Abstract
Background
Recent studies have revealed that circular RNAs are involved in the biological process of some kinds of human cancers. However, little is known about their diagnostic values and functions in colorectal cancer (CRC).
Methods
The expression levels of hsa_circ_0000567 in 102 paired CRC tissues and adjacent noncancerous tissues, 5 CRC cell lines, and a normal colorectal epithelial cell line were detected by quantitative real‐time polymerase chain reaction (qRT‐PCR). The correlations between hsa_circ_0000567 expression levels and the clinicopathological factors of patients with CRC were analyzed. Furthermore, the loss‐of‐function assay was performed to investigate the functions of hsa_circ_0000567 in vitro. Finally, a receiver operating characteristic (ROC) curve was established to evaluate the diagnostic value of hsa_circ_0000567.
Results
Hsa_circ_0000567 expression was significantly downregulated in CRC tissues and CRC cell lines. In addition, the decreased hsa_circ_0000567 expression in CRC was negatively correlated with tumor size (P = .011), lymph metastasis (P = .003), distal metastasis (P < .0001), and tumor–node–metastasis (TNM) stage (P = .003) in CRC. Moreover, knockdown of hsa_circ_0000567 promoted CRC cells proliferation and migration in vitro. Importantly, the area under the ROC curve (AUC) was 0.8653, which indicates hsa_circ_0000567 can serve as a diagnostic biomarker.
Conclusion
Hsa_circ_0000567 may be a novel suppressor and a potential diagnosis biomarker in CRC.
Keywords: biomarker, circular RNA, colorectal cancer, hsa_circ_0000567
Abbreviations
- AUC
area under the ROC curve
- circRNAs
circular RNAs
- CRC
colorectal cancer
- lncRNA
long noncoding RNA
- miRNA
microRNA
- ncRNA
noncoding RNA
- qRT‐PCR
quantitative real‐time polymerase chain reaction
- ROC
receiver operating characteristics
- siRNA
small interfering RNA
1. INTRODUCTION
Circular RNAs (circRNAs) are a novel type of endogenous noncoding RNAs (ncRNAs), which are more stable, highly conserved and often exhibit tissue type‐specific and developmental stage‐specific expression.1, 2 Unlike linear RNAs, such as microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and messenger RNAs (mRNAs), circRNAs can resist to the digestion induced by exonucleolytic RNA for their closed‐loop structure.3 These properties provide circRNAs with the potential to become ideal diagnostic biomarkers in diseases.
Emerging evidence has indicated that circRNAs are involved in many pathophysiological processes in many human diseases.4, 5, 6, 7 In the study field of tumorigenesis and progression, recent studies have revealed that certain circRNAs are deregulated in many kinds of human cancers through high‐throughput RNA sequencing, such as gastric cancer, breast cancer, basal cell carcinoma, and cutaneous squamous cell carcinoma.8, 9, 10, 11 Furthermore, several aberrant circRNAs have been identified as oncogenes or tumor suppressors.12, 13, 14 However, the expression and functions of most deregulated circRNAs remain largely unknown, and the mechanisms are rarely uncovered.
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in men and the second in women worldwide. It is the fourth leading cause of cancer death worldwide.15, 16 Although the diagnostic methods and treatments have been developed, CRC is still a stable contributor to cancer deaths for lack of sensitive, specific diagnostic biomarkers and effective targets. Therefore, exploring the molecular mechanism underlying CRC carcinogenesis and metastasis may provide novel insight into CRC diagnosis and therapy. As is known, ncRNAs, such as miRNAs, lncRNAs are involved in the development and progression of CRC.17 However, as a novel identified ncRNAs, the relationship between circRNAs and CRC development is still unknown.18
Based on the previous studies,19, 20 which identified the deregulated circRNAs in CRC samples and normal colorectal tissues by high‐throughput RNA sequencing, we focused on hsa_circ_0000567 (http://circbase.org/cgi-bin/simplesearch.cgi) for it is one of the most obviously downregulated circRNAs in CRC tissues. Hsa_circ_0000567 is located at chr14: 99924616‐99932150, and its spliced mature sequence length is 683 nt. Hsa_circ_0000567 is derived from exon 2‐6 of SET domain‐containing 3 (SETD3). The expression and function of hsa_circ_0000567 in CRC have not yet been known. In this study, we first elucidated hsa_circ_0000567 was downregulated in CRC, and knockdown of hsa_circ_0000567 promoted CRC cells proliferation and migration in vitro. In addition, the diagnostic value of hsa_circ_0000567 was assessed. Our findings indicate that hsa_circ_0000567 may be related to the growth and metastasis of CRC and can serve as a promising diagnostic biomarker for CRC.
2. MATERIALS AND METHODS
2.1. Cell culture
Normal human colorectal epithelial cell line (FHC) and 5 CRC cell lines (SW480, RKO, CACO2, SW620, and HCT116) were purchased from the American Type Culture Collection (ATCC, USA). All cell lines were cultured in RPMI‐1640 Medium (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA) at 37°C with 5% CO2.
2.2. Tissue samples collection
Total 102 pairs of fresh CRC tissues and adjacent noncancerous tissues were obtained from Yijishan hospital, a subsidiary of Wannan Medical College (Wuhu, China). After being removed from the bodies, the tissue samples were immediately frozen in liquid nitrogen and stored at −80°C. All tissues were confirmed to be adenocarcinoma by 2 experienced pathologists. No radiotherapy or chemotherapy was carried out before operation. The Human Research Ethics Committee from College approved all aspects of this study, and verbal consent was obtained from all patients.
2.3. RNA extraction and reverse transcription (RT)
Total RNA from tissues or cells was extracted using RNAiso Plus reagent (Takara, Japan) following the manufacturer's instructions. Then, chloroform was added to separate organic phase from inorganic phase. The isopropanol was used for the precipitation of total RNA. RNA reverse transcription reaction was performed with Reverse Transcription Kit (Takara, Japan).
2.4. Quantitative real‐time polymerase chain reaction (qRT‐PCR)
qRT‐PCR was performed using Sybr Green qPCR master mix (DBI Bioscience, Germany) according to the manufacturer's instructions. The threshold cycle (C t) value for each sample was obtained through ABI 7500 software v2.0.6, and the ΔC t value was calculated by normalizing to the GAPDH mRNA or 18S rRNA appropriately (ΔC t = ‐ or ΔC t = . The relative RNA expression was calculated using the method.21 = (Cancer) /(Normal) . The primers were synthesized by RiboBio (Guangzhou, China). The primer sequences: hsa_circ_0000567, forward, 5′‐AGTTCGGTATCTTCAGT CCACAC‐3′, reverse, 5′‐TCTTACCCATTTTTCTGACTGGATG‐3′. GAPDH: forward, 5′‐GGAGCGAGATCCCTCCAAAAT‐3′, reverse, 5′‐GGCTGTTGTCA TACTTCTCATGG‐3′, 18S rRNA, forward, 5′‐ACACGGACAGGATTGACAGA‐3′, reverse, 5′‐GGACATCTAAGGGCATCACA‐3′.
2.5. RNase R digestion and Actinomycin D treatment
RNA samples were degraded using RNase R (2 U/μg RNA, Epicentre Biotechnologies, USA) for 0, 5, 15, 20 minutes at 37°C and then subjected to qRT‐PCR as described above. In addition, SW480 cells were treated with 50 ng/mL Actinomycin D (Amresco, USA) to block new RNA synthesis for 24 hours, and then, total RNA was extracted and qRT‐PCR was performed.
2.6. Small interfering RNA (siRNA) and transfection
The siRNA used to inhibit hsa_circ_0000567 expression was purchased from RiboBio (Guangzhou, China), and the sequences are TAAAGTCATCCAGTCAGAA. Cells were transfected with siRNA oligonucleotides using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer's instructions.
2.7. MTT proliferation assay
About 1 × 103 cells suspended in 100 μL culture medium were seeded in 96‐well plates, and 50 μL 1× MTT (KeyGen, China) per well was added at 1, 2, 3, 4, 5 days. After incubated at 37°C for a further 4 hours, MTT was removed, and 150 μL DMSO (Sigma, Japan) was added to dissolve the formazan. Absorbance was detected at 570 nm.
2.8. Cell migration assay
Transwell chamber (BD Company, USA) was used for migration assay. About 1 × 105 cells suspended in 250 μL serum‐free medium were placed in the upper chamber, and 600 μL medium with 20% FBS was added to the lower chamber. After cultured for 36 hours, the nonmigrated cells were erased, and the migrated cells were stained using Giemsa (KeyGen, China). The cells were captured in 5 fields (× 200) under a microscope and counted.
2.9. Statistical analysis
Statistical analyses were performed using SPSS 20.0 software (IBM, USA) and GraphPad Prism version 6.0. Data were presented as mean ± standard deviation (SD). The differences between groups were tested using a two‐tailed Student's t test. The relationships between hsa_circ_0000567 expression levels and clinicopathological factors of patients with CRC were analyzed by a two‐tailed Student's t test or one‐way analysis of variance (ANOVA). The receiver operating characteristic (ROC) curve was built using GraphPad Prism. Differences were considered significant if P < .05. *P < .05, **P < .01, ***P < .001, ****P < .0001.
3. RESULTS
3.1. Validation of hsa_circ_0000567 in CRC tissues and CRC cell lines
Hsa_circ_0000567 is derived from exon 2‐6 of SETD3 gene locus by back‐splicing. We designed the divergent primers crossing the back‐splice junction for detecting hsa_circ_0000567 expression (Figure 1A). The melt curve analysis of qRT‐PCR yielded only a single peak, demonstrating the amplified product was specific without nonspecific amplification or primer dimmers (Figure 1B). Subsequently, the Sanger sequencing of the amplified product confirmed the existence of back‐splice junction (Figure 1C). These results demonstrate that hsa_circ_0000567 exists in CRC and can be detected specifically by qRT‐PCR.
Figure 1.
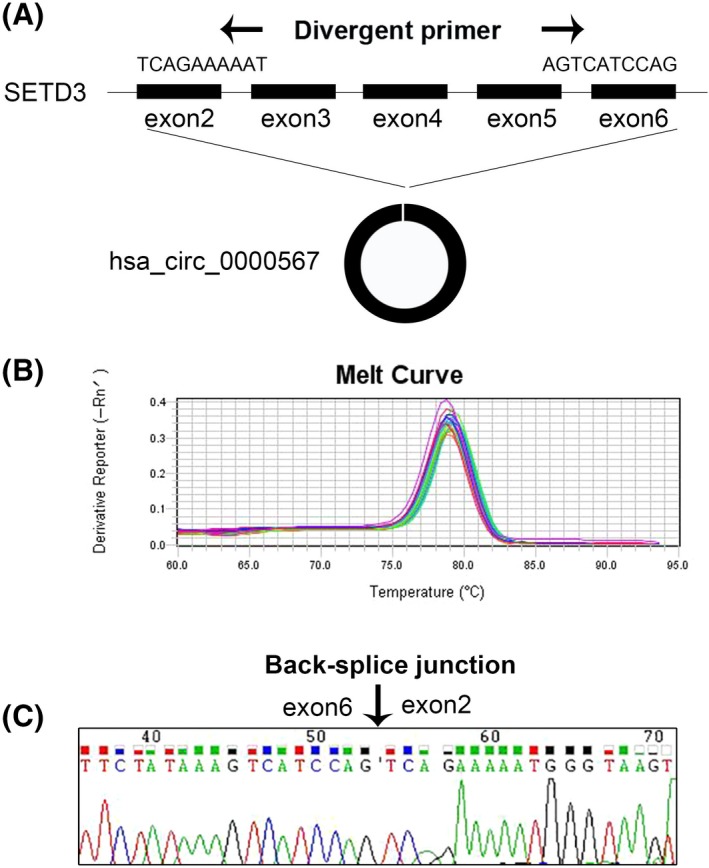
Validation of hsa_circ_0000567 in human CRC tissues and CRC cell lines. (A) Hsa_circ_0000567 is generated from exon 2‐6 of SETD3 gene locus by back slicing. Black arrows indicate the divergent primers. (B) Melt curves of qRT‐PCR amplified products. (C) Sanger sequencing of a PCR amplified product resulting from divergent primers indicating the head‐to‐tail junction of exon 2 and exon 6. Black arrow denotes the back‐splice junction
3.2. Hsa_circ_0000567 was more stable than linear RNA
As circRNAs can resist the digestion of RNase R for lack of free 5′‐ and 3′‐ends, we used RNase R to test the stability of hsa_circ_0000567. Results indicated hsa_circ_0000567 was more stable than the endogenous linear GAPDH mRNA (Figure 2A). Additionally, SW480 cells were treated with Actinomycin D to block new RNA synthesis for 24 hours. As a result, hsa_circ_0000567 levels were not obviously changed, while GAPDH mRNAs were significantly decreased (P < .05) (Figure 2B).
Figure 2.
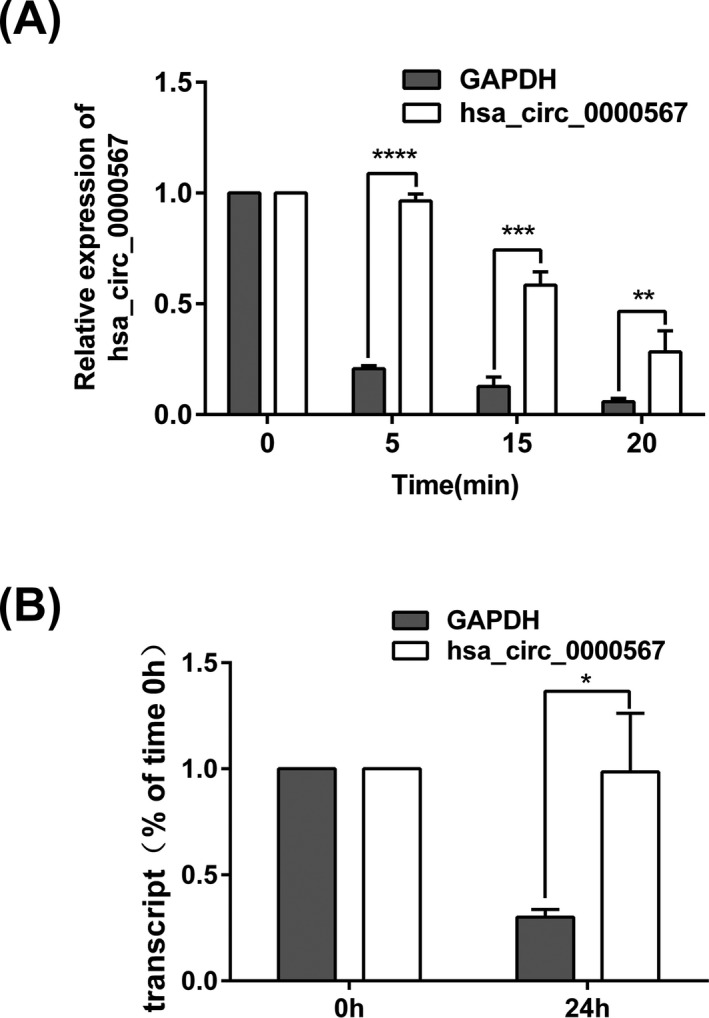
Hsa_circ_0000567 was more stable than linear RNA. (A) qRT‐PCR was performed to detect hsa_circ_0000567 and GAPDH mRNA after digested with RNase R (2 U/μg RNA) for the indicated time points. (B) The levels of hsa_circ_0000567 and GAPDH mRNA were measured by qRT‐PCR normalizing to 18S rRNA in SW480 cells after treated with Actinomycin D (50 ng/mL) for 24 h. *P < .05, **P < .01, ***P < .001, ****P < .0001
3.3. Hsa_circ_0000567 was downregulated in CRC and associated with tumor growth and metastasis of patients with CRC
We detected the expression of hsa_circ_0000567 in 102 pairs of CRC tissues and adjacent noncancerous tissues using qRT‐PCR. The results showed that hsa_circ_0000567 was lower in 96 of 102 cases of CRC samples compared with those of the counterparts (P < .0001) (Figure 3A,B).
Figure 3.
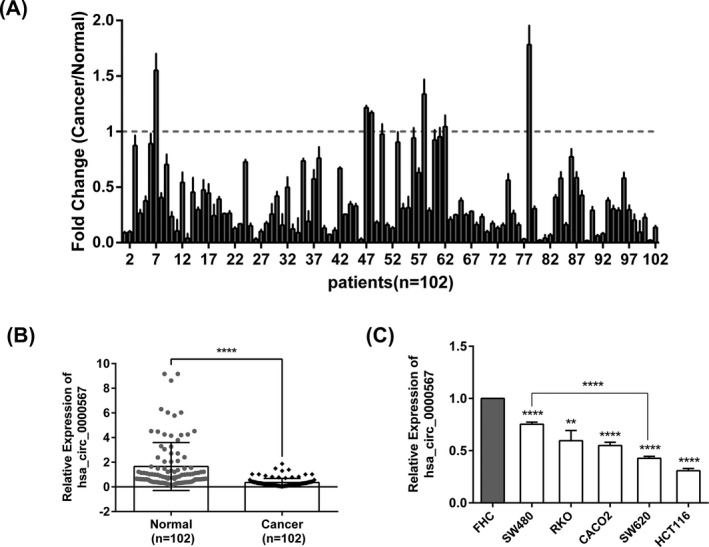
Hsa_circ_0000567 was downregulated in CRC tissues and CRC cell lines. (A) qRT‐PCR was performed to detect hsa_circ_0000567 expression in 102 paired CRC tissues and adjacent noncancerous tissues, and the results were presented as the fold change in CRC cancer tissues relative to the matched normal tissues. (B) The relative expression of hsa_circ_0000567 in 102 pairs of human CRC tissues (Cancer) and the adjacent noncancerous tissues (Normal) by qRT‐PCR analysis. (C) Hsa_circ_0000567 was decreased in 5 CRC cell lines (SW480, RKO, CACO2, SW620, and HCT116) compared with the normal colorectal epithelium cell line FHC by qRT‐PCR analysis. **P < .01, ****P < .0001
To further investigate the clinical significance of hsa_circ_0000567 in CRC, we analyzed the relationship between the expression of hsa_circ_0000567 and the clinicopathological features of patients with CRC. As shown in Table 1, hsa_circ_0000567 expression was negatively correlated with tumor size (P = .011), lymph metastasis (P = .003), distal metastasis (P < .0001), and tumor–node–metastasis (TNM) stage (P = .003) in CRC.
Table 1.
Relationship between hsa_circ_0000567 relative expression () in CRC tissues and clinicopathological factors of patients with CRC
| Characteristics | No. of patients | Mean ± SD | P value |
|---|---|---|---|
| Age (years)a | |||
| ≤60 | 55 | 0.385 ± 0.375 | .324 |
| >60 | 47 | 0.319 ± 0.273 | |
| Gender | |||
| Male | 58 | 0.378 ± 0.362 | .417 |
| Female | 44 | 0.324 ± 0.288 | |
| Cancer location | |||
| Colon | 58 | 0.345 ± 0.360 | .728 |
| Rectum | 44 | 0.368 ± 0.294 | |
| Diameter (cm)b | |||
| ≤4.5 | 51 | 0.438 ± 0.394 | .011 |
| >4.5 | 51 | 0.272 ± 0.232 | |
| Differentiation | |||
| Well | 19 | 0.432 ± 0.377 | .403 |
| Moderate | 60 | 0.354 ± 0.350 | |
| Poor | 23 | 0.292 ± 0.226 | |
| Serosal invasion | |||
| No | 31 | 0.376 ± 0.328 | .675 |
| Yes | 71 | 0.346 ± 0.336 | |
| Lymphatic metastasis | |||
| No | 45 | 0.474 ± 0.424 | .003 |
| Yes | 57 | 0.260 ± 0.192 | |
| Distal metastasis | |||
| No | 82 | 0.389 ± 0.358 | <.0001 |
| Yes | 20 | 0.216 ± 0.116 | |
| TNM classification | |||
| I‐II | 54 | 0.476 ± 0.429 | .003 |
| III‐IV | 48 | 0.263 ± 0.192 | |
Group of age was performed according to median.
Tumor size was grouped according to median.
The bold values denote the differences between groups are significant.
We also examined hsa_circ_0000567 expression levels in CRC cell lines. Consistent with the findings in CRC tissues, hsa_circ_0000567 expression levels in 5 CRC cell lines (SW480, RKO, CACO2, SW620, and HCT116) were all significantly downregulated compared with normal colorectal epithelial cell line FHC. Moreover, hsa_circ_0000567 expression was significantly lower in highly metastatic CRC cell line SW620 compared with the low metastasis cell line SW480 (P < .0001, Figure 3C). These results demonstrate hsa_circ_0000567 may play a suppressor role in CRC growth and metastasis.
3.4. Knockdown of hsa_circ_0000567 promoted CRC cells proliferation and migration in vitro
To further verify the inhibitory role of hsa_circ_0000567 in CRC, we performed the loss‐of‐function assay in vitro. We chose SW480 and CACO2 cells because hsa_circ_0000567 expression could be knocked down more efficiently in them than in the other cell lines using siRNA. qRT‐PCR revealed hsa_circ_0000567 expression was significantly reduced in SW480 and CACO2 cells after transfection with hsa_circ_0000567 siRNA (Figure 4A). MTT assay showed that cell proliferation ability was enhanced after knockdown of hsa_circ_0000567 expression (Figure 4B). Furthermore, repression of hsa_circ_0000567 expression promoted the migration of CRC cells (Figure 4C).
Figure 4.
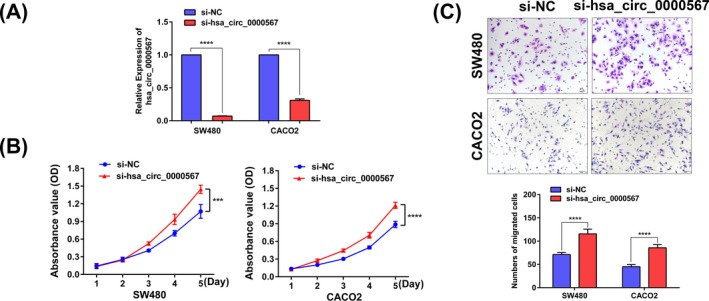
Knockdown of hsa_circ_0000567 promoted CRC cells proliferation and migration in vitro. (A) qRT‐PCR analysis of hsa_circ_0000567 expression in SW480 and CACO2 cells after transfection of hsa_circ_0000567 siRNA and NC siRNA. (B) Decreased expression of hsa_circ_0000567 enhanced CRC cells proliferation. (C) Inhibition of hsa_circ_0000567 expression increased cells migration as determined by transwell assay. Bars represent the migrated cell numbers. ***P < .001, ****P < .0001
3.5. Potential diagnostic value of hsa_circ_0000567 in CRC
We have verified hsa_circ_0000567 was deregulated in CRC and more stable than linear RNA, which suggests hsa_circ_0000567 may serve as a promising biomarker. To evaluate the diagnostic value of hsa_circ_0000567 in CRC, we built a ROC curve using the expression of hsa_circ_0000567 in CRC tissues and adjacent noncancerous tissues. The calculated area under the ROC curve (AUC) was 0.8653, which means the higher diagnostic value of hsa_circ_0000567 in CRC. The sensitivity, specificity, Youden's index, and cut‐off value were 0.8333, 0.7647, 0.598, and 0.4714, respectively (Figure 5). In addition, we found that hsa_circ_0000567 in human blood exosome from the patients with CRC was also lower than that from the normal individuals in exoRBase (Figure S1), which suggests the circulating hsa_circ_0000567 may also be used as a diagnostic biomarker. However, future work is needed to validate the diagnostic value of hsa_circ_0000567 in blood exosome.
Figure 5.
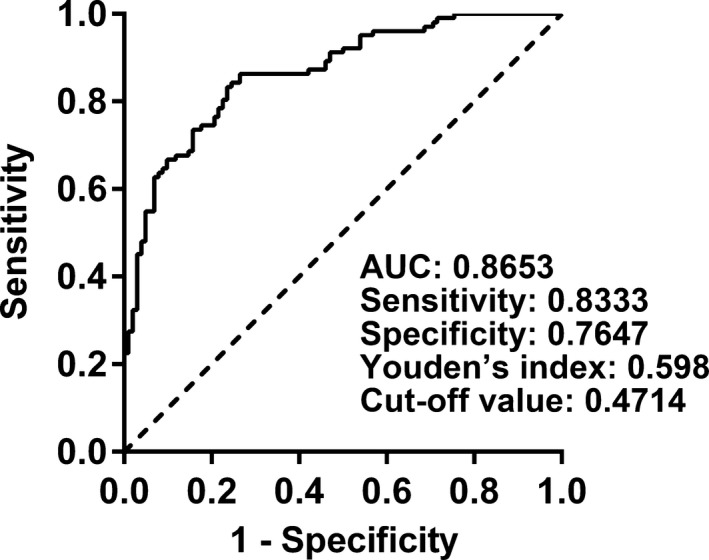
ROC curve was constructed to evaluate the potential diagnostic value of hsa_circ_0000567. The area under the ROC curve (AUC) was 0.8653 (95% CI = 0.8166‐0.9141; P < .0001). The sensitivity, specificity, Youden's index, and cut‐off value were 0.8333, 0.7647, 0.598, and 0.4714, respectively
4. DISCUSSION
CircRNAs are covalently closed continuous loop structure without 5′ caps or 3′ tails. They were first observed in eukaryotic cells using electron microscopy more than three decades ago but originally perceived as by‐products of splicing.22 With the development of next‐generation sequencing technology and novel computational approaches, thousands of circRNAs have been identified and cataloged. The majority of circRNAs are derived from exons of protein‐coding genes, which are termed as exonic circRNAs (ecircRNAs). In addition, they can also arise from the introns or intron‐containing exons to form intronic circRNAs (ciRNA) and exon‐intron circRNAs (EIciRNAs).23 Studies demonstrated that ecircRNAs are predominantly located in the cytoplasm functioning as miRNA sponges or binding to RNA‐binding proteins,24, 25 while ciRNA and EIciRNAs could regulate transcription of their parent genes in the nucleus.26, 27
To our best knowledge, this is the first study to elucidate the expression, function, and diagnostic value of hsa_circ_0000567 in CRC. We validated the existent of hsa_circ_0000567 in CRC tissues and CRC cell lines by Sanger sequencing. Further studies revealed hsa_circ_0000567 was more stable than linear RNA after treated with RNase R or Actinomycin D (Figure 2). The biological stability of hsa_circ_0000567 makes it more suitable as biomarker than miRNA, lncRNA, or other linear RNAs. To date, several circRNAs have emerged as new biomarkers in cancers. For example, hsa_circ_0000190 could serve as a biomarker for gastric cancer diagnosis, hsa_circ_0005986 was used as a biomarker in the diagnosis of HCC, and circular RNA ciRS‐7 was identified as a prognostic biomarker in colorectal cancer.28, 29, 30 In this study, we investigated that hsa_circ_0000567 was downregulated in CRC tissues and CRC cell lines (Figure 3). Furthermore, we confirmed that hsa_circ_0000567 expression in CRC tissues can be used to diagnose CRC by building a ROC curve (Figure 5). In addition, body fluids are convenient in the diagnosis of human diseases, and certain circRNAs were abundant and stable in plasma or exosomes.31, 32 We found that hsa_circ_0000567 in blood exosomes from patients with CRC was also decreased compared with that from normal persons in exoRBase, indicating the circulating hsa_circ_0000567 may be used as a biomarker for CRC screening (Figure S1). However, future work still needs to be done to examine hsa_circ_0000567 level in exosomes from clinical samples.
Previous studies have reported that circRNAs play a crucial role in cancer biological processes, such as proliferation, migration, and metastasis.33, 34, 35, 36 Our study revealed that the decreased hsa_circ_0000567 was negatively correlated with tumor size, lymph metastasis, distal metastasis, and TNM stage in patients with CRC (Table 1). Consistent with the findings in CRC tissue samples, hsa_circ_0000567 expression in CRC cell lines was also associated with the metastasis ability of cells (Figure 3C). These results suggest hsa_circ_0000567 may be involved in the growth and metastasis of cells. Subsequently, the hypothesis was verified by the loss‐of‐function assay in vitro (Figure 4).
As hsa_circ_0000567 is generated from exons of SETD3 gene, as an ecircRNAs, it is possibly located in the cytoplasm and may function as miRNA sponges or bind to proteins to regulate gene expression at the post‐transcriptional level according to previous studies.24, 25 Future work is needed to uncover the mechanisms of hsa_circ_0000567 in CRC progression.
In summary, our data identified hsa_circ_0000567 plays a suppressor role in CRC proliferation and metastasis. More importantly, hsa_circ_0000567 may serve as a potential novel biomarker for the diagnosis of CRC.
Supporting information
ACKNOWLEDGMENTS
This work was supported by the Natural Science Foundation of the Anhui Provincial High‐Education Institutions of China (No. KJ2017A250) and the Natural Science Foundation for Young Scholars of Wannan Medical College (No. WK2016Z01, WK201619).
Wang J, Li X, Lu L, He L, Hu H, Xu Z. Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J Clin Lab Anal. 2018;32:e22379 10.1002/jcla.22379
REFERENCES
- 1. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333‐338. [DOI] [PubMed] [Google Scholar]
- 2. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suzuki H, Tsukahara T. A view of pre‐mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331‐9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu Q, Zhang X, Hu X, et al. Emerging roles of circRNA related to the mechanical stress in human cartilage degradation of osteoarthritis. Mol Ther Nucleic Acids. 2017;7:223‐230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cui X, Niu W, Kong L, et al. hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in Major depressive disorder. Biomark Med. 2016;10:943‐952. [DOI] [PubMed] [Google Scholar]
- 6. Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR‐141 by targeting TGF‐beta1. Biochem Biophys Res Comm. 2017;487:769‐775. [DOI] [PubMed] [Google Scholar]
- 7. Wu N, Jin L, Cai J. Profiling and bioinformatics analyses reveal differential circular RNA expression in hypertensive patients. Clin Exp Hypertens 2017;39:454‐459. [DOI] [PubMed] [Google Scholar]
- 8. Sui W, Shi Z, Xue W, et al. Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncol Rep. 2017;37:1804‐1814. [DOI] [PubMed] [Google Scholar]
- 9. Sand M, Bechara FG, Sand D, et al. Circular RNA expression in basal cell carcinoma. Epigenomics. 2016;8:619‐632. [DOI] [PubMed] [Google Scholar]
- 10. Sand M, Bechara FG, Gambichler T, et al. Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2016;83:210‐218. [DOI] [PubMed] [Google Scholar]
- 11. Nair AA, Niu N, Tang X, et al. Circular RNAs and their associations with breast cancer subtypes. Oncotarget. 2016;7:80967‐80979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta‐catenin pathway. Oncotarget. 2015;6:6001‐6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non‐coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919‐3931. [DOI] [PubMed] [Google Scholar]
- 14. Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339‐2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359‐E386. [DOI] [PubMed] [Google Scholar]
- 16. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87‐108. [DOI] [PubMed] [Google Scholar]
- 17. Ragusa M, Barbagallo C, Statello L, et al. Non‐coding landscapes of colorectal cancer. World J Gastroenterol. 2015;21:11709‐11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taborda MI, Ramirez S, Bernal G. Circular RNAs in colorectal cancer: possible roles in regulation of cancer cells. World J Gastrointest Oncol. 2017;9:62‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bachmayr‐Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmittgen TD, Livak KJ. Analyzing real‐time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101‐1108. [DOI] [PubMed] [Google Scholar]
- 22. Hsu MT, Coca‐Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339‐340. [DOI] [PubMed] [Google Scholar]
- 23. Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta. 2016;1859:163‐168. [DOI] [PubMed] [Google Scholar]
- 24. Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6:563‐579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846‐2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Z, Huang C, Bao C, et al. Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256‐264. [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792‐806. [DOI] [PubMed] [Google Scholar]
- 28. Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167‐171. [DOI] [PubMed] [Google Scholar]
- 29. Fu L, Chen Q, Yao T, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR‐129‐5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget. 2017;8:43878‐43888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weng W, Wei Q, Toden S, et al. Circular RNA ciRS‐7‐A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23:3918‐3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981‐984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE. 2015;10:e0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jin H, Jin X, Zhang H, Wang W. Circular RNA hsa‐circ‐0016347 promotes proliferation, invasion and metastasis of osteosarcoma cells. Oncotarget. 2017;8:25571‐25581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li P, Chen H, Chen S, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116:626‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS‐7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17‐27. [DOI] [PubMed] [Google Scholar]
- 36. Zhu M, Xu Y, Chen Y, Yan F. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138‐144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


