Abstract
Background
Anti‐HCV assays are widely used as a screening tool for HCV infection. However, diagnostic performances and effective signal‐to‐cut‐off ratios (S/COs) for predicting true HCV infections would vary according to the assays used. Thus, we evaluated the diagnostic performances of the new Elecsys Anti‐HCV assay.
Methods
A total of 41 694 cases tested by the Elecsys Anti‐HCV II assay (Roche Diagnostics, Germany) during January 2013 to December 2015 were retrospectively analyzed by comparing with the diagnosis on HCV infections determined by patients’ medical records and results of laboratory tests.
Results
Excluding 62 cases with unclear history of HCV infection, 430 and 41 202 cases were respectively assorted as “true infection” and “no evidence of infection,” and 99.85% of the initial results by the Elecsys assay were concordant with the diagnosis on HCV infection. Sensitivity, specificity, positive and negative predictive values were respectively 99.30%, 99.86%, 88.04%, and 99.99%, where the prevalence of the HCV infection was 1.0%. The area under the receiver operating characteristics curve value of the Elecsys assay was 0.9980 (95% confidence interval [CI]=0.9944 to 1.0017). The S/CO by the Elecsys assay for predictive of a true‐positive ≥95% of the time was 19.0 (95% CI=15.0 to 25.1).
Conclusion
The Elecsys Anti‐HCV II assay showed excellent diagnostic performances, particularly in terms of sensitivity, specificity, and NPV. However, the results obtained by this assay with S/CO less than a certain value would need to be retested by HCV RNA PCR or another anti‐HCV assay.
Keywords: anti‐HCV, diagnostic performance, Elecsys Anti‐HCV II assay, hepatitis C virus, signal‐to‐cut‐off ratio
1. Introduction
Hepatitis C virus (HCV) is one of the leading causes of chronic liver disease, affecting around 170 million persons or 3% among total population worldwide.1 In the United States, approximately 4.1 million individuals have been infected with HCV, and an estimated 3.2 million among them are living with HCV infection.2
Anti‐HCV assays are widely used as a screening tool for HCV infection. As a rule, screening tests for the diagnosis of infectious diseases need to have high sensitivities to detect all or nearly all of true‐positive cases. As a consequence, screening tests generally produce more false‐positive results than confirmatory tests, but this sacrifice of specificity would be tolerated when a good confirmatory test is available and the consequences of the false‐positive results are also be tolerable. In these circumstances, the recombinant immunoblot assay (RIBA) had been widely used as a confirmatory tool for anti‐HCV positive cases owing to its high specificity,3, 4 although this assay is labor‐intensive and time‐consuming. However, reactive results from an anti‐HCV assay cannot discriminate persons with resolved past HCV infection from those who are currently infected with HCV.
The United States Centers for Disease Control and Prevention (CDC) had recommended that an individual would be considered to have serologic evidence of HCV infection only after a positive result of anti‐HCV has been confirmed by RIBA or HCV RNA PCR, particularly in populations with a lower prevalence of disease, to verify false‐positive screening test results.3 However, the majority of laboratories report positive anti‐HCV results based on a positive screening assay alone. The testing algorithm includes an option for using the signal‐to‐cut‐off ratio (S/CO) of a positive result from an anti‐HCV assay as a screening test. This can be an alternative to RIBA or PCR in some instances, reducing the necessity for supplemental testing and providing additional clue on the subject's true anti‐HCV antibody status.
In 2013, CDC published an updated guidance for clinicians and laboratorians on testing for HCV infection.5 In this guideline, a single nonreactive anti‐HCV result indicates no HCV antibody detected, and a reactive result imply current HCV infection, or resolved past HCV infection, or false‐positivity. In addition, a reactive result is recommended to be followed by HCV RNA PCR but not by RIBA owing to the discontinuation of widely used RIBA HCV. Consequently, high specificity as well as high sensitivity of an anti‐HCV assay became more important. To trade‐off sensitivity of an assay against specificity, appropriate cut‐off for the determination of results would be needed since low cut‐off can guarantee higher sensitivity while the specificity would be sacrificed.6
Regardless of the anti‐HCV prevalence or characteristics of the tested population, a specific S/CO can be used to predict a true anti‐HCV‐positive result determined by supplemental tests such as RIBA for ≥95% of the time.3 For instance, the S/CO predictive of a true‐positive ≥95% of the time for the Vitros Anti‐HCV assay was suggested as 8.0. However, methods and molecules used for generating and detecting signals as well as epitopes and specificities of antigens and antibodies in the assay reagents are different between the assays.7 Thus, effective cut‐off values and diagnostic performances would vary according to the assay,7, 8 and need to be validated before used in clinical setting. CDC provides S/COs predictive of a true‐positive for some commercially available anti‐HCV assays,9 but those values for the Elecsys assays have not been suggested yet.
Recently, an improved version of the Elecsys Anti‐HCV assay was developed and introduced to clinical laboratories. Few recent studies evaluated the performances of this new assay.10, 11, 12 To evaluate the diagnostic performances of the new Elecsys assay and to determine its effective S/CO, we retrospectively analyzed results by this assay for large population tested during recent 3 years.
2. Materials and Methods
2.1. Subjects and case definition
Between January 2013 and December 2015, a total of 41 694 cases excluding duplicated patients were tested in a single general hospital by using the Elecsys Anti‐HCV II assay (Roche Diagnostics GmbH, Mannheim, Germany). As a policy of our laboratory on routine testing for anti‐HCV, samples showing initial S/CO equal to or greater than 0.80 were retested when there was neither obvious history of HCV infection nor previous results for laboratory tests such as anti‐HCV, RIBA, and HCV RNA PCR.
For this study, medical records for the subjects including previous and follow‐up laboratory tests regarding HCV infection were retrospectively reviewed in duplicates by two or more physicians independently to determine each case as one with or without HCV infection, when the initial S/CO by the Elecsys assay was above 0.80.
When accorded with one of the following criteria, the case was defined as “true HCV infection” which includes current HCV infection or resolved past HCV infection:
The follow‐up (within 3 months) and/or previous anti‐HCV were consistently positive more than once.
One or more of the results among follow‐up (within 3 months) or previous tests including RIBA and HCV RNA PCR were positive.
The patient had obvious history of HCV infection in the medical records and showed one or more positive results for follow‐up (within 3 months) or previous laboratory tests including anti‐HCV, RIBA, and HCV RNA PCR.
The case was defined as “no evidence of HCV infection” when the subjects corresponded to one of the following criteria:
The initial result by the Elecsys Anti‐HCV II assay was negative, and the patient showed normal aminotransferase levels, and he/she already had negative previous results for anti‐HCV or had no history of HCV infection in the medical records.
When the initial and repeated results of the Elecsys Anti‐HCV assay were discrepant but the aminotransferase levels of the patient were within normal, and he/she had no history of HCV infection and one or more of the results from previous or follow‐up anti‐HCV and/or RIBA were all negative.
When a case was not able to be classified as either “true HCV infection” or “no evidence of HCV infection,” the subject was assorted as the group with unclear history of HCV infection and excluded from further analysis.
This study was approved by the Institutional Review Board of Ilsan Hospital.
2.2. Assays
Anti‐HCV was detected by using cobas e 601 analyzer with the Elecsys Anti‐HCV II assay kit (Roche Diagnostics GmbH). This assay utilizes the electrochemiluminescence immunoassay (ECLIA) principle. An S/CO equal to or greater than 1.00 is suggested to be positive for anti‐HCV by the manufacturer. In this study, cases with initial S/CO equal to or greater than 0.80 by the Elecsys assay were retested when there was neither obvious history of HCV infection nor previous results by at least one of the tests for HCV infection including anti‐HCV assay, RIBA, and HCV RNA PCR. RIBA and HCV RNA PCR assays were performed using MP diagnostics HCV blot 3.0 (MP biomedicals SAS, Singapore) and Roche COBAS AmpliPrep/COBAS TaqMan HCV Quantitation Test, version 2.0 (Roche Diagnostics GmbH) at the Green Cross Reference Laboratory (Yongin‐si, Gyeonggi‐do, Republic of Korea) and Seoul Medical Science Institute (Yongin‐si, Gyeonggi‐do, Republic of Korea). RIBA results were classified into “negative,” “positive” or “indeterminate” following the manufacturer's instructions.
2.3. Statistical analysis
All statistical analyses were performed by Analyse‐it for Microsoft Excel Method Evaluation Edition version 4.60.2 (Analyse‐it Software, Ltd., Leeds, UK) or IBM SPSS Statistics 23 (IBM Corp., Armonk, NY, US). Correlation coefficient between the S/COs by different assays was calculated by Spearman rank test and Kruskal‐Wallis test were used for a comparison between the groups. Sensitivity, specificity, positive and negative predictive values (PPV and NPV), and their 95% confidence intervals (95% CIs) of the Elecsys Anti‐HCV II assay were estimated by comparing initial results by the Elecsys assay with the diagnosis on HCV infection determined by review of medical records. Receiver operating characteristics curve analysis was performed to calculate the area under the curve (AUROC) of the Elecsys assay for predicting “true HCV infection.” The optimal S/CO was defined as the S/CO showing maximum Youden index. To determine the S/CO for predictive of a true‐positive ≥95% of the time, cases showing the same S/CO by the Elecsys assay were pooled, and the true‐positive rates for the respective S/COs were calculated by dividing the number of cases with “true HCV infection” showing certain S/CO by the number of total cases with the same S/CO. Then, probit regression analysis was performed by plotting a graph of the response rates against respective S/CO values.
3. Results
3.1. Distribution of the S/COs
The initial S/COs by the Elecsys Anti‐HCV II assay are summarized in the Table 1. Among a total of 41 147 cases with initial S/COs less than 1.00, 99.0% and 99.9% showed S/COs below 0.22 and 0.69 respectively. In addition, 50.0% and 90.0% among the initial positive 547 cases showing S/CO equal to or greater than 1.00 demonstrated S/COs less than 36.3 and 112.1 respectively.
Table 1.
Distribution of the initial signal‐to‐cut‐off ratios by the Elecsys Anti‐HCV II assay
| Initial result | Signal‐to‐cut‐off ratio | Frequency (n) | Relative frequency (%) | Cumulative frequency (n) | Cumulative relative frequency (%) |
|---|---|---|---|---|---|
| Negative | <0.10 | 36 418 | 88.5 | 36 418 | 88.5 |
| ≥0.10 to <0.20 | 4254 | 10.3 | 40 672 | 98.8 | |
| ≥0.20 to <0.30 | 236 | 0.6 | 40 908 | 99.4 | |
| ≥0.30 to <0.40 | 96 | 0.2 | 41 004 | 99.7 | |
| ≥0.40 to <0.50 | 49 | 0.1 | 41 053 | 99.8 | |
| ≥0.50 to <0.60 | 29 | 0.1 | 41 082 | 99.8 | |
| ≥0.60 to <0.70 | 28 | 0.1 | 41 110 | 99.9 | |
| ≥0.70 to <0.80 | 19 | 0.0 | 41 129 | 100.0 | |
| ≥0.80 to <0.90 | 7 | 0.0 | 41 136 | 100.0 | |
| ≥0.90 to <1.00 | 11 | 0.0 | 41 147 | 100.0 | |
| Positive | ≥1.00 to <20.0 | 156 | 28.5 | 156 | 28.5 |
| ≥20.0 to <40.0 | 142 | 26.0 | 298 | 54.5 | |
| ≥40.0 to <60.0 | 109 | 19.9 | 407 | 74.4 | |
| ≥60.0 to <80.0 | 45 | 8.2 | 452 | 82.6 | |
| ≥80.0 to <100.0 | 30 | 5.5 | 482 | 88.1 | |
| ≥100.0 to <120.0 | 15 | 2.7 | 497 | 90.9 | |
| ≥120.0 to <140.0 | 16 | 2.9 | 513 | 93.8 | |
| ≥140.0 to <160.0 | 14 | 2.6 | 527 | 96.3 | |
| ≥160.0 to <180.0 | 6 | 1.1 | 533 | 97.4 | |
| ≥180.0 to <200.0 | 8 | 1.5 | 541 | 98.9 | |
| ≥200.0 to <220.0 | 1 | 0.2 | 542 | 99.1 | |
| ≥220.0 to <240.0 | 3 | 0.5 | 545 | 99.6 | |
| ≥240.0 to <260.0 | 1 | 0.2 | 546 | 99.8 | |
| ≥260.0 to <280.0 | 0 | 0.0 | 546 | 99.8 | |
| ≥280.0 to <300.0 | 1 | 0.2 | 547 | 100.0 |
Meanwhile, the median S/CO of the 62 cases with unclear history of HCV infection was 24.6 (1st to 3rd quartiles=7.9‐47.5). Excluding those, a total of 41 632 cases were divided into 430 subjects with “true HCV infection” and 41 202 with “no evidence of HCV infection” (Table 2). Median S/COs by the Elecsys assay were 41.9 (1st to 3rd quartiles=24.9‐69.8) for the “true HCV infection” group and 0.05 (1st to 3rd quartiles=0.04‐0.08) for the “no evidence of HCV infection” group (Figure 1).
Table 2.
Concordance of the initial results by the Elecsys Anti‐HCV II assay with the clinical diagnosis on HCV infection
| HCV infection | Anti‐HCV | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 427 | 3 | 430 |
| Negative | 58 | 41 144 | 41 202 |
| Unclear | 62 | 0 | 62 |
| Total | 547 | 41 147 | 41 694 |
Excluding 63 cases with unclear HCV infection history, 99.85% among the 41 632 initial results by the Elecsys Anti‐HCV II assay were consistent with the diagnosis on HCV infection.
Figure 1.
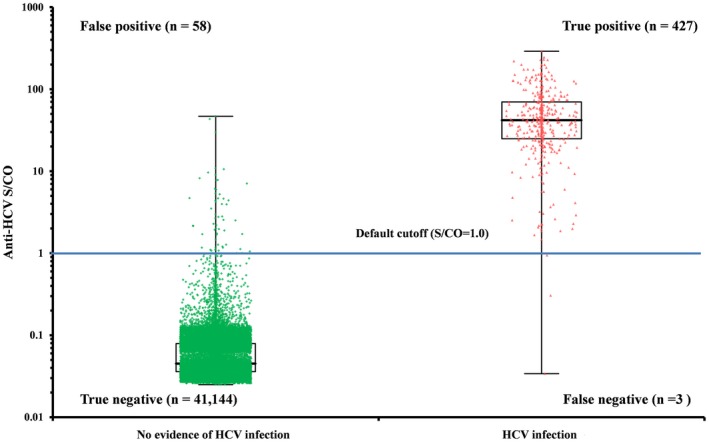
Distribution of the signal‐to‐cut‐off ratio (S/CO) by the Elecsys Anti‐HCV II assay according to the diagnosis on the HCV infection. Median S/COs by the Elecsys assay were 41.9 (1st to 3rd quartiles=24.9‐69.8) in the “true HCV infection” group (n=430) and 0.05 (1st to 3rd quartiles=0.04‐0.08) in the “no evidence of HCV infection” group (n=41 202)
3.2. Diagnostic performances of the Elecsys Anti‐HCV II assay
Excluding 62 cases with unclear HCV infection history, 99.85% among the 41 632 initial results by the Elecsys Anti‐HCV II assay were concordant with the diagnosis on HCV infection (Table 2). Sensitivity, specificity, PPV, and NPV of the Elecsys assay were respectively 99.30%, 99.86%, 88.04%, and 99.99%, where the prevalence of the HCV infection was 1.0% (Table 3).
Table 3.
Diagnostic performances of the Elecsys Anti‐HCV II assay
| Parameter | Value (%) | 95% CI (%) |
|---|---|---|
| Sensitivity | 99.30 | 97.97 to 99.76 |
| Specificity | 99.86 | 99.82 to 99.89 |
| PPVa | 88.04 | 85.06 to 90.50 |
| NPVa | 99.99 | 99.98 to 100.00 |
CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value.
The prevalence of HCV infection was 1.0%.
In addition, the AUROC of the Elecsys assay for detecting “true HCV infection” cases was 0.9980 (95% CI=0.9944 to 1.0017), and the optimal S/CO cut‐off showing maximum diagnostic performances was 0.93 with sensitivity of 99.53% and specificity of 99.85%.
3.3. Comparison of the Elecsys Anti‐HCV II assay and RIBA
A total of 97 cases were confirmed by the RIBA. Of these cases, 67.0% (n=65), 8.2% (n=8), 24.8% (n=24) were positive, indeterminate and negative for RIBA. Median S/COs by the Elecsys assay were 51.7 (1st to 3rd quartiles=34.3‐72.9) for the “RIBA positive,” 27.0 (1st to 3rd quartiles=8.5‐50.3) for the “RIBA indeterminate” and 2.2 (1st to 3rd quartiles=0.5‐5.7) for the “RIBA negative.” Distribution of the S/CO of Elecsys Anti‐HCV II assay results according to the RIBA are summarized in the Table 4.
Table 4.
Distribution of the S/CO of Elecsys Anti‐HCV II assay results according to the RIBA
| Anti‐HCV S/CO | No. of samples | No. (%) of RIBA result | ||
|---|---|---|---|---|
| Negative | Indeterminate | Positive | ||
| 0.0‐1.0 | 7 | 6 (89) | 0 (0) | 1 (11) |
| 1.0‐5.0 | 13 | 11 (84) | 1 (8) | 1 (8) |
| 5.0‐20.0 | 11 | 4 (36) | 3 (28) | 4 (36) |
| 20.0‐50.0 | 29 | 1 (3) | 2 (7) | 26 (90) |
| 50.0‐100.0 | 26 | 2 (8) | 2 (8) | 22 (84) |
| >100.0 | 11 | 0 (0) | 0 (0) | 11 (100) |
| Total | 97 | 24 (25) | 8 (8) | 65 (67) |
S/CO, signal‐to‐cut‐off ratio.
3.4. Correlation between the results of anti‐HCV assay and HCV RNA PCR
A total of 323 cases were evaluated by HCV RNA PCR. Of these cases, 19 (5.9%) cases were negative for both anti‐HCV and HCV RNA, 136 (42.1%) were negative for HCV RNA but positive for anti‐HCV. The remaining 168 (52.0%) cases were positive for both anti‐HCV and HCV RNA, thus all HCV RNA‐positive also showed positive for anti‐HCV. The correlation coefficient between the S/CO of anti‐HCV assay and the viral loads in the HCV RNA‐positive samples was 0.0908 (95% CI=−0.0742 to 0.2509) and was not statistically significant (P=.2661). Median S/COs by the Elecsys assay were 43.3 (1st to 3rd quartiles=28.7‐67.0) for the HCV RNA‐positive cases and 22.5 (1st to 3rd quartiles=2.2‐57.2) for the HCV RNA negative cases, and they were statistically significant (P<.0001).
3.5. The S/CO for predictive of a true positive
Results of the probit analysis are summarized in the Table 5 and Figure 2. The S/CO by the Elecsys Anti‐HCV II assay in prediction of a true‐positive ≥95% of the time was 19.0 (95% CI=15.0 to 25.1), whereas the S/CO for predictive of a true‐positive in 50% of cases was 4.3 (95% CI=3.6 to 5.1).
Table 5.
The signal‐to‐cut‐off ratios by the Elecsys Anti‐HCV II assay according to the probability of HCV infection
| Probability | Anti‐HCV S/CO | 95% CI |
|---|---|---|
| 1% | 0.5 | 0.4 to 0.7 |
| 5% | 0.9 | 0.7 to 1.2 |
| 10% | 1.3 | 1.1 to 1.6 |
| 25% | 2.3 | 1.9 to 2.7 |
| 50% | 4.2 | 3.6 to 5.1 |
| 75% | 7.9 | 6.5 to 9.7 |
| 90% | 13.7 | 11.0 to 17.5 |
| 95% | 19.0 | 15.0 to 25.1 |
| 99% | 35.4 | 26.7 to 49.6 |
S/CO, signal‐to‐cut‐off ratio; CI, confidence interval.
Figure 2.
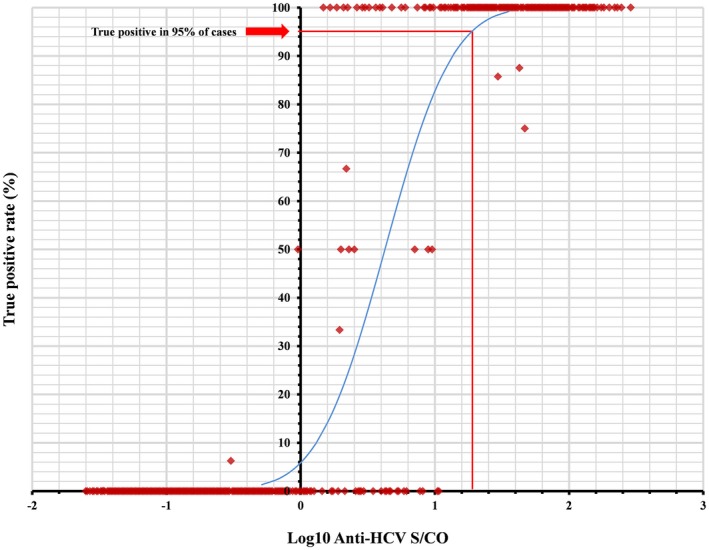
A schematic diagram of the probit regression analysis. The signal‐to‐cut‐off ratio (S/CO) by the Elecsys Anti‐HCV II assay in prediction of a true‐positive ≥95% of the time was 19.0 (95% confidence interval=15.0 to 25.1). Each dot was drawn by plotting the response rate against respective S/CO value, and the regression line was drawn only for illustrative purposes
4. Discussion
High sensitivities of screening tests are always demanded not to miss even a single affected individual, particularly when the test is intended to detect anyone infected with highly contagious agents. Our study estimated the diagnostic performances and effective cut‐off for S/CO of the new Elecsys Anti‐HCV II assay. Consequently, the assay showed high sensitivity of 99.30% and specificity of 99.86%, corresponding with the true anti‐HCV status assumed by reviewing patients’ medical records and laboratory results in 99.85% of all available cases during 3 years. Our previous evaluation on the performances of the Elecsys Anti‐HCV II assay also demonstrated sensitivity of 98.0% and specificity of 100.0% by comparing other anti‐HCV assays using 500 specimens.11 Other recent study also reported the sensitivity and specificity of the new Elecsys assay as 100.00% and 99.64%, respectively, using 859 routine clinical samples.10 This assay also demonstrated improved specificities ranging from 99.15% to 99.95% compared with those of previous version and other commercial anti‐HCV assays.12
Meanwhile, the prevalence of anti‐HCV in our data was 1.0%, and similar to this, the prevalence of HCV infection in Korea was reported to be 0.78%.13 In this situation of relatively low prevalence, PPV of an anti‐HCV assay as a screening tool would be not high owing to false‐positive results. Actually, PPV of the Elecsys Anti‐HCV II assay in our results was 88.04%, even though the sensitivity and specificity of the assay were 99.30% and 99.86% respectively. Similarly, PPV of the same assay in a recent study was 85.71% where the prevalence of anti‐HCV was 2.1%, although only 18 true‐positive cases were included in the evaluation.10
The S/CO predictive of a true‐positive ≥95% of the time for the Elecsys assay was estimated to be 19.0 (95% CI=15.0 to 25.1) from our data. Those for other assays including the Abbott Architect, the Ortho Vitros, and the Siemens Advia Centaur anti‐HCV assays were suggested as 5.0, 8.0, and 11.0 respectively.9 Differences between those values would not reflect differences between analytical performances of the respective assays but the values themselves would be characteristics of the assays owing to the differences in the methods and molecules utilized for signal generation and detection as well as epitopes and specificities of antigens and antibodies in the reagents. Applying the cut‐off S/CO of 19.0 from our results, PPV of the Elecsys assay would increase to 99.18%, retaining high NPV of 99.84%. In addition, cut‐off values for predicting various probability of true anti‐HCV status were estimated by probit analysis and could be referred in the result interpretation.
The definitive method to determine the true anti‐HCV status has been regarded as RIBA. We compared S/CO of the new Elecsys Anti‐HCV II assay to the RIBA results. The S/CO values among the groups classified by the results of RIBA showed statistically significant differences (P<.0001). In a previous study, 332 samples with S/CO of between 1 and 20 by the VITROS anti‐HCV assay were tested with RIBA, and none of the 163 samples with S/CO less than 5 was RIBA positive, while 89% of the 57 cases with S/CO between 16 and 20 by the same assay were positive for RIBA.14 Reagents for RIBA are now discontinuing and unavailable, and the procedure of RIBA is labor‐intensive and time‐consuming. Therefore, clinical laboratories would need to apply their own algorithms without using RIBA to confirm the positive results of an anti‐HCV assay. Although we used patients’ medical records and laboratory results including RIBA and HCV RNA assay as a source to determine true anti‐HCV status, we utilized data from large population to enhance the validity of study results. With the results of our study, we also suggest a method to establish a laboratory's own effective cut‐off value for S/CO from an anti‐HCV assay for oneself.
In conclusion, the new version of the Elecsys Anti‐HCV assay showed excellent diagnostic performances, particularly in terms of superior sensitivity, specificity, and NPV. Like other anti‐HCV assays, the results by the Elecsys assay showing S/CO less than a certain cut‐off would be retested by HCV RNA PCR or another anti‐HCV assay, and a clinical laboratory could need to establish its own effective cut‐off for S/CO by using the method suggested in this study as an example.
Kim B, Ahn HJ, Choi MH, Park Y. Retrospective analysis on the diagnostic performances and signal‐to‐cut‐off ratios of the Elecsys Anti‐HCV II assay. J Clin Lab Anal. 2018;32:e22165 10.1002/jcla.22165
References
- 1. Memon MI, Memon MA. Hepatitis C: an epidemiological review. J Viral Hepat. 2002;9:84–100. [DOI] [PubMed] [Google Scholar]
- 2. Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, Alter MJ. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med. 2006;144:705–714. [DOI] [PubMed] [Google Scholar]
- 3. Alter MJ, Kuhnert WL, Finelli L, Centers for Disease Control and Prevention . Guidelines for laboratory testing and result reporting of antibody to hepatitis C virus. MMWR Recomm Rep. 2003;52:1, 13, 15; quiz CE1‐4. [PubMed] [Google Scholar]
- 4. Ponde RA. Enzyme‐linked immunosorbent/chemiluminescence assays, recombinant immunoblot assays and nucleic acid tests in the diagnosis of HCV infection. Eur J Clin Microbiol Infect Dis. 2013;32:985–988. [DOI] [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention (CDC) . Testing for HCV infection: an update of guidance for clinicians and laboratorians. MMWR Morb Mortal Wkly Rep. 2013;62:362–365. [PMC free article] [PubMed] [Google Scholar]
- 6. Moretti M, Pieretti B, Masucci A, Sisti D, Rocchi M, Delprete E. Role of signal‐to‐cutoff ratios in hepatitis C virus antibody detection. Clin Vaccine Immunol. 2012;19:1329–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alborino F, Burighel A, Tiller FW, et al. Multicenter evaluation of a fully automated third‐generation anti‐HCV antibody screening test with excellent sensitivity and specificity. Med Microbiol Immunol. 2011;200:77–83. [DOI] [PubMed] [Google Scholar]
- 8. Kim S, Kim JH, Yoon S, Park YH, Kim HS. Clinical performance evaluation of four automated chemiluminescence immunoassays for hepatitis C virus antibody detection. J Clin Microbiol. 2008;46:3919–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Centers for Disease Control and Prevention (CDC) . Signal‐to‐cutoff ratios for commercially available assays. http://www.cdc.gov/hepatitis/HCV/LabTesting.htm. Accessed June 1, 2016.
- 10. Yang R, Guan W, Wang Q, Liu Y, Wei L. Performance evaluation and comparison of the newly developed Elecsys Anti‐HCV II assay with other widely used assays. Clin Chim Acta. 2013;426:95–101. [DOI] [PubMed] [Google Scholar]
- 11. Park Y, Park JY, Kim MJ, Kim HS. Comparison of the diagnostic performance of Elecsys Anti‐HCV II and Elecsys and Vitros anti‐HCV assays. J Lab Med Qual Assur. 2012;34:51–56. [Google Scholar]
- 12. Esteban JI, van Helden J, Alborino F, et al. Multicenter evaluation of the Elecsys(R) anti‐HCV II assay for the diagnosis of hepatitis C virus infection. J Med Virol. 2013;85:1362–1368. [DOI] [PubMed] [Google Scholar]
- 13. Kim do Y, Kim IH, Jeong SH, et al. A nationwide seroepidemiology of hepatitis C virus infection in South Korea. Liver Int. 2013;33:586–594. [DOI] [PubMed] [Google Scholar]
- 14. Oethinger M, Mayo DR, Falcone J, Barua PK, Griffith BP. Efficiency of the ortho VITROS assay for detection of hepatitis C virus‐specific antibodies increased by elimination of supplemental testing of samples with very low sample‐to‐cutoff ratios. J Clin Microbiol. 2005;43:2477–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]


