Abstract
Objectives
This study was designed to unveil the association of GPR174 rs3827440, PTPN22 rs3789604, and RNASET2 rs9355610 with the onset of liver damage (LD) among the Graves’ disease (GD) patients.
Methods
A total of 120 GD patients were divided into the none‐LD and LD groups. Several indicators were detected for assessing liver functions, and genotypes of single nucleotide polymorphisms (SNPs) were identified. Logistic regression was introduced for investigating the relationship between risk SNPs and LD‐associated hyperthyroidism in GD patients.
Results
Significant differences were identified between LD and none‐LD groups regarding genotype distributions of rs3827440, rs3789604, and rs9355610. Results from logistic regression indicted that among the GD patients, C carriers of PTPN22 rs3789604 were associated with a higher risk of LD‐associated hyperthyroidism, while C carriers of rs3827440 (GPR174) and G carriers of rs9355610 (RNASET2) were associated with a reduced risk of LD‐associated hyperthyroidism.
Conclusions
The C allele of rs3789604 (PTPN22) was a significant risk factor for LD‐associated hyperthyroidism in GD patients, whereas C allele of GPR174 rs3827440 and G allele of RNASET2 rs9355610 appeared to be a protective factor for this disease.
Keywords: gene polymorphism, GPR174, liver damage, PTPN22, RNASET2
1. INTRODUCTION
Graves’ disease (GD) is the most usual cause of hyperthyroidism, and its prevalence is high at 0.25%~1.09% in China and up to 1.3% in the US.1, 2 The GD patients produce auto‐antibodies which bound to the thyroid‐stimulating hormone receptor (TSHR), leading to hyperthyroidism and diffuse hyperplasia of the thyroid gland.3, 4, 5 Approximately 70%‐80% of untreated GD patients are estimated to have biochemical liver damage (LD).5 LD will add difficulties to the selection of appropriate GD treatments, particularly for assessing the anti‐thyroid drugs (ATDs).5 What's more, a large number of studies have confirmed that metabolites of ATDs, particularly propylthiouracil, might contribute to severe hepatotoxicity within GD patients.6
To date, some evidence has revealed that GD is a multi‐factorial disease, which may result from interactions between susceptible genes and environmental hazards.3, 7 As suggested by family and twin studies, about 79% of predisposition to GD could be attributed to genetic factors.3 Thus, identifying susceptible genes and loci that are related to hepatic dysfunctions may be critical to discovering novel interventions for GD. Recent studies have screened out a series of genes as potential thyroid disease susceptibility loci, including Ribonuclease T2 (RNASET2), G protein‐coupled receptor 174 (GPR174), and protein tyrosine phosphatase non‐receptor type 22 (PTPN22).4, 8, 9
RNASET2 is discovered as the member of the Rh/T2/S family and is able to encode extra‐cellular RNases.10 The abnormal RNASET2 expression is probably due to its sub‐cellular distribution and the presence of splice variants.11 Previous studies have implied that RNASET2 may participate in the immune response involved in GD.12 Investigations on Chinese Han populations have dug out evidence indicating that RNASET2 polymorphisms especially the loci rs9355610 is significantly associated with the susceptibility to GD.4 GPR174 is an Xq21 hypothetical purinergic receptor, and it is extensively expressed in immune and thyroid tissues.9, 13 Furthermore, T allele of GPR174 rs3827440 has been suggested to be associated with a higher risk of GD in both Chinese and Polish populations.14 PTPN22 is located on chromosome 1 (1p13.1−lp13.3) and it encodes human lymphoid tyrosine phosphatase (LYP).8, 15 More importantly, PTPN22 has been discovered to have significant associations with a series of autoimmune diseases, such as GD, rheumatoid arthritis (RA), systemic lupus erythematosus, and type 1 diabetes.8, 16 Aside from that, PTPN22 is highly expressed in immune and hematopoietic cells, and it has a negative impact on both T‐cell activation and signal transduction of Csk proteins.3, 16 As suggested by Ichimura et al., rs3789604 was a functional SNP of R620W within PTPN22, and it was related to GD among the Japanese population.17 Besides, it has been reported that the gain‐of‐function variants of PTPN22 might be associated with the rejection in liver transplantation, suggesting that PTPN22 could also function to regulate the liver damages derived from hyperthyroidism of GD.18
Since few studies have clarified the association between genetic risk factors and LD‐associated hyperthyroidism in GD patients, this prospective cohort study was specifically designed in order to provide solid evidence for such a relationship. Additionally, investigation of gene polymorphism within RNASET2, GPR174, and PTPN22 may contribute to discovering new interventions for LD‐associated hyperthyroidism in GD patients.
2. MATERIALS AND METHODS
2.1. Subjects
Altogether 120 GD patients were recruited from the First Affiliated Hospital of Nanchang University from March 2014 to April 2015. They were further divided into the none‐LD group (n=79) and the LD group (n=41). All the participants were individuals of Han ethnicities, and they all have signed the informed consents. This study was approved by the ethics committee of the First Affiliated Hospital of Nanchang University.
2.2. Diagnostic criteria of GD
GD was diagnosed based on the following criteria: (1) symptoms or signs of high metabolism were observed; (2) diffuse enlargement of thyroid gland was existent; (3) laboratory examination showed increased free triiodothyronine (FT3) and free thyroxine (FT4), declined thyroid stimulating hormone (TSH) and positive thyroid stimulating hormone receptor (TSHR) antibody; (4) potential symptoms of exophthalmos and other invasive diseases, such as pretibial mucous edema, were present; (5) nodular goiter toxicity, highly functional thyroid adenoma and other diseases that could also cause hyperthyroidism were excluded.
2.3. Diagnostic criteria of LD in GD patients
The diagnosis of GD‐induced LD was based on the following diagnostic criteria: (1) with untreated GD hyperthyroidism; (2) presence of at least one indicator of LD including increase in alanine aminotransferase (ALT) and aspartate transaminase (AST), total bilirubin (TBIL), gamma glutamyltranspeptidase (GGT), and alkaline phosphatase (ALP), declined total protein (TP), serum albumin (ALB), and symptoms of hepatomegaly and jaundice.
2.4. Collection of specimens
Venous blood samples were collected from patients prior to the therapy. The anticoagulant citrate dextrose (ACD) solution was used for anti‐coagulation. The extraction kit for DNA (Teck biological technology limited corporation, Beijing, China) was applied to extract the genome of subjects. Nanodrop2000 (Thermo Scientific, Waltham, MA, USA) was applied for detecting the concentration and purity of DNA, and the purity of sample DNA (A260/A280) was between 1.8 and 2.0. The automatic biochemical analyzer (Sinothinker Technology, Shenzhen, China) was used to evaluate several GD and LD indicators for assistance of diagnosis and patient characterization. Genomic DNA was extracted from samples based on the instructions of DNA extraction kit (Thermo Fisher Scientific, Waltham, MA, USA) and then preserved in an −20°C refrigerator.
2.5. Genotyping
SNPs related to GD were chosen and investigated in this study. Three SNPs (rs3827440 within GPR174, rs3789604 within PTPN22 and rs9355610 within RNASET2) were included according to their association with GD pathogenicity. Samples were sent to Microread Genetics Go, Ltd (Beijing, China) for direct sequencing and genotyping. They used TaqMan probes synthesized by American ABI corporation, and genotyping was conducted based on the Fluidigm EP1 platform of Fluidigm 96×96 Dynamic Array. The results were shown in Figure 1.
Figure 1.
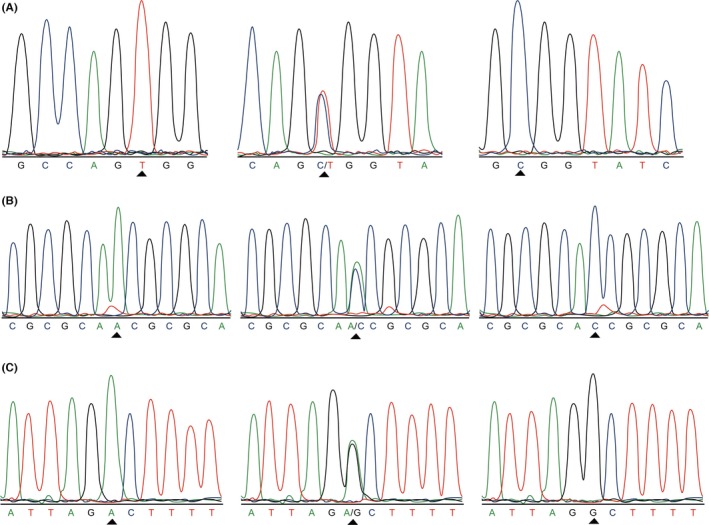
Direct sequencing and genotyping for GPR174,PTPN22 and RNASET2 among the GD patients with LD. (A) Direct sequencing results of rs3827440 polymorphisms in GPR174: T, C/T and C. (B) Direct sequencing results of rs3789604 polymorphisms (A/C) in PTPN22: A, A/C and C. (C) Direct sequencing results of rs9355610 polymorphisms (A/G) in RNASET2: A, A/G and G
2.6. Statistical analysis
All statistics in this study were analyzed by SPSS 21.00 statistical software. Mann‐Whitney test was performed in order to analyze continuous data that were expressed as the mean±SD between two groups. Chi‐square test was carried out in order to analyze categorical data. Logistic regression which incorporated the OR and the corresponding 95% confidence interval was specifically used to assess the influence of risk loci for LD associated with the hyperthyroidism in GD patients. The power calculation was performed with the software of ps power. P‐value of less than .05 provided evidence for statistical significance.
3. RESULTS
3.1. Characteristics of study participants
The sample size was found to be robust after power calculation with the software of ps power. As shown in Table 1, 120 GD patients were enrolled, including 41 LD cases. The LD group consisted of 11 males and 30 females with an average age of (38.1±10.9) years old. The none‐LD group included 20 males and 59 females with the average age of (41.6±11.4) years old. In addition, gender, age, as well as the levels of GD indicators, including FT3, FT4 and TSH, showed little discrepancy between the LD and non‐LD groups (P>.05). As for LD indicators, ALT, AST, GGT, TBIL and ALP presented significantly increased levels in LD patients, while TP and ALB displayed declined levels in the LD group in comparison with those in the non‐LD group (P<.05).
Table 1.
The basic characteristics of GD patients in none‐LD group and LD group
| Group | None‐LD group (n=79) | LD group (n=41) | P value |
|---|---|---|---|
| Age | 41.6±11.4 | 38.1±10.9 | .328a |
| Gender | |||
| Male | 20 | 11 | .858b |
| Female | 59 | 30 | |
| ALT (0~40 U/L) | 25.2±6.5 | 68.4±35.7 | <.001a |
| AST (29~35 U/L) | 30.0±7.0 | 50.5±19.1 | <.001a |
| GGT (3~50 U/L) | 37.0±20.1 | 62.9±27.2 | <.001a |
| TBIL (3.4~17.1 μmol/L) | 14.7±5.3 | 19.9±8.2 | <.001a |
| ALP (40~150 U/L) | 127.3±58.7 | 171.2±57.1 | <.001a |
| TP (60~80 g/L) | 60.4±1.8 | 54.8±5.2 | <.001a |
| ALB (35~55 g/L) | 38.7±2.1 | 33.7±2.9 | <.001a |
| FT3 (3~9 pmol/L) | 14.3±6.8 | 16.8±8.9 | .089a |
| FT4 (9~25 pmol/L) | 52.6±20.7 | 59.6±18.9 | .073a |
| TSH (0.04‐2.5 mIU/L) | 0.016±0.003 | 0.015±0.004 | .126a |
GD, Graves’ disease; none‐LD group, GD patients without LD were included; LD group, GD patients with LD were included; ALT, Alanine aminotransferase; AST, Aspartate transaminase; GGT: Gamma‐glutamyl transferase; TBIL, Total bilirubin; ALP, Alkaline phosphatase; TP, Total protein; ALB, Serum albumin; FT3, Free triiodothyronine; FT4, Free thyroxine; TSH, thyroid stimulating hormone.
Represents that P value was related to none‐LD and LD group and was analyzed by Mann‐Whitney test.
Represents that P value was related to none‐LD and LD group and was analyzed by Chi‐square test.
3.2. Association between risk SNPs and LD in GD patients
The detailed genotype and allele frequency distribution of GPR174 rs3827440, PTPN22 rs3789604, and RNASET2 rs9355610 were shown in Table 2, and the approach of logistic regression was applied to identify the risk factors for the onset of LD‐associated hyperthyroidism among GD patients. After adjusting age, gender and other confounding factors, the results of logistic regression suggested that the frequency of rs3827440 C allele in LD patients was significantly lower than that in the non‐LD group (TC vs TT: OR=0.13; 95% CI: 0.04‐0.48; P=.002; CC vs TT: OR=0.12; 95% CI: 0.02‐0.69; P=.018; TC+CC vs TT: OR=0.24; 95% CI: 0.08‐0.67, P=.007; C vs T: OR=0.31; 95% CI: 0.14‐0.68; P=.004), which provided an evidence that C allele of rs3827440 might act as a protective factor in preventing GD patients from the onset of LD. As for rs3789604 of PTPN22, it was indicated in the form of raw ORs that the C allele might increase the risk of LD in GD patients (AC vs AA: OR=1.36, 95% CI: 0.57‐3.25, P=.483; CC vs AA: OR=8.81; 95% CI: 2.49‐31.25, P<.001; AC+CC vs AA: OR=2.30, 95% CI: 1.06‐4.97, P=.033; C vs A: OR=2.79, 95% CI: 1.57‐4.94, P<.001). In addition, the adjusted ORs also attained a similar trend as the raw ORs (AC vs AA: OR=1.80, 95% CI: 0.57‐5.69, P=.315; CC vs AA: OR=35.15, 95% CI: 5.41‐228.56, P<.001; AC+CC vs AA: OR=3.09, 95% CI: 1.13‐8.47, P=.028; C vs A: OR=5.82, 95% CI: 2.66‐12.77, P<.001). Under all genetic models, it was also revealed that the G allele of RNASET2 rs9355610 might play a protective role against LD in GD patients (GA vs GG: OR=0.10, 95% CI: 0.03‐0.31, P<.001; AA vs GG: OR=0.09, 95% CI: 0.02‐0.52, P=.007; GA+AA vs GG: OR=0.17, 95% CI: 0.07‐0.42, P<.001; A vs G: OR=0.25, 95% CI: 0.13‐0.50, P<.001).
Table 2.
The genotype distribution of three SNPs in none‐LD group and LD group
| Gene | Genotype | LD group (n=41) | None‐LD group (n=79) | P valuea | Raw OR (95% CI) | P valueb | Adjusted OR (95% CI) |
|---|---|---|---|---|---|---|---|
| GPR174 | rs3827440 | ||||||
| TT | 23 | 34 | Ref. | Ref. | Ref. | Ref. | |
| TC | 12 | 28 | .296 | 0.63 (0.27‐1.50) | .002 | 0.13 (0.04‐0.48) | |
| CC | 6 | 17 | .230 | 0.52 (0.18‐1.53) | .018 | 0.12 (0.02‐0.69) | |
| TC+CC | 18 | 45 | .276 | 0.59 (0.28‐1.27) | .007 | 0.24 (0.08‐0.67) | |
| T | 58 | 96 | Ref. | Ref. | Ref. | Ref. | |
| C | 24 | 62 | .127 | 0.64 (0.36‐1.50) | .004 | 0.31 (0.14‐0.68) | |
| PTPN22 | rs3789604 | ||||||
| AA | 16 | 47 | Ref. | Ref. | Ref. | Ref. | |
| AC | 13 | 28 | .483 | 1.36 (0.57‐3.25) | .315 | 1.80 (0.57‐5.69) | |
| CC | 12 | 4 | <.001 | 8.81 (2.49‐31.25) | <.001 | 35.15 (5.41‐228.56) | |
| AC+CC | 25 | 32 | .033 | 2.30 (1.06‐4.97) | .028 | 3.09 (1.13‐8.47) | |
| A | 45 | 122 | Ref. | Ref. | Ref. | Ref. | |
| C | 37 | 36 | <.001 | 2.79 (1.57‐4.94) | <.001 | 5.82 (2.66‐12.77) | |
| RNASET2 | rs9355610 | ||||||
| GG | 27 | 20 | Ref. | Ref. | Ref. | Ref. | |
| GA | 12 | 41 | <.001 | 0.22 (0.09‐0.52) | <.001 | 0.10 (0.03‐0.31) | |
| AA | 2 | 18 | <.001 | 0.08 (0.02‐0.40) | .007 | 0.09 (0.02‐0.52) | |
| GA+AA | 14 | 59 | <.001 | 0.18 (0.08‐0.40) | <.001 | 0.17 (0.07‐0.42) | |
| G | 66 | 81 | Ref. | Ref. | Ref. | Ref. | |
| A | 16 | 77 | <.001 | 0.26 (0.14‐0.48) | <.001 | 0.25 (0.13‐0.50) |
GD, Graves’ disease; LD, Liver damage; OR, Odds ratios; CI, Confidence interval; Ref., Reference.
P value for raw OR.
P value for adjusted OR.
4. DISCUSSION
GD is an autoimmune disorder where autoantibodies target antigens expressed in thyroid tissues and hence hyperthyroidism is promoted.14, 19, 20 Consensus has been reached that several genetic and environmental factors would affect GD.2, 14 On the other hand, there has been evidence suggesting that part of untreated GD patients tend to suffer from LD.5 Thus, we attempted to investigate the relation of genetic polymorphisms and the onset of LD‐associated hyperthyroidism among GD patients.
It has been suspected that GD may be affected by various susceptible genes, 3 and sequencing enabled us to discover that different genotypes were identified in rs3827440 (GPR174), rs3789604 (PTPN22) and rs9355610 (RNASET2). More importantly, the frequency of GPR174 rs3827440 C/T polymorphism, PTPN22 rs3789604 A/C polymorphism and RNASET2 rs9355610 G/A polymorphism significantly differed among the LD and non‐LD groups. For rs3827440, T allele carriers were associated with a higher risk of hyperthyroid LD compared with C allele carriers. Similarly, C allele carriers of rs3789604 may experience a higher risk of hyperthyroid LD when compared with the A allele carriers, whereas G allele carriers of rs9355610 were related with an increased risk of hyperthyroid LD in comparison to the A allele carriers. As suggested by recent studies, lysophosphatidylserine (LysoPS), a ligand of GPR174, had an important role in regulating the immune system.21, 22 Moreover, LysoPS was able to stimulate intracellular cyclic adenosine monophosphate (cAMP) by interacting with GPR174.22 The elevated cAMP could inhibit the production of T‐helper 1 (Th1) cytokines, and stabilize or enhance the level of T‐helper 2 (Th2) cytokines.23 Graves's disease was a Th2‐related disorder, and increased cAMP may contribute to the polarization of Th2 and hence facilitation of GD‐derived LD pathogenesis.9 Another research revealed that GPR174 was extensively expressed in immune‐related tissues, and was moderately expressed in thyroid tissues.9 Besides that, variants of PTPN22 have been considered as a significant risk factor for GD, since they triggered irregular antigen presentation and T‐cell activation in immunological synapses.24
In this study, C allele of rs3789604 was a significant risk factor for LD, while C allele of rs3827440 and G allele of rs9355610 appeared to be a protective factor for LD‐associated hyperthyroidism in GD. Several susceptible genes that are related with GD have been discovered in different ethnic populations.3 PTPN22 rs2476601 was initially discovered as a susceptible locus for autoimmune diabetes. Studies on Asian populations have suggested that rs2476601 may play a significant role in GD.25, 26, 27, 28 However, genome wide association study (GWAS) has provided contradictory results that SNPs of PTPN22 were not significantly associated with the autoimmune diseases among Asians.3
So far, a large number of researches on GD were devoted to unveiling the correlation between Graves’ disease and rs2476601.29, 30 Some studies indicated that rs2476601 rarely influenced the onset of GD in the Chinese Han population,3 while others provided evidence that rs3789604 also had an association with GD.31 Meanwhile, rs3789604 was strongly associated with GD among a Japanese population and such an association was confirmed by a study on the Chinese population.25, 26 Since this study investigated GD‐derived LD, it was reasonable to believe that the aforementioned SNPs were associated with GD‐derived LD.
Several limitations in this study should be addressed carefully. This study only indicated that the distribution of GPR174 rs3827440 polymorphisms significantly differed among the three groups. However, whether this trend was related with GD‐derived LD remained vague, hence studies on the mechanisms underlying the correlation between genotypes and GD‐derived LD were encouraged in the future.
In summary, GPR174 rs3827440, PTPN22 rs3789604, and RNASET2 rs9355610 were significantly associated with altered GD‐derived LD risk. Ongoing researches should be further devoted to this so that potential interventions for LD resulted from GD may be discovered.
5. CONCLUSION
The C allele of rs3789604 (PTPN22) was a significant risk factor for LD associated with the hyperthyroidism in GD patients, whereas C allele of rs3827440 (GPR174) and G allele of rs9355610 (RNASET2) appeared to be a protective factor for this disease. The result of this research would provide several new loci for the diagnosis and targeted therapy of LD associated with the hyperthyroidism in GD in clinical application.
Zhang Q, Liu S, Guan Y, Chen Q, Zhang Q, Min X. RNASET2, GPR174, and PTPN22 gene polymorphisms are related to the risk of liver damage associated with the hyperthyroidism in patients with Graves’ disease. J Clin Lab Anal. 2018;32:e22258 10.1002/jcla.22258
REFERENCES
- 1. Burch HB, Cooper DS. Management of Graves disease: a review. JAMA. 2015;314:2544‐2554. [DOI] [PubMed] [Google Scholar]
- 2. Zhao SX, Xue LQ, Liu W, et al. Robust evidence for five new Graves’ disease risk loci from a staged genome‐wide association analysis. Hum Mol Genet. 2013;22:3347‐3362. [DOI] [PubMed] [Google Scholar]
- 3. Xue L, Pan C, Gu Z, et al. Genetic heterogeneity of susceptibility gene in different ethnic populations: refining association study of PTPN22 for Graves’ disease in a Chinese Han population. PLoS ONE. 2013;8:e84514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen XJ, Gong XH, Yan N, et al. RNASET2 tag SNP but not CCR6 polymorphisms is associated with autoimmune thyroid diseases in the Chinese Han population. BMC Med Genet. 2015;16:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He K, Hu Y, Xu XH, Mao XM. Hepatic dysfunction related to thyrotropin receptor antibody in patients with Graves’ disease. Exp Clin Endocrinol Diabetes 2014;122:368‐372. [DOI] [PubMed] [Google Scholar]
- 6. Li C, Tan J, Zhang G, et al. Risk factors of hyperthyroidism with hepatic function injury: a 4‐year retrospective study. Horm Metab Res 2015;47:209‐213. [DOI] [PubMed] [Google Scholar]
- 7. Vos XG, Endert E, Tijssen JG, Wiersinga WM. Genotypes in relation to phenotypic appearance and exposure to environmental factors in Graves’ hyperthyroidism. Eur J Endocrinol. 2012;167:783‐792. [DOI] [PubMed] [Google Scholar]
- 8. Maine CJ, Marquardt K, Cheung J, Sherman LA. PTPN22 controls the germinal center by influencing the numbers and activity of T follicular helper cells. J Immunol. 2014;192:1415‐1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chu X, Shen M, Xie F, et al. An X chromosome‐wide association analysis identifies variants in GPR174 as a risk factor for Graves’ disease. J Med Genet. 2013;50:479‐485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Acquati F, Bertilaccio S, Grimaldi A, et al. Microenvironmental control of malignancy exerted by RNASET2, a widely conserved extracellular RNase. Proc Natl Acad Sci USA. 2011;108:1104‐1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel S, Chen H, Monti L, et al. RNASET2–an autoantigen in anaplastic large cell lymphoma identified by protein array analysis. J Proteomics. 2012;75:5279‐5292. [DOI] [PubMed] [Google Scholar]
- 12. Chu X, Pan CM, Zhao SX, et al. A genome‐wide association study identifies two new risk loci for Graves’ disease. Nat Genet. 2011;43:897‐901. [DOI] [PubMed] [Google Scholar]
- 13. Napier C, Mitchell AL, Gan E, Wilson I, Pearce SH. Role of the X‐linked gene GPR174 in autoimmune Addison's disease. J Clin Endocrinol Metab. 2015;100:E187‐E190. [DOI] [PubMed] [Google Scholar]
- 14. Szymanski K, Miskiewicz P, Pirko K, et al. rs3827440, a nonsynonymous single nucleotide polymorphism within GPR174 gene in X chromosome, is associated with Graves’ disease in Polish Caucasian population. Tissue Antigens. 2014;83:41‐44. [DOI] [PubMed] [Google Scholar]
- 15. Ostanek L, Ostanek‐Panka M, Bobrowska‐Snarska D, et al. PTPN22 1858C>T gene polymorphism in patients with SLE: association with serological and clinical results. Mol Biol Rep. 2014;41:6195‐6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ivashkiv LB. PTPN22 in autoimmunity: different cell and different way. Immunity. 2013;39:91‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taniyama M, Maruyama T, Tozaki T, Nakano Y, Ban Y. Association of PTPN22 haplotypes with type 1 diabetes in the Japanese population. Hum Immunol. 2010;71:795‐798. [DOI] [PubMed] [Google Scholar]
- 18. Dullin R, Koch M, Sterneck M, Nashan B, Thude H. Association between a gain‐of‐function variant of PTPN22 and rejection in liver transplantation. Transplantation. 2015;99:431‐437. [DOI] [PubMed] [Google Scholar]
- 19. Manji N, Carr‐Smith JD, Boelaert K, et al. Influences of age, gender, smoking, and family history on autoimmune thyroid disease phenotype. J Clin Endocrinol Metab. 2006;91:4873‐4880. [DOI] [PubMed] [Google Scholar]
- 20. Park S, Kim TY, Sim S, et al. Thyrotoxic periodic paralysis and polymorphisms of the ADRB2, AR, and GABRA3 genes in men with Graves’ disease. Endocrinol Metab. 2016;31:142‐146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Inoue A, Ishiguro J, Kitamura H, et al. TGFalpha shedding assay: an accurate and versatile method for detecting GPCR activation. Nat Methods. 2012;9:1021‐1029. [DOI] [PubMed] [Google Scholar]
- 22. Sugita K, Yamamura C, Tabata K, Fujita N. Expression of orphan G‐protein coupled receptor GPR174 in CHO cells induced morphological changes and proliferation delay via increasing intracellular cAMP. Biochem Biophys Res Commun. 2013;430:190‐195. [DOI] [PubMed] [Google Scholar]
- 23. Mosenden R, Tasken K. Cyclic AMP‐mediated immune regulation–overview of mechanisms of action in T cells. Cell Signal. 2011;23:1009‐1016. [DOI] [PubMed] [Google Scholar]
- 24. Huber A, Menconi F, Corathers S, Jacobson EM, Tomer Y. Joint genetic susceptibility to type 1 diabetes and autoimmune thyroiditis: from epidemiology to mechanisms. Endocr Rev. 2008;29:697‐725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gu LQ, Zhu W, Zhao SX, et al. Clinical associations of the genetic variants of CTLA‐4, Tg, TSHR, PTPN22, PTPN12 and FCRL3 in patients with Graves’ disease. Clin Endocrinol. 2010;72:248‐255. [DOI] [PubMed] [Google Scholar]
- 26. Ichimura M, Kaku H, Fukutani T, et al. Associations of protein tyrosine phosphatase nonreceptor 22 (PTPN22) gene polymorphisms with susceptibility to Graves’ disease in a Japanese population. Thyroid. 2008;18:625‐630. [DOI] [PubMed] [Google Scholar]
- 27. Ban Y, Tozaki T, Taniyama M, Tomita M, Ban Y. The codon 620 single nucleotide polymorphism of the protein tyrosine phosphatase‐22 gene does not contribute to autoimmune thyroid disease susceptibility in the Japanese. Thyroid. 2005;15:1115‐1118. [DOI] [PubMed] [Google Scholar]
- 28. Purcell S, Neale B, Todd‐Brown K, et al. PLINK: a tool set for whole‐genome association and population‐based linkage analyses. Am J Hum Genet. 2007;81:559‐575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Velaga MR, Wilson V, Jennings CE, et al. The codon 620 tryptophan allele of the lymphoid tyrosine phosphatase (LYP) gene is a major determinant of Graves’ disease. J Clin Endocrinol Metab. 2004;89:5862‐5865. [DOI] [PubMed] [Google Scholar]
- 30. Smyth D, Cooper JD, Collins JE, et al. Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes. 2004;53:3020‐3023. [DOI] [PubMed] [Google Scholar]
- 31. Clayton D. Testing for association on the X chromosome. Biostatistics. 2008;9:593‐600. [DOI] [PMC free article] [PubMed] [Google Scholar]


