||
| Male | 38 (31.7) | 0.4753 ± 0.8222 | .834 |
| Female | 82 (68.3) | 0.5029 ± 0.5899 | |
| Age(years) | |||
| <45 | 59 (49.2) | 0.4302 ± 0.5834 | .305 |
| ≥45 | 61 (50.8) | 0.5560 ± 0.7416 | |
| Extrathyroidal extension | |||
| Yes | 39 (32.5) | 0.2288 ± 0.2322 | <.001a |
| No | 81 (67.5) | 0.6219 ± 0.7673 | |
| T stage | |||
| T1‐2 | 80 (66.7) | 0.6278 ± 0.7703 | <.001a |
| T3‐4 | 40 (33.3) | 0.2268 ± 0.2295 | |
| Lymph node metastasis | |||
| Yes | 84 (70.0) | 0.3898 ± 0.5779 | .022a |
| No | 36 (30.0) | 0.7376 ± 0.8008 | |
| TNM stage | |||
| I‐II | 79 (65.8) | 0.5380 ± 0.6441 | .321 |
| III‐IV | 41 (34.2) | 0.4096 ± 0.7143 | |
| Multifocality | |||
| Yes | 64 (53.3) | 0.4983 ± 0.7215 | .943 |
| No | 56 (46.7) | 0.4894 ± 0.6092 | |
| Microcarcinoma | |||
| Yes | 35 (29.2) | 0.8046 ± 0.9174 | .011a |
| No | 85 (70.8) | 0.3663 ± 0.4859 | |
| Tumor size (cm) | |||
| ≤2 | 77 (64.2) | 0.6448 ± 0.7760 | <.001a |
| >2 | 43 (35.8) | 0.2245 ± 0.2478 | |
Significant association.
3.2. Hsa_circ_0137287 is downregulated in PTC
The expression levels of hsa_circ_0137287 were detected in 120 PTC and 60 adjacent noncancerous thyroid tissues by qRT‐PCR. The expression of hsa_circ_0137287 was significantly downregulated in PTC tissues compared with normal tissues (P < .0001, Figure 1). The mean fold change was 7.33.
Figure 1.
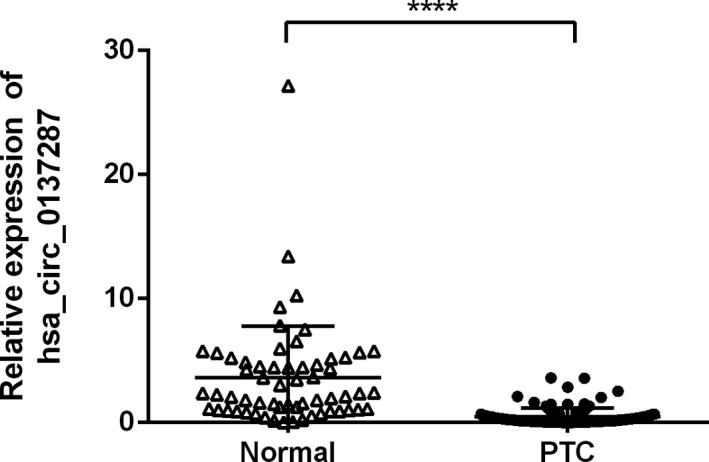
The expression of hsa_circ_0137287 in 120 PTC tissues and 60 normal thyroid tissues, **** P < 0.0001
3.3. Downregulation of hsa_circ_0137287 correlates with aggressive clinicopathologic characteristics of PTC
We also analyzed the association between the expression level of hsa_circ_0137287 and clinicopathologic characteristics of patients with PTC. The results (Table 1) showed that hsa_circ_0137287 was significantly associated with extrathyroidal extension (P < .001), T stage (P < .001), lymph node metastasis (P = .022), microcarcinoma (P = .011), and tumor size (P < .001). In particular, there was a negative correlation between the expression of hsa_circ_0137287 and tumor size (r = −.438, P < .001, Figure 2). However, there were no significantly associations between the expression of hsa_circ_0137287 and other clinicopathologic parameters, such as gender, age, TNM stage, and multifocality.
Figure 2.
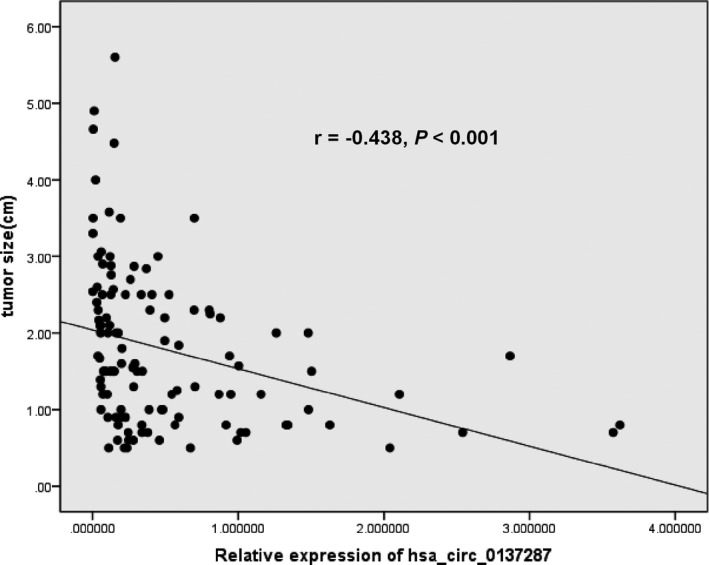
The correlation between the expression of hsa_circ_0137287 and tumor size (r = −0.438, P < .001)
3.4. Potential diagnostic value of hsa_circ_0137287 in PTC
We further performed an analysis to explore the diagnostic value of hsa_circ_0137287 as a potential biomarker for PTC. Three ROC curves were built to evaluate whether hsa_circ_0137287 could predict malignancy, extrathyroidal extension, and lymph node metastasis, respectively. As shown in Figure 3, the area under curve (AUC) was 0.8973 (95%CI = 0.8452‐0.9494, P < .0001), suggesting that hsa_circ_0137287 could serve as a diagnostic biomarker for PTC. When using the cutoff value of 0.7032, the Youden index, sensitivity, and specificity were 0.6917, 79.2%, and 90.0%, respectively. Additionally, we also found that hsa_circ_0137287 might be a promising biomarker for predicting extrathyroidal extension (Figure 3). The AUC was 0.6885 (95%CI = 0.5908‐0.7862, P = .0009). The Youden index, sensitivity and specificity were 0.2953, 64.1% and 65.4%, respectively, when taking 0.1924 as the cutoff value. Finally, as a potential diagnostic tool for lymph node metastasis (Figure 3), the AUC was 0.6691(95%CI = 0.5641‐0.7742, P = .0034).The Youden index, sensitivity, and specificity were 0.2817, 89.3%, and 38.9%, respectively, when taking 0.8368 as the cutoff value.
Figure 3.
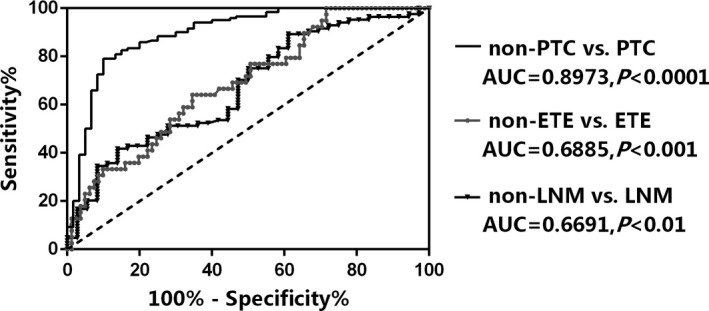
The ROC curves of hsa_circ_0137287. ETE, extrathyroidal extension; LNM, lymph node metastasis
4. DISCUSSION
circRNAs were first found to be by‐products of mRNA splicing with no functions.18 They are characterized by the distinct loop structures without 5′ caps and 3′ tails compared to mRNAs.19 Due to the application of next‐generation sequencing and bioinformatics, circRNAs were now discovered to be widely expressed in cells, exceeding the abundance of the linear mRNAs.5, 10, 20 circRNAs have now become a new star in RNA research field, as recent reports revealed their critical roles in various biological processes.12, 21, 22, 23, 24 They can mediate gene expression by sponging miRNAs and RBPs as well as regulating transcription and alternative splicing. Notably, recent evidence also shows that some circRNAs can be translated into proteins,25, 26 which expands the definition and function of circRNA. Moreover, circRNAs are more stable than linear RNAs due to the lack of free 3′ or 5′ ends, which makes them resistant to RNA exonuclease. Thus, they can serve as potential biomarkers for cancers. A growing number of studies have shown that circRNAs are implicated in a variety of cancers, such as gastric cancer,27, 28, 29 hepatocellular carcinoma,30, 31 colorectal cancer,32 esophageal squamous cell carcinoma,33 and lung cancer.34, 35 However, researchers have yet to reveal a definite relationship between circRNA and PTC.
To date, only one study reported that circRNAs were dysregulated in PTC and hsa_circRNA_100395/miR‐141‐3p/miR‐200a‐3p axis was predicted to be involved in the pathogenesis of PTC through microarray profiling and bioinformatic analysis.36 Here, we find that hsa_circ_0137287 is significantly downregulated in PTC and is potentially valuable in the diagnosis of PTC with a high value of AUC. As the 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer suggested, extrathyroidal extension, lymph node, and tumor size are important factors in determining treatment and prognosis of patients with PTC. In this study, we also find that downregulation of hsa_circ_0137287 is significantly associated with extrathyroidal extension, lymph node, and larger tumor size of PTC, which indicates that it may also serve as a promising tool in evaluating aggressive clinicopathologic characteristics of PTC.
However, a number of limitations exist in the current study. First, the present study only detected the expression of hsa_circ_0137287 in tissue samples. It would be of more value if the hsa_circ_0137287 can be detected in the circulation of patients with PTC. Second, it is necessary to explore the further function and mechanism of hsa_circ_0137287 in PTC through in vitro and in vivo studies. Future studies on hsa_circ_0137287 will eventually improve our understanding of the mechanisms underlying PTC tumorigenesis and progression.
In conclusion, in this study we first find that hsa_circ_0137287 is downregulated in PTC and the expression of hsa_circ_0137287 is correlated with aggressive clinicopathologic characteristics such as extrathyroidal extension, lymph node metastasis, advanced T stage, and larger tumor size. These findings suggest that hsa_circ_0137287 may serve as a potential diagnostic biomarker for PTC. However, the molecular mechanisms of hsa_circ_0137287 that involve in PTC need to be further studied.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (Grant nos. 81702645, 81672642, 81702653, and 81602349), Medical and Health Research Program of Zhejiang Province (Grant No. 2015DTA003) and Zhejiang Province Natural Science Foundation of China (Grant No. LY17H070003).
Lan X, Cao J, Xu J, et al. Decreased expression of hsa_circ_0137287 predicts aggressive clinicopathologic characteristics in papillary thyroid carcinoma. J Clin Lab Anal. 2018;32:e22573 10.1002/jcla.22573
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7‐30. [DOI] [PubMed] [Google Scholar]
- 2. Morris LG, Tuttle RM, Davies L. Changing trends in the incidence of thyroid cancer in the United States. JAMA Otolaryngol Head Neck Surg. 2016;142:709‐711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lim H, Devesa SS, Sosa JA, Check D, Kitahara CM. Trends in thyroid cancer incidence and mortality in the United States, 1974‐2013. JAMA. 2017;317:1338‐1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haugen BR, Alexander EK, Bible KC, et al. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid. 2016;26:1‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qu S, Liu Z, Yang X, et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301‐309. [DOI] [PubMed] [Google Scholar]
- 6. Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454‐2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829‐1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333‐338. [DOI] [PubMed] [Google Scholar]
- 10. Chen B, Huang S. Circular RNA: an emerging non‐coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41‐50. [DOI] [PubMed] [Google Scholar]
- 11. Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018. 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 13. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA‐9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151‐1164. [DOI] [PubMed] [Google Scholar]
- 14. Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339‐2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rybak‐Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870‐885. [DOI] [PubMed] [Google Scholar]
- 17. Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666‐1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell. 1991;64:607‐613. [DOI] [PubMed] [Google Scholar]
- 19. Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome‐wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131‐3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J Hematol Oncol. 2018;11:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Piwecka M, Glazar P, Hernandez‐Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526. [DOI] [PubMed] [Google Scholar]
- 22. Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR‐223. Eur Heart J. 2016;37:2602‐2611. [DOI] [PubMed] [Google Scholar]
- 23. Yang C, Yuan W, Yang X, et al. Circular RNA circ‐ITCH inhibits bladder cancer progression by sponging miR‐17/miR‐224 and regulating p21. PTEN expression. Mol Cancer. 2018;17:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang WJ, Wang Y, Liu S, et al. Silencing circular RNA hsa_circ_0000977 suppresses pancreatic ductal adenocarcinoma progression by stimulating miR‐874‐3p and inhibiting PLK1 expression. Cancer Lett. 2018;422:70‐80. [DOI] [PubMed] [Google Scholar]
- 25. Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018. 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Legnini I, Di Timoteo G, Rossi F, et al. Circ‐ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22‐37 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xie Y, Shao Y, Sun W, et al. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomark Med. 2018;12:11‐20. [DOI] [PubMed] [Google Scholar]
- 28. Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167‐171. [DOI] [PubMed] [Google Scholar]
- 29. Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32:e22281 10.1002/jcla.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161‐169. [DOI] [PubMed] [Google Scholar]
- 31. Fu L, Yao T, Chen Q, et al. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405‐58416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8:16020‐16025. [PMC free article] [PubMed] [Google Scholar]
- 33. Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu X, Wang X, Wei S, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284:2170‐2182. [DOI] [PubMed] [Google Scholar]
- 35. Yao JT, Zhao SH, Liu QP, et al. Over‐expression of CircRNA_100876 in non‐small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453‐456. [DOI] [PubMed] [Google Scholar]
- 36. Peng N, Shi L, Zhang Q, et al. Microarray profiling of circular RNAs in human papillary thyroid carcinoma. PLoS ONE. 2017;12:e0170287. [DOI] [PMC free article] [PubMed] [Google Scholar]


