Abstract
Background
Clostridium difficile is a major pathogen responsible for nosocomial infectious diarrhea. We explored optimal laboratory strategies for diagnosis of C. difficile infection (CDI) in our clinical settings, a 1400‐bed tertiary care hospital.
Methods
Using 191 fresh stool samples from adult patients, we evaluated the performance of Xpert C. difficile (Xpert CD), C. diff Quik Chek Complete (which simultaneously detects glutamate dehydrogenase [GDH] and C. difficile toxins [CDT]), toxigenic culture, and a two‐step algorithm composed of GDH/CDT as a screening test and Xpert CD as a confirmatory test.
Results
Clostridium difficile was detected in 35 samples (18.3%), and all isolates were toxigenic strains. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value of each assay for detecting CDI were as follows: Quik Chek Complete CDT (45.7%, 100%, 100%, 89.1%), Quik Chek Complete GDH (97.1%, 99.4%, 97.1%, 99.4%), Xpert CD (94.3%, 100%, 100%, 98.7%), and toxigenic culture (91.4%, 100%, 100%, 98.1%). A two‐step algorithm performed identically with Xpert CD assay.
Conclusion
Our data showed that most C. difficile isolates from adult patients were toxigenic. We demonstrated that a two‐step algorithm based on GDH/CDT assay followed by Xpert CD assay as a confirmatory test was rapid, reliable, and cost effective for diagnosis of CDI in an adult patient setting with high prevalence of toxigenic C. difficile.
Keywords: adult, Clostridium difficile, diagnosis, glutamate dehydrogenase, multistep algorithm, tertiary care hospital
1. Introduction
Clostridium difficile is a major pathogen responsible for infectious diarrhea in health care settings. Toxin A (tcdA, enterotoxin) and toxin B (tcdB, cytotoxin) are primary virulence factors of C. difficile and C. difficile infection (CDI) is associated with considerable morbidity and risk of mortality.1, 2 During the past decade, the incidence of CDI has markedly increased in most countries.3 In United States, the health care costs attributable to hospital‐acquired CDI have been estimated at $500 million to $1.5 billion per year.4, 5, 6 To reduce hospital stays and health care costs associated with CDI, laboratories have been challenged to provide rapid, accurate, and cost‐effective results. The reference method for the detection of C. difficile is toxigenic culture or cell culture cytotoxin neutralization assay.7, 8 Given the test's labor‐intensive and time‐consuming nature, however, its use in clinical laboratories has been limited.9 In recent years, rapid diagnostic tests, performed directly on the stool samples, have been introduced and are beginning to replace the traditional culture‐based methods. Enzyme immune assay (EIA) targeting C. difficile toxin A or B is fast and inexpensive, but its low sensitivity limits its use as a stand‐alone diagnostic test. The current Infectious Diseases Society of America guidelines recommend two‐step testing that uses EIA detection of glutamate dehydrogenase (GDH), a C. difficile common antigen, as initial screening and then confirmatory test based on culture method.9 Nucleic acid amplification methods detect toxin B (tcdB) and/or toxin A (tcdA) genes within the pathogenicity locus (PaLoc) of C. difficile.10 They are rapid, sensitive, and specific, and fully automated on‐demand PCR assays, such as Xpert C. difficile (Xpert CD; Cepheid, Sunnyvale, CA, USA) or BD MAX Cdiff Assay (BD Diagnostics, Franklin Lakes, NJ, USA) became available in clinical laboratories. However, molecular methods usually can be quite costly and the clinical significance of detecting low‐level, non‐viable C. difficile needs to be elucidated.11
Two‐ or three‐step approaches for the laboratory diagnosis of CDI, which are composed of one screening test followed by a confirmatory test, have been suggested to overcome the limitation of single rapid test.9, 12, 13, 14, 15 At an institutional level, diagnostic algorithms that optimize test combinations should reflect the clinical setting of the institution. The prevalence of toxin‐producing C. difficile is one of the most important factors to be considered, because this will inevitably have an impact upon the positive and negative predictive values of any diagnostic tests.16, 17 The goal of this study was to investigate the prevalence of CDI in our institution, a 1400‐bed tertiary care hospital, and to select the optimal testing strategy based on our clinical setting. Two different rapid tests, a lateral‐flow membrane immunoassay and one real‐time PCR assay, were compared with the toxigenic culture method.
2. Materials and Methods
2.1. Samples
This prospective study was conducted between November 2013 and March 2014 at a 1400‐bed tertiary care hospital in Korea. During the study period, we examined 191 consecutive diarrheal stool samples from adult patients (median 65 years) for diagnosis of CDI. Samples from pediatric patients were excluded because of insufficient sample volume. Following routine testing, residual stool samples were used for additional molecular studies and GDH assay. The samples were tested daily or stored at 4°C and tested within 24‐48 h of collection. This study protocol was approved by the Institutional Review Board of Gachon University Gil Medical Center.
2.2. Toxigenic culture
All stool samples were inoculated on C. difficile selective chromogenic agar plate (ChromID C.difficile agar; bioMerieux SA, Craponne, France) and incubated anaerobically at 37°C for 48 h. Gray‐to‐black colonies suspicious for C. difficile were identified using Gram stain and VITEK 2 ANC card (bioMerieux). The identified C. difficile isolates were used to detect tcdA, tcdB, and triose phosphate isomerase (tpi; a C. difficile‐specific internal fragment of a housekeeping gene) genes by multiplex PCR using published primer pairs.18
2.3. C. diff Quik Chek Complete assay
C. diff Quik Chek Complete assay (GDH/CDT; TechLab, Blacksburg, VA, USA), a lateral‐flow membrane immunoassay which detect GDH antigen and toxin A and B concurrently, was performed according to the manufacturer's instructions. GDH antigen and/or toxins were reported as positive, if a corresponding band was seen on the device display window.
2.4. Xpert C. difficile assay
Xpert CD assay (Cepheid), the fully automated real‐time PCR platform, was performed on the GeneXpert DX system to detects tcdB, binary toxin genes (cdtA and cdtB), and tcdCΔ117 (deletion in the negative regulator of toxin production). Binary toxin gene specific PCR and tcdC sequencing were performed in accordance with published methods for isolates that tested positive for binary toxin genes.19, 20
2.5. Interpretation of results and statistical analysis
Given the reported recovery rate on ChromID C. difficile agar (about 98%),21, 22 true positive was defined as positivity by toxigenic culture or positivity by both Xpert CD and GDH or CDT. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated for the GDH/CDT, Xpert CD and toxigenic culture, and by using an algorithm in which GDH/CDT followed by Xpert CD to resolve discrepant samples (GDH+/CDT−). Xpert CD invalid results were excluded from the performance characterization.
3. Results
One hundred and ninety‐one fresh stool samples were tested for CDI and 35 samples (18.3%) were determined to be positive by our interpretation criteria (Table 1). Thirty‐five samples (18. 3%) were positive for GDH with 34 samples (97.1%) giving a true positive result. Among 35 GDH positive samples, 16 (45.7%) were also positive for CDT, and subsequently confirmed by toxigenic culture except for one sample. There was no false positive result for CDT according to our criteria. Thirty‐two (16.8%) samples were positive by toxigenic culture including one sample positive by toxigenic culture only. Three CDI positive samples were missed by toxigenic culture. Xpert CD assay was positive for 33 samples (17.3%). There were three invalid results by Xpert CD assay. Two of them were resolved after repeated tests, however, the remaining one was invalid repeatedly. We found no C. difficile ribotype 027 in our study. The sensitivity, specificity, PPV, and NPV for each method are described in Table 2. Overall, the performances of GDH/CDT assay and Xpert CD assay were comparable with toxigenic culture for diagnosis of CDI.
Table 1.
Results of three laboratory assays for 191 stool samples
| Result | No. of samples | |||
|---|---|---|---|---|
| GDH | CDT | Xpert CD | Toxigenic culture | |
| P | P | P | P | 15 |
| P | P | P | N | 1 |
| P | N | P | P | 15 |
| P | N | P | N | 2 |
| P | N | N | P | 1 |
| P | N | N | N | 1 |
| N | N | N | P | 1 |
| N | N | N | N | 154 |
| N | N | I | N | 1 |
GDH, glutamate dehydrogenase; CDT, C. difficile toxin; P, positive; N, negative; I, invalid.
Table 2.
Performance characteristics of individual assays and two step algorithms for detecting toxigenic C. difficile compared to toxigenic culture as a reference method
| % sensitivity (95% CI) | % specificity (95% CI) | % PPV (95% CI) | % NPV (95% CI) | |
|---|---|---|---|---|
| GDH | 97.1 (85.1‐99.9) | 99.4 (96.5‐100) | 97.1 (85.1‐99.9) | 99.4 (96.5‐100) |
| CDT | 45.7 (28.8‐63.4) | 100 (97.7‐100) | 100 (79.4‐100) | 89.1 (83.6‐93.3) |
| Xpert CDa | 94.3 (80.8‐99.3) | 100 (97.7‐100) | 100 (89.4‐100) | 98.7 (95.5‐99.9) |
| Toxigenic culture | 91.4 (76.9‐89.2) | 100 (97.7‐100) | 100 (89.1‐100) | 98.1 (94.6‐99.6) |
| GDH/CDT + Xpert CD | 94.3 (80.8‐99.3) | 100 (97.7‐100) | 100 (89.4‐100) | 98.7 (95.5‐99.9) |
CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; GDH, glutamate dehydrogenase; CDT, C. difficile toxin.
Invalid result was excluded from analysis.
We simulated the two‐step testing algorithm by employing C. diff Quik Chek Complete assay as a first line test, followed by Xpert CD assay (Figure 1). About half of CDI positive samples (45.7%; 16/35) with GDH+/CDT+ needed no further test for diagnosis of CDI if C. diff Quik Chek Complete assay has been used as a screening test. Overall, 171(89.5%; 16 GDH+/CDT+ and 155 GDH−/CDT−) of samples were correctly classified using GDH/CDT assay only. C. diff Quik Chek Complete/Xpert CD algorithm identified three CDI+ samples which were negative by toxigenic culture, and 2 CDI+ samples were missed by this algorithm (Table 1, Figure 1). In conclusion, our two‐step algorithm for diagnosis of CDI showed performance equal to that of Xpert CD assay alone, and only 19 GDH+/CDT− samples (9.9%) needed additional Xpert CD assay as a confirmatory test (Table 2, Figure 1).
Figure 1.
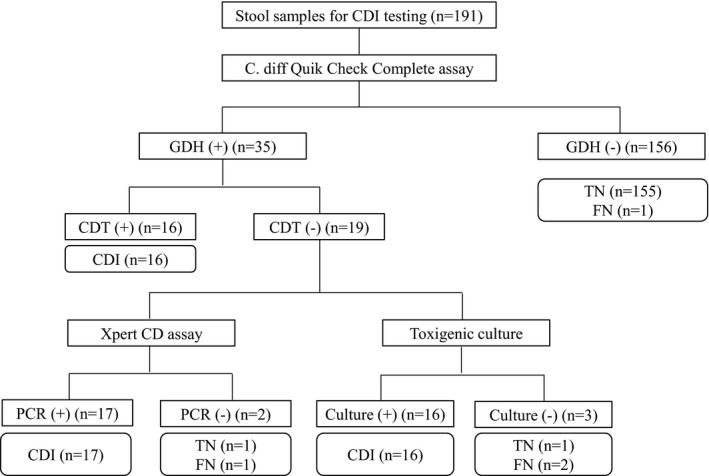
Results of two step algorithm testing composed of C. diff Quik Chek Complete assay followed by Xpert CD assay for diagnosis of CDI
4. Discussion
In this study, we found that most C. difficile isolates from adult patients in our institution were toxin‐producing strains. GDH assay and Xpert CD assay performed comparably with toxigenic culture for the diagnosis of CDI. GDH assay is a screening method that is preferable in two‐ or three‐step algorithms over previous studies for CDI diagnosis, because of its high sensitivity and excellent NPV.23, 24, 25 Two meta‐analyses on the performance of GDH assay for detection of CDI, however, showed that PPV of GDH varied from ~30% to ~100% in a range of CDI prevalence from 5% to 50%.26, 27 Such wide‐range variability in the performance of GDH assay arises from the target detected. A positive GDH assay indicates the presence of the C. difficile in stool samples and is not indicative of CDI itself. Such a limitation mandates its use to a screening test rather than stand‐alone test. Usually, GDH assay is coupled with a confirmatory test indicating the presence of toxin, such as toxigenic culture, cytotoxin assay, toxin PCR, or toxin EIA. In our study, the prevalence of CDI was 18.3% (35/191) and PPV (97.1%) of GDH assay was higher than the findings of others who worked under similar CDI prevalence.17, 25, 28, 29, 30 Such a high PPV may result from the finding that most C. difficile isolates from adult patients in our institution were toxigenic strains. The frequency was quite high compared to that of pediatric patients during the same period (32%, 16 of 50 C. difficile isolates confirmed by toxigenic culture, data not shown in this article).
A variety of nucleic acid amplification methods for detection of CDI have been extensively studied and, compared to other non‐culture‐based methods, are reported to be the most sensitive methods.31 Xpert CD assay is the first on‐demand molecular diagnostic test to detect CDI and its use as a stand‐alone test or a component of two‐step algorithms has been evaluated well.32 In this study, the diagnostic performance of Xpert CD assay was comparable to toxigenic culture. Xpert CD assay identified two positive samples, in agreement with the findings of GDH assay, but negative by toxigenic culture. These discrepancies might result from the presence of low‐level or non‐cultivable organisms in the stool samples. The clinical significance of detecting low‐level or non‐cultivable C. difficile by molecular methods, however, needs to be clarified.11 Definitive results were not obtained upon first analysis for three samples tested with Xpert CD assay, most probably due to PCR inhibition or a technical error. After repeated testing as recommended by the manufacturers, only one sample remained invalid.
Toxin detection by C. diff Quik Chek Complete assay showed poor diagnostic sensitivity compared to other methods, and all CDT positive samples were concurrently GDH positive. Combined with GDH, CDT results showed 100% PPV for diagnosis of CDI. Toxin EIAs are usually highly specific; however, our data shows that a more sensitive assay is preferable for CDI diagnosis in our clinical settings where toxigenic C. difficile accounts for majority of C. difficile isolates.
In our virtual simulation of a two‐step testing algorithm, C. diff Quik Chek Complete assay alone detected a half of CDI positive samples without further tests (Figure 1). When Xpert CD assay was used as a confirmatory test, two false negative results were observed. Novak‐Weekley et al.32 argued that Xpert CD assay alone was superior to other individual tests and multistep testing algorithms in terms of sensitivity and NPV, while decreasing turnaround time significantly. Our data showed similar results. However, the two‐step algorithm with C. diff Quik Chek Complete assay followed by Xpert CD assay seemed to be more cost effective because only 19 Xpert CD assays (9.9%; 19/191) were needed by this algorithm while maintaining the clinical performance similar to toxigenic culture or Xpert CD assay alone. Delay in reporting time remains a concern under this two step algorithm, but Xpert CD assay needs an additional hour to obtain final results, and it means that the reporting of the results would not be delayed significantly.
A limitation to our study is the small sample size (191 samples), and the exclusion of pediatric stool samples due to limited sample volume. The low sensitivity and NPV of GDH assay in pediatric population has been reported.33, 34 It means that more confirmatory tests would be needed for pediatric samples under C. diff Quik Chek Complete/Xpert CD algorithm. Epidemiologic characteristics of CDI in pediatric population from our clinical setting should be delineated to optimize the diagnostic algorithm.
In summary, we demonstrated that a two‐step algorithm based on C. diff Quik Chek Complete assay followed by Xpert CD assay as a confirmatory test was rapid, reliable and cost effective for diagnosis of CDI in adult patient setting with high prevalence of toxigenic C. difficile. As a screening test, C. diff Quik Chek Complete assay showed superior sensitivity to culture method in detection of C. difficile.
Seo JY, Jeong JH, Kim KH, Ahn J‐Y, Park P‐W, and Seo Y‐H. Laboratory diagnosis of Clostridium difficile infection: Comparison of Techlab C. diff Quik Chek Complete, Xpert C. difficile, and multistep algorithmic approach. J Clin Lab Anal. 2017;31:e22135 10.1002/jcla.22135
References
- 1. Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. The role of toxin A and toxin B in Clostridium difficile infection. Nature. 2010;467:711–713. [DOI] [PubMed] [Google Scholar]
- 2. Hall AJ, Curns AT, McDonald LC, Parashar UD, Lopman BA. The roles of Clostridium difficile and norovirus among gastroenteritis‐associated deaths in the United States, 1999‐2007. Clin Infect Dis. 2012;55:216–223. [DOI] [PubMed] [Google Scholar]
- 3. Freeman J, Bauer MP, Baines SD, et al. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev. 2010;23:529–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kyne L, Hamel MB, Polavaram R, Kelly CP. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile . Clin Infect Dis. 2002;34:346–353. [DOI] [PubMed] [Google Scholar]
- 5. McGlone SM, Bailey RR, Zimmer SM, et al. The economic burden of Clostridium difficile . Clin Microbiol Infect. 2012;18:282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zimlichman E, Henderson D, Tamir O, et al. Health care‐associated infections: a meta‐analysis of costs and financial impact on the US health care system. JAMA Intern Med. 2013;173:2039–2046. [DOI] [PubMed] [Google Scholar]
- 7. Chang TW, Bartlett JG, Gorbach SL, Onderdonk AB. Clindamycin‐induced enterocolitis in hamsters as a model of pseudomembranous colitis in patients. Infect Immun. 1978;20:526–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bouza E, Pelaez T, Alonso R, Catalan P, Munoz P, Creixems MR. “Second‐look” cytotoxicity: an evaluation of culture plus cytotoxin assay of Clostridium difficile isolates in the laboratory diagnosis of CDAD. J Hosp Infect. 2001;48:233–237. [DOI] [PubMed] [Google Scholar]
- 9. Cohen SH, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA). Infect Control Hosp Epidemiol. 2010;31:431–455. [DOI] [PubMed] [Google Scholar]
- 10. Braun V, Hundsberger T, Leukel P, Sauerborn M, von Eichel‐Streiber C. Definition of the single integration site of the pathogenicity locus in Clostridium difficile . Gene. 1996;181:29–38. [DOI] [PubMed] [Google Scholar]
- 11. Naaber P, Stsepetova J, Smidt I, et al. Quantification of Clostridium difficile in antibiotic‐associated‐diarrhea patients. J Clin Microbiol. 2011;49:3656–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gilligan PH. Is a two‐step glutamate dehyrogenase antigen‐cytotoxicity neutralization assay algorithm superior to the premier toxin A and B enzyme immunoassay for laboratory detection of Clostridium difficile? J Clin Microbiol. 2008;46:1523–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Larson AM, Fung AM, Fang FC. Evaluation of tcdB real‐time PCR in a three‐step diagnostic algorithm for detection of toxigenic Clostridium difficile . J Clin Microbiol. 2010;48:124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Culbreath K, Ager E, Nemeyer RJ, Kerr A, Gilligan PH. Evolution of testing algorithms at a university hospital for detection of Clostridium difficile infections. J Clin Microbiol. 2012;50:3073–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alcala L, Reigadas E, Marin M, Fernandez‐Chico A, Catalan P, Bouza E. Comparison of GenomEra C. difficile and Xpert C. difficile as confirmatory tests in a multistep algorithm for diagnosis of Clostridium difficile infection. J Clin Microbiol. 2015;53:332–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Planche T, Aghaizu A, Holliman R, et al. Diagnosis of Clostridium difficile infection by toxin detection kits: a systematic review. Lancet Infect Dis. 2008;8:777–784. [DOI] [PubMed] [Google Scholar]
- 17. Eastwood K, Else P, Charlett A, Wilcox M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real‐time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol. 2009;47:3211–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lemee L, Dhalluin A, Testelin S, et al. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (Toxin A), and tcdB (Toxin B) genes for toxigenic culture of Clostridium difficile . J Clin Microbiol. 2004;42:5710–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akerlund T, Persson I, Unemo M, et al. Increased sporulation rate of epidemic Clostridium difficile Type 027/NAP1. J Clin Microbiol. 2008;46:1530–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Terhes G, Urban E, Soki J, Hamid KA, Nagy E. Community‐acquired Clostridium difficile diarrhea caused by binary toxin, toxin A, and toxin B gene‐positive isolates in Hungary. J Clin Microbiol. 2004;42:4316–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han SB, Chang J, Shin SH, et al. Performance of chromID Clostridium difficile agar compared with BBL C. difficile selective agar for detection of C. difficile in stool specimens. Ann Lab Med. 2014;34:376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang JJ, Nam YS, Kim MJ, et al. Evaluation of a chromogenic culture medium for the detection of Clostridium difficile . Yonsei Med J. 2014;55:994–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reller ME, Lema CA, Perl TM, et al. Yield of stool culture with isolate toxin testing versus a two‐step algorithm including stool toxin testing for detection of toxigenic Clostridium difficile . J Clin Microbiol. 2007;45:3601–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quinn CD, Sefers SE, Babiker W, et al. C. Diff Quik Chek complete enzyme immunoassay provides a reliable first‐line method for detection of Clostridium difficile in stool specimens. J Clin Microbiol. 2010;48:603–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharp SE, Ruden LO, Pohl JC, Hatcher PA, Jayne LM, Ivie WM. Evaluation of the C.Diff Quik Chek Complete Assay, a new glutamate dehydrogenase and A/B toxin combination lateral flow assay for use in rapid, simple diagnosis of Clostridium difficile disease. J Clin Microbiol. 2010;48:2082–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crobach MJ, Dekkers OM, Wilcox MH, Kuijper EJ. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile‐infection (CDI). Clin Microbiol Infect. 2009;15:1053–1066. [DOI] [PubMed] [Google Scholar]
- 27. Shetty N, Wren MW, Coen PG. The role of glutamate dehydrogenase for the detection of Clostridium difficile in faecal samples: a meta‐analysis. J Hosp Infect. 2011;77:1–6. [DOI] [PubMed] [Google Scholar]
- 28. Landry ML, Topal J, Ferguson D, Giudetti D, Tang Y. Evaluation of biosite triage Clostridium difficile panel for rapid detection of Clostridium difficile in stool samples. J Clin Microbiol. 2001;39:1855–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vanpoucke H, De Baere T, Claeys G, Vaneechoutte M, Verschraegen G. Evaluation of six commercial assays for the rapid detection of Clostridium difficile toxin and/or antigen in stool specimens. Clin Microbiol Infect. 2001;7:55–64. [DOI] [PubMed] [Google Scholar]
- 30. Snell H, Ramos M, Longo S, John M, Hussain Z. Performance of the TechLab C. DIFF CHEK‐60 enzyme immunoassay (EIA) in combination with the C. difficile Tox A/B II EIA kit, the Triage C. difficile panel immunoassay, and a cytotoxin assay for diagnosis of Clostridium difficile‐associated diarrhea. J Clin Microbiol. 2004;42:4863–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carroll KC, Loeffelholz M. Conventional versus Molecular Methods for the Detection of Clostridium difficile . J Clin Microbiol. 2011;49(9 Supplement):S49–S52. [Google Scholar]
- 32. Novak‐Weekley SM, Marlowe EM, Miller JM, et al. Clostridium difficile testing in the clinical laboratory by use of multiple testing algorithms. J Clin Microbiol. 2010;48:889–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hart J, Putsathit P, Knight DR, Sammels L, Riley TV, Keil A. Clostridium difficile infection diagnosis in a paediatric population: comparison of methodologies. Eur J Clin Microbiol Infect Dis. 2014;33:1555–1564. [DOI] [PubMed] [Google Scholar]
- 34. Ota KV, McGowan KL. Clostridium difficile testing algorithms using glutamate dehydrogenase antigen and C. difficile toxin enzyme immunoassays with C. difficile nucleic acid amplification testing increase diagnostic yield in a tertiary pediatric population. J Clin Microbiol. 2012;50:1185–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]


