Abstract
Background
Circular RNAs (circRNAs) are recently found involved in cancer occurrence and development. However, their values in the diagnosis of gastric cancers are largely unknown. In this study, we analyzed the values of hsa_circ_0000181 in the diagnosis of gastric cancer.
Methods
Using divergent primers, hsa_circ_0000181 expression levels in fresh gastric cancer tissues and paired adjacent non‐tumorous tissues, and plasmas from patient with gastric cancer and health people were detected by real‐time quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). The association between hsa_circ_0000181 levels and the clinicopathologic features of patients with gastric cancer was further analyzed. Finally, to evaluate the diagnostic value, receiver operating characteristic (ROC) curve was established.
Results
Hsa_circ_0000181 levels in gastric cancer tissues and plasma from gastric cancer patients were significantly decreased than those in paired adjacent non‐tumorous tissues (P < .001) and healthy people (P < .001), respectively. Furthermore, hsa_circ_0000181 expression in gastric cancer tissues was significantly correlated with tumor diameter (P = .027), lymphatic metastasis (P = .044), distal metastasis (P = .023), and carbohydrate antigen 19‐9 (P = .031). Its decreased levels in patients' plasma were significantly associated with differentiation (P = .038) and carcinoembryonic antigen (P = .037). The areas under ROC curve were 0.756. The specificity of tissue hsa_circ_0000181 and sensitivity of plasma hsa_circ_0000181 were 85.2% and 99.0%, respectively.
Conclusions
Thanks to the high stability, tissue and plasma hsa_circ_0000181 may be a novel biomarker for the diagnosis of gastric cancer.
Keywords: biomarker, circular RNA, gastric cancer, hsa_circ_0000181, molecular diagnosis
1. INTRODUCTION
Circular RNAs (circRNAs) are a class of noncoding RNAs with stable structure and high tissue/developmental‐stage specificity.1 They were first found in viroidal plant pathogens and then found commonly in mammalian cells.2 CircRNAs are generated by back‐splicing events as loops without a free 3′ or 5′ ends. They are hard to be degraded by RNase and are more stable than linear RNAs in blood and tissue samples.3 Recently, several circRNAs have been identified. For example, cerebellar degeneration‐related protein 1 antisense (Cdr1as), which encodes cerebellar degeneration‐related protein 1, may serve as microRNA (miRNA) sponges to bind miR‐7.1 Other researchers found that circRNAs can be biomarkers of aging in Drosophila and diseases' biomarkers in human saliva.4, 5 However, the role of circRNAs in gastric cancer is still poorly understood.
Gastric cancer is the third leading cause of cancer‐related death worldwide.6, 7 Although the total mortality of patients with gastric cancer has been decreasing on account of the application of radical surgery and the development of early detection technology, numerous patients are still diagnosed at late stages resulting in a 5‐year survival rate of only 30%.8, 9 Nowadays, endoscopic biopsy following by pathologic examination is still the golden standard for the diagnoses of gastric cancer. However, as is known to all, making a gastroscopy inspection will sometimes make patients uncomfortable and is at a high cost. In addition, its results mostly depend on the experiences of endoscopists.10 Meanwhile, using serologic biomarkers is another way to screen gastric cancer. Serologic test is painless, noninvasive, cheap, and of great convenience. Nevertheless, traditional serum biomarkers such as carcinoembryonic antigen (CEA), carbohydrate antigen 19‐9 (CA19‐9) are not effective in the diagnosis of gastric cancer. They have a relatively low sensitivity and specificity.11 Therefore, it is vital to identify ideal biomarkers to improve diagnosis and treatment of gastric cancer.
Since circRNAs can be used as biomarkers in some diseases, in this study we focus on hsa_circ_0000181 (http://circbase.org/cgi-bin/simplesearch.cgi). Its gene is located at chr1:212977661‐212981190, and its associated‐gene symbol is TATDN3 (TatD DNase domain containing 3). The reason why we choose hsa_circ_0000181 is that it is a gastric cancer associated circRNA according to our previous microarray screening (GEO No. GSE89143, Guo, 2016: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE89143).12 In this study, we first verified that hsa_circ_0000181 levels were significantly changed in gastric cancer tissues and plasmas from gastric cancer patients. Moreover, we found that its expression levels were significantly associated with several clinicopathologic features of patients with gastric cancer. The results indicated that hsa_circ_0000181 might be used as a novel biomarker of gastric cancer.
2. MATERIALS AND METHODS
2.1. Patients and sample collection
A total of 115 fresh gastric cancer tissues and paired adjacent non‐tumorous tissues were collected from Affiliated Hospital of Ningbo University and Yinzhou People's Hospital, China from June 2012 to December 2016. As soon as taken from patients' bodies, the tissue samples were soaked in RNA‐fixer Reagent (Bioteke, Beijing, China) at −80°C until use. The adjacent tissues were about 5 cm from cancer edge. They were assessed by two pathologists and confirmed non‐tumorous tissues. What's more, no radiotherapy or chemotherapy was applied to these patients. Before any treatment was applied, we collected 102 patients' fasting peripheral plasmas (3 mL) with ethylenediaminetetraacetic acid (EDTA) as the anticoagulant. According to the age‐and gender‐matched standard, we further collected fresh fasting plasmas from 105 healthy people at Ningbo No. 2 Hospital, China, at February 2016. Immediately obtained, the plasma was centrifuged and then stored at −80°C until use as previously described.13
Tumors were staged according to the tumor‐node‐metastasis (TNM) staging system of the International Union Against Cancer.13 Histological grade was assessed following the National Comprehensive Cancer Network (NCCN) clinical practice guideline of oncology (V.1.2011).14
The Human Research Ethics Committee from Ningbo University approved all parts of this research (IRB No.20100303). All subjects provided written informed consent.
2.2. Total RNA extraction
According to the manufacturer's instructions, total RNA of tissue samples and plasma samples were first extracted by TRIzol reagent and TRIzol LS Reagent (Invitrogen, Karlsruhe, Germany), respectively. Then, the concentration and purity of total RNA were measured by DeNovix DS‐11 Spectrophotometer (DeNovix, Wilmington, DE, USA). Finally, RNA was stored at −80°C till use.
2.3. Reverse transcription
Using the GoScript RT System (Promega, Madison, WI, USA), cDNA was transcripted as previously described.15
2.4. qRT‐PCR detection of hsa_circ_0000181
The polymerase chain reaction (PCR) was performed using GoTaq qPCR master mix (Promega) on the Mx3005P QPCR System (Stratagene, La Jolla, CA, USA) following the manufacturer's instructions. The sequences of the divergent primers overlapped the splice junction for the detection of hsa_circ_0000181 by qRT‐PCR were 5′‐GAACTGAATGGGCTTGCTATGAAA‐3′ and 5′‐CAGCTGCACTGAGAACATCTCTGA‐3′. Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) was amplified to normalize hsa_circ_0000181 levels. The convergent primer sequences of GAPDH were 5′‐TCGACAGTCAGCCGCATCTTCTTT‐3′ and 5′‐ACCAAATCCGTTGACTCCGACCTT‐3′. These primers were synthesized by KareBay Biochem (Ningbo, China). ΔC q method was used to analyze the data.16 Through two independent experiments, all results were expressed as the mean ± SD.
2.5. Statistical analysis
We use Statistical Product and Service Solutions (SPSS) software 22.0 (SPSS lnc, Chicago, IL, USA) and GraghPad Prism (GraphPad Software, La Jolla, CA, USA) to analyze the experimental data. Using the t‐test, we analyzed the differences of hsa_circ_0000181 expression levels between cancer tissues and adjacent non‐tumor tissues, and between plasma samples from gastric cancer patients and healthy controls. By one‐way analysis of variance (ANOVA), we further evaluated the correlation between hsa_circ_0000181 levels and clinicopathologic factors of patients with gastric cancer. The receiver operating characteristic (ROC) curve was established to evaluate the diagnostic value. P < .05 was considered to have significant difference.
3. RESULTS
3.1. Hsa_circ_0000181 expression was downregulated in gastric cancer tissues and plasma of patients with gastric cancer
By qRT‐PCR, we measured hsa_circ_0000181 levels in tissues and plasma. The results showed that its levels in gastric cancer tissues were significantly lower than those in adjacent non‐tumorous tissues (P < .001; Figure 1A). Moreover, we found that hsa_circ_0000181 levels were decreased in plasma from patients with gastric cancer comparing with controls (P = .001; Figure 1B).
Figure 1.
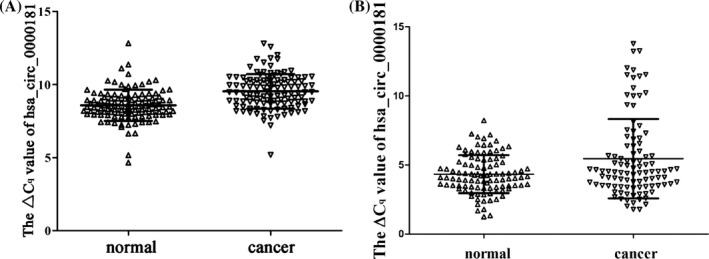
Hsa_circ_0000181 expression levels in gastric cancer tissues (A) and plasma (B) from patients with gastric cancers. The qRT‐PCR method was first used to detect hsa_circ_0000181 and GAPDH levels. Then, ΔC q method was used to analyze the relative level of hsa_circ_0000181. P < .001, n = 115. Higher ΔC q value indicates lower expression
3.2. Diagnostic values of hsa_circ_0000181 in gastric cancer
As what was shown in Figure 1, we found that hsa_circ_0000181 was significantly downregulated in gastric cancer tissues as well as in plasma. As a result, we further evaluated its diagnostic values for gastric cancer.
As what is shown in Table 1, hsa_circ_0000181 expression levels in gastric cancer tissues were significantly related to tumor diameter (P = .027), distal metastasis (P = .023), lymphatic metastasis (P = .044), and CA19‐9 level (P = .031). To better assess the diagnostic values, a ROC curve was made. The area under the ROC curve (AUC) in tissues was 0.756 (P < .0001; Figure 2). And the specificity and sensitivity were 85.2% and 53.9%, respectively. The false‐positive rate and false‐negative rate were 14.8% and 46.1%, respectively. The negative predictive value (NPV) and positive predictive value (PPV) were 64.9% and 78.5%, respectively. The cutoff value was 9.40.
Table 1.
The relationship of hsa_circ_0000181 expression levels (ΔC q) in gastric cancer tissues with clinicopathologic factors of patients with gastric cancer
| Factors | No. of patients (%) | Mean ± SD | P value |
|---|---|---|---|
| Age (y) | |||
| <60 | 31 (27.0) | 9.46 ± 1.32 | .710 |
| ≥60 | 84 (73.0) | 9.56 ± 1.14 | |
| Gender | |||
| Male | 80 (69.6) | 9.60 ± 1.12 | .362 |
| Female | 35 (30.4) | 9.38 ± 1.33 | |
| Diameter (cm) | |||
| ≥5 | 53 (46.1) | 9.82 ± 1.18 | .027 |
| <5 | 62 (54.9) | 9.33 ± 1.14 | |
| Differentiation | |||
| Well | 20 (17.4) | 9.26 ± 0.86 | .673 |
| Moderate | 58 (50.4) | 9.63 ± 1.02 | |
| Poor | 37 (32.2) | 9.54 ± 1.19 | |
| Lymphatic metastasis | |||
| N0 | 52 (45.2) | 9.61 ± 1.31 | .044 |
| N1 | 19 (16.5) | 10.08 ± 0.86 | |
| N2 | 13 (11.3) | 9.06 ± 0.94 | |
| N3 | 31 (27.0) | 9.26 ± 1.11 | |
| Distal metastasis | |||
| M1 | 12 (10.4) | 8.80 ± 1.19 | .023 |
| M0 | 103 (89.6) | 9.62 ± 1.16 | |
| Invasion | |||
| Tis & T1 | 31 (27.0) | 9.47 ± 1.29 | .743 |
| T2 | 12 (10.4) | 9.23 ± 0.79 | |
| T3 | 4 (3.5) | 9.75 ± 0.63 | |
| T4 | 68 (59.1) | 9.60 ± 1.22 | |
| TNM stage | |||
| 0 & I | 35 (30.4) | 9.46 ± 1.26 | .100 |
| II | 23 (20.0) | 9.78 ± 1.26 | |
| III | 45 (39.1) | 9.66 ± 1.03 | |
| IV | 12 (10.4) | 8.80 ± 1.19 | |
| CEA | |||
| Positive | 106 (92.2) | 9.55 ± 1.20 | .921 |
| Negative | 9 (7.8) | 9.60 ± 1.34 | |
| CA19‐9 | |||
| Positive | 70 (60.9) | 9.36 ± 1.13 | .031 |
| Negative | 45 (39.1) | 9.87 ± 1.27 | |
Figure 2.
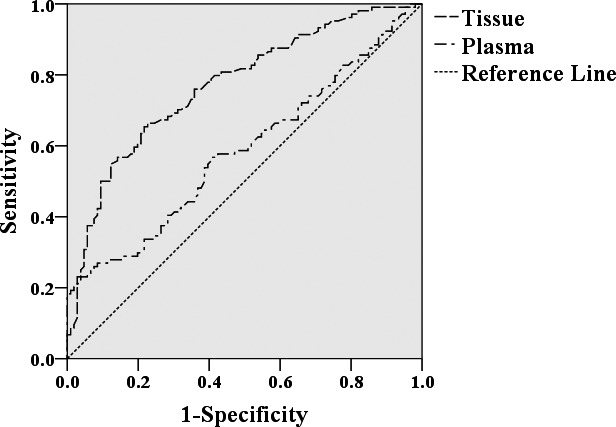
ROC curve of hsa_circ_0000181
As shown in Table 2, hsa_circ_0000181 levels in plasma of patients with gastric cancer were associated with CEA level (P = .037) and differentiation (P = .038). These findings indicated that patients with positive CEA expression had higher hsa_circ_0000181 levels. In addition, the AUC of hsa_circ_0000181 in plasma was 0.582 (P < .05; Figure 2). The specificity and sensitivity were 20.6% and 99.0%, respectively. The false‐negative rate and false‐positive rate were 1% and 79.4%, respectively. The NPV and PPV were 99.5% and 11.1%, respectively. The cutoff value was 7.27.
Table 2.
The relationship of hsa_circ_0000181 expression levels (ΔC q) in plasma from patients with gastric cancer and clinicopathologic factors of patients with gastric cancer
| Factors | No. of patients (%) | Mean ± SD | P value |
|---|---|---|---|
| Age (y) | |||
| <60 | 29 (28.4) | 4.99 ± 2.64 | .310 |
| ≥60 | 73 (71.6) | 5.63 ± 2.96 | |
| Gender | |||
| Male | 72 (70.6) | 5.34 ± 2.85 | .573 |
| Female | 30 (29.4) | 5.70 ± 2.98 | |
| Diameter (cm) | |||
| ≥5 | 49 (48.0) | 5.46 ± 2.93 | .943 |
| <5 | 53 (52.0) | 5.50 ± 2.85 | |
| Differentiation | |||
| Well | 8 (7.8) | 4.56 ± 0.83 | .038 |
| Moderate | 54 (52.9) | 4.98 ± 2.32 | |
| Poor | 40 (39.2) | 6.48 ± 3.42 | |
| Lymphatic metastasis | |||
| N0 | 41 (40.2) | 5.59 ± 2.85 | .892 |
| N1 | 19 (18.6) | 5.14 ± 2.50 | |
| N2 | 12 (11.8) | 5.22 ± 2.92 | |
| N3 | 30 (29.4) | 5.73 ± 3.23 | |
| Distal metastasis | |||
| M1 | 10 (9.8) | 5.28 ± 3.19 | .819 |
| M0 | 92 (90.2) | 5.50 ± 2.86 | |
| Invasion | |||
| Tis & T1 | 16 (15.7) | 5.93 ± 3.10 | .691 |
| T2 | 13 (12.7) | 5.32 ± 2.32 | |
| T3 | 9 (8.8) | 6.37 ± 3.68 | |
| T4 | 64 (62.7) | 5.28 ± 2.85 | |
| TNM stage | |||
| 0 & I | 24 (23.5) | 5.80 ± 2.92 | .267 |
| II | 23 (22.5) | 4.61 ± 1.66 | |
| III | 47 (46.1) | 5.92 ± 3.31 | |
| IV | 8 (7.8) | 4.75 ± 2.58 | |
| CEA | |||
| Positive | 73 (77.7) | 5.22 ± 2.77 | .037 |
| Negative | 21 (22.3) | 6.71 ± 3.07 | |
| CA19‐9 | |||
| Positive | 47 (50.0) | 5.28 ± 3.12 | .357 |
| Negative | 47 (50.0) | 5.83 ± 2.64 | |
4. DISCUSSION
CircRNAs were first discovered in RNA viruses as early as the 1970s using electron microscopy.17 In the following years, due to the development of high‐throughput RNA sequencing (RNA‐Seq) technology and bioinformatics methods, more and more circRNAs have been found. According to their origin, we can divide circRNAs into three major categories: intron‐derived circRNAs, back‐spliced exons formed circRNA, and exon‐intron circRNA or EIciRNA.18, 19, 20
CircRNAs are expressed in human cells in a tissue/developmental‐stage specificity, and sometimes their expression levels can be 10 times higher than their associated linear isoforms.1, 21 CircRNAs have two most important properties: highly conserved and high stability.22 With these two significant properties, circRNAs may be served as better biomarkers in diagnosing cancer in comparison with other noncoding RNAs.
By now, functions of some circRNAs have been revealed. For example, ciRS‐7 (circular RNA sponge for miR‐7) contains about 70 conserved miRNA target sites with miR‐7, which can forcefully suppress miR‐7 activity.23 Some researchers found that miR‐7 suppressed the growth of non‐small‐cell lung cancer (NSCLC) cells24; others found that miR‐7 acted as a therapeutic target in Parkinson's disease.25 In addition, miR‐7 inhibitors promoted pancreatic β‐cell replication through mTOR signaling pathway.26 Thus, acting as a miR‐7 sponge, ciRS‐7 may directly or indirectly participate in occurrence and development of human diseases.
Another famous circRNA is circular antisense noncoding RNA in the INK4 locus (cANRIL). The single nucleotide polymorphism (SNP) in cANRIL gene is associated with the risk of atherosclerotic vascular disease (ASVD).27 This means that circRNAs may play an important role in the progression of tumors or other diseases.
Although more and more circRNAs have been identified, the diagnostic values of most circRNAs are still largely unknown. In this study, for the first time, we found that hsa_circ_0000181 was downregulated in the gastric cancer tissues and plasma from gastric cancer patients (Figure 1). Furthermore, we found its potential diagnostic value in gastric cancer (Figure 2).
The lymphatic metastasis, tumor size, and distal metastasis are important factors affecting prognosis of gastric cancer.28, 29, 30, 31 Patients with lymphatic metastasis have lower 5‐year survival rate.30 Larger tumor size contributes to a poor survival rate of 5 years of patients with gastric cancer.28 Distal metastasis is also a factor of decreasing 5‐year survival rate.31 In our study, we found that low expression of hsa_circ_0000181 in gastric cancer tissue was related to tumor size, distal metastasis, and lymphatic metastasis (Table 1). Patients having higher hsa_circ_0000181 expression levels have higher rate of distal metastasis (Table 1). As a tissue‐based biomarker, hsa_circ_0000181 had a higher specificity (85.2%), as well as a high PPV (78.5%) and NPV (64.9%).
It was reported that differentiation also contributes to 5‐year survival rate.32 Interestingly, we found that hsa_circ_0000181 levels were correlated with differentiation (Table 2). Patients with lower differentiation had lower hsa_circ_0000181 levels. In addition, hsa_circ_0000181 levels also associated with blood CEA levels (Table 2). These mean that as a plasma‐based biomarker, hsa_circ_0000181 should be considered as a noninvasive biomarker of gastric cancer.
Nowadays, CEA and CA19‐9 have been widely used as plasma‐based biomarkers for the diagnosis of gastric cancer and other tumors.9, 11, 33 However, they have low sensitivity and specificity. A study indicated that CEA's sensitivity and specificity were 68.6% and 59.3%, respectively; and for CA19‐9, 60.5% and 55.9%, respectively.34 In comparison with CEA and CA19‐9, hsa_circ_0000181 had a higher sensitivity in screening gastric cancer (Figure 2).
The limitation of this study is that it is only a relative small sample study. It would be better if initial validation on a smaller cohort of patients (Training set) followed by a larger cohort of patients (Validation set).
In conclusion, our data suggest that hsa_circ_0000181 may be served as a novel biomarker in the diagnosis of gastric cancer.
ACKNOWLEDGMENTS
This work was supported by The Applied Research Project on Nonprofit Technology of Zhejiang Province (No. 2016C33177), Zhejiang Provincial High‐Education Teaching Reform Project (No. jg2015047), The Scientific Innovation Team Project of Ningbo (No. 2017C1100019), National Natural Science Foundation of China (No. 81772279), and the K. C. Wong Magna Fund in Ningbo University.
Zhao Q, Chen S, Li T, Xiao B, Zhang X. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J Clin Lab Anal. 2018;32:e22333 10.1002/jcla.22333
REFERENCES
- 1. Yao T, Chen Q, Fu L, Guo J. Circular RNAs: biogenesis, properties, roles, and their relationships with liver diseases. Hepatol Res. 2017;47:497‐504. [DOI] [PubMed] [Google Scholar]
- 2. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single‐stranded covalently closed circular RNA molecules existing as highly base‐paired rod‐like structures. Proc Natl Acad Sci USA. 1976;73:3852‐3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li P, Chen H, Chen S, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116:626‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Westholm J, Miura P, Olson S, et al. Genome‐wide analysis of drosophila, circular RNA reveals their structural and sequence properties and age‐dependent neural accumulation. Cell Rep. 2011;9:1966‐1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lin X, Lo HC, Wong DT, Xiao X. Noncoding RNAs in human saliva as potential disease biomarkers. Front Genet. 2015;6:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang J, Qu J, Li Z, et al. Pretreatment platelet‐to‐lymphocyte ratio is associated with the response to first‐line chemotherapy and survival in patients with metastatic gastric cancer. J Clin Lab Anal. 2017. 10.1002/jcla.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xiang F, Ni Z, Zhan Y, Xu J, Wu R, Kang X. Association of 758 G/A polymorphism of 3′untranslated region of prohibitin with risk of gastric cancer. J Clin Lab Anal. 2017. 10.1002/jcla.22182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7:8601‐8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hashad D, Elbanna A, Ibrahim A, Khedr G. Evaluation of the role of circulating long non‐coding RNA H19 as a promising novel biomarker in plasma of patients with gastric cancer. J Clin Lab Anal. 2016;30:1100‐1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park JY, von Karsa L, Herrero R. Prevention strategies for gastric cancer: a global perspective. Clin Endosc. 2014;47:478‐489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marlet J, Bernard M. Comparison of LUMIPULSE (®) G1200 with kryptor and modular E170 for the measurement of seven tumor markers. J Clin Lab Anal. 2016;30:5‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shao Y, Li J, Lu R, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173‐1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shao Y, Ye M, Li Q, et al. LncRNA‐RMRP promotes carcinogenesis by acting as a miR‐206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget. 2016;7:37812‐37824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132‐136. [DOI] [PubMed] [Google Scholar]
- 15. Yang Y, Shao Y, Zhu M, et al. Using gastric juice lncRNA‐ABHD11‐AS1 as a novel type of biomarker in the screening of gastric cancer. Tumour Biol. 2016;37:1183‐1188. [DOI] [PubMed] [Google Scholar]
- 16. Pang Q, Ge J, Shao Y, et al. Increased expression of long intergenic non‐coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol. 2014;35:5441‐5447. [DOI] [PubMed] [Google Scholar]
- 17. Hsu M, Cocaprados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339‐340. [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792‐806. [DOI] [PubMed] [Google Scholar]
- 19. Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;56:103‐111. [DOI] [PubMed] [Google Scholar]
- 20. Li Z, Huang C, Bao C, et al. Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256‐264. [DOI] [PubMed] [Google Scholar]
- 21. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472‐480. [PMC free article] [PubMed] [Google Scholar]
- 23. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384‐388. [DOI] [PubMed] [Google Scholar]
- 24. Cao Q, Mao ZD, Shi YJ, et al. MicroRNA‐7 inhibits cell proliferation, migration and invasion in human non‐small cell lung cancer cells by targeting FAK through ERK/MAPK signaling pathway. Oncotarget. 2016;7:77468‐77481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha‐synuclein expression and toxicity by microRNA‐7. Proc Natl Acad Sci USA. 2009;106:13052‐13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y, Liu J, Liu C, Naji A, Stoffers DA. MicroRNA‐7 regulates the mTOR pathway and proliferation in adult pancreatic β‐cells. Diabetes. 2013;62:887‐895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burd CE, Jeck WR, Liu Y, et al. Expression of linear and novel circular forms of an INK4/ARF‐associated non‐coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamamura Y, Nakajima T, Ohta K, et al. Determining prognostic factors for gastric cancer using the regression tree method. Gastric Cancer. 2002;5:201‐207. [DOI] [PubMed] [Google Scholar]
- 29. Sun W, Yang Y, Xu C, Xie Y, Guo J. Roles of long noncoding RNAs in gastric cancer and their clinical applications. J Cancer Res Clin Oncol. 2016;142:2231‐2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Siewert JR, Böttcher K, Stein HJ, Roder JD. Relevant prognostic factors in gastric cancer: ten‐year results of the German Gastric Cancer Study. Ann Surg. 1998;228:449‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim JP, Lee JH, Kim SJ, Yu HJ, Yang HK. Clinicopathologic characteristics and prognostic factors in 10783 patients with gastric cancer. Gastric Cancer. 1998;1:125‐133. [DOI] [PubMed] [Google Scholar]
- 32. Shen ZL, Song KY, Ye YJ, et al. Significant differences in the clinicopathological characteristics and survival of gastric cancer patients from two cancer centers in China and Korea. J Gastric Cancer. 2015;15:19‐28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peng HX, Yang L, He BS, et al. Combination of preoperative NLR, PLR and CEA could increase the diagnostic efficacy for I‐III stage CRC. J Clin Lab Anal. 2016. 10.1002/jcla.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu J, Li G, Wang Z, et al. Circulating microRNA‐21 is a potential diagnostic biomarker in gastric cancer. Dis Markers. 2014;2015:435656. [DOI] [PMC free article] [PubMed] [Google Scholar]


