Abstract
Background
H2BK12ac is an important histone acetylation pattern of H2B, which has been reported in several cancers. However, whether H2BK12ac joins in Ras-ERK1/2 activation-induced osteosarcoma (OS) cell behaviors remain unclear. The study explored this peradventure and revealed the underlying mechanism.
Methods
MG-63 cells were transfected with pEGFP-N1, pEGFP-RasWT and pEGFP-K-RasG12V/T35S, H2BK12ac and ERK1/2 expression levels were analyzed by Western blot. Effects of H2BK12ac on cell viability, migration, colony formation and cell cycle were investigated by MTT, Transwell, soft-agar colony formation and flow cytometry assays. RT-qPCR and ChIP were performed to study the effect of H2BK12ac and CBP on ERK1/2-downstream gene transcriptions.
Results
H2BK12ac was specifically down-regulated by Ras-ERK1/2 activation in MG-63 cells. Down-regulated H2BK12ac participated in regulating cell proliferation and migration of MG-63 cells, meanwhile, affected the transcription of ERK1/2-downstream genes. Additionally, silence of HDAC1 up-regulated H2BK12ac expression, and inhibited the promoting effect of Ras-ERK1/2 on MG-63 cells' proliferation, migration and RNA expression levels of ERK1/2-downstream genes. Further, the degradation of CBP mediated by MDM2 was discovered to be linked to Ras-ERK1/2 activation-induced H2BK12ac down-regulation.
Conclusion
These findings from the study demonstrated that Ras-ERK1/2 signaling could promote the development of OS via regulating H2BK12ac through MDM2-mediated CBP degradation.
Keywords: osteosarcoma, Ras-ERK1/2 signaling, H2BK12ac, HDAC1, CBP, MDM2
Plain Language Summary
H2BK12ac is specifically down-regulated by Ras-ERK1/2 signaling pathway;
H2BK12ac inhibition is involved in regulating MG-63 cells' phenotype;
H2BK12ac joins in regulating the transcription of ERK1/2-downstream genes;
HDAC1 knockdown regulates H2BK12ac and inhibits the carcinogenic role of Ras-ERK1/2;
Ras-ERK1/2 regulates H2BK12ac by MDM2-mediated CBP degradation.
Introduction
Osteosarcoma (OS) is a common malignant bone tumor in children and adolescents under the age of 20, which accounts for about 5 percent of pediatric tumors.1 The prominent symptom of OS is intense pain at the site of the tumor, and the lump can be found in the pain area with the development of OS.2 An increasing body of evidence suggests that extracellular signal-regulated kinase (ERK) signaling pathway is closely associated with the occurrence and development of OS.3 In vivo animal experiment revealed that the ERK1/2 signaling pathway is related to the lung metastasis of OS.4 Other recent research hinted that activation of ERK1/2 signaling pathway could mediate OS cells' apoptosis.5 Besides, the abnormal of ERK signaling pathway has been reported to regulate tumor proliferation and migration in OS similar to other cancers.6,7 However, the exact regulation mechanism of ERK1/2 signaling pathway in OS is still unclear.
Ras is the expression product of proto-oncogene c-Ras, which is an important signaling regulator in the upstream of ERK1/2 signaling pathway.8 It is a molecular switch of ERK1/2 signaling pathway, which can mediate a variety of extracellular signal transduction, and its active state can affect cell growth, differentiation and cytoskeleton.9,10 The previous studies have demonstrated that the gene mutation of Ras is closely associated with the occurrence of malignancy.11,12 An interesting study demonstrated that Ras-ERK1/2 activation played a crucial role during tumor maintenance and which might be particularly important for the treatment of Ras-driven cancers.13 However, how the mutation of Ras affects the development of OS is still needed for further research.
Histone modification is a key epigenetic mechanism for transcriptional mediation in eukaryotes, which includes methylation, acetylation, phosphorylation and ubiquitination.14 Histone modification is not only closely linked to chromosomal remodeling, but also plays an important role in determining cell fate, cell growth and carcinogenesis.15 Existing evidence has demonstrated that the abnormal changes in histone modification participated in regulating the biological processes of tumor cells.16 Histone H2B acetylated on lysine 12 (H2BK12ac) is a crucial histone acetylation pattern, which has been reported to be connected with the recurrence of lung cancer.17 However, the effect of H2BK12ac on the development of OS is still unclear.
In the present study, we aimed to explore the relationship between the Ras-ERK1/2 activation and H2BK12ac, as well as uncovered the regulatory mechanism of H2BK12ac on Ras-ERK1/2 activation-induced MG-63 cells' phenotype. In this process, the effect of histone deacetylases 1 (HDAC1), CREB binding protein (CBP) and murine double minute 2 (MDM2) were also studied. These findings might provide a novel sight for the treatment of OS.
Materials And Methods
Plasmids Construction
The coding regions of H2B, CBP, HDAC1, MDM2 and wild-type K-Ras (RasWT) were amplified by using PCR. These PCR products were sub-cloned into pEGFP-N1 plasmid and HA-tag (Invitrogen, Carlsbad, CA, USA). The pEGFP-K-RasG12V/T35S was mutated using pEGFP-K-RasG12V construct as a template. The siRNAs that were specific for MDM2 and HDAC1 were synthetized from GenePharma (Shanghai, China). The pEGFP-H2BK12Q was constructed using the TaKaRa MutanBEST Kit (TaKaRa, Dalian, China) based on the kit instructions.
Cell Culture And Treatments
The OS cell line of MG-63 was obtained from American Type Culture Collection (ATCC, Rockville, MD, USA), which was grown in the common culture medium of Dulbecco’s modified Eagle’s medium (DMEM, Gibco-BRL, Carlsbad, CA, USA), and the culture medium contains 10% fetal bovine serum (FBS, Gibco-BRL), 100 units/mL penicillin and 100 µg/mL streptomycin (Gibco-BRL). The cells were incubated in a humidified atmosphere of 95% air and 5% CO2 at 37°C.
Cell Transfection
MG-63 cells (5 × 105 per well) were seeded in 6-well plates and incubated overnight. The plasmids or siRNA were transfected alone or co-transfected into MG-63 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Following 48 hrs transfection, cells were harvested and were analyzed using Western blot or RT-qPCR assay.
Cell Viability Assay
MG-63 cells (5 × 103 cells/well) were incubated in 96-well plates containing the complete DMEM (100 µL/well) for 24 hrs. After transfection for 48 hrs, 20 µL of 3-(4, 5-dimethylthiazol-2-yl)-2 5-diphenyl-2Htetrazolium bromide solution (5 mg/mL, MTT; Sigma-Aldrich, St. Louis, MO, USA) was added to each well of 96-well plates. After incubation for 4 hrs at 37°C, 100 µL of dimethyl sulfoxide (DMSO, Sigma-Aldrich) was added to each well to lyse the formazan crystals. The absorbance was measured at 570 nm wavelength by using a Microplate Reader (Bio-Rad, Hercules, CA, USA).
Soft-Agar Colony Formation Assay
After transfection with GFP, H2B, Ras and H2BK12Q, MG-63 cells were collected and suspended in DMEM containing 0.35% low-melting agarose. Cells were then solidified in DMEM with 0.6% agarose in 6-well culture plate at a density of 1 × 103 cells/well. After incubation for three weeks, the number of the colonies was observed under a microscopically (Nikon, Tokyo, Japan).
Flow Cytometric Analysis Of Cell Cycle Distribution
MG-63 cells transfected with Ras, GFP, si-HDAC1-1, si-HDAC1-2 and si-NC were collected, and washed with PBS for 2 times, as well as fixed in 70% ethanol for 2 hrs at 4°C. Then, cells were stained in propidium iodide (PI) solution (0.2 mg/mL, Roche, Basel, Switzerland) for 30 mins at room temperature. Total of 4000 events for each sample were acquired by using an Epics XL flow cytometer (Beckman Coulter, Fullerton, CA, USA). The percentages of cells in the G0/G1, S and G2/M phases were analyzed according to propidium-DNA fluorescence using the system software (Beckman Coulter, Fullerton, CA, USA).
Transwell Migration Assay
The transwell assay with 8-µm pore filters (Costar, Boston, MA) was performed to assess cell migration of MG-63 cells. After transfection, 1 × 104 MG-63 cells in serum-free media were supplemented into the upper chamber, and the lower chamber was filled with the complete culture medium. After incubation for 24 hrs in the conventional culture conditions, the transwell culture chamber was taken out, and the cells that migrated to the bottom surface of the membrane were fixed with methanol. Subsequently, cells were stained with 0.5% crystal violet for 20 mins, and counted by using a microscope (Nikon, Japan). The OD values were measured at 570 nm using the microplate reader (Bio-Rad).
Reverse Transcription-Quantitative PCR (RT-qPCR)
MG-63 cells were lysed directly in cell culture plates, and total RNA from the cells were examined using TRIzol reagent (Invitrogen). The first-strand cDNA was synthetized using M-MuLV reverse transcriptase (Fermentas) and oligo-dT primers (Invitrogen). RT-qPCR was performed using a QuantiTect SYBR Green PCR Kit (Qiagen). GAPDH served as endogenous control, and the relative expression levels all of the samples were calculated by using the 2−ΔΔCT method.18
Chromatin Immunoprecipitation (ChIP)
After transfection, cells (3 × 106 cells/sample) were cross-linked in 1% formaldehyde for 10 mins at room temperature, and then washed with PBS for two times and lysed with SDS lysis buffer (Upstate Biotechnology, Lake Placid, NY, USA). The lysates were then subjected to an ultrasonic bath for sonication (Bioruptor, Diagenode). After centrifugation at 15,000 × g for 10 mins, the supernatants were diluted with ChIP dilution buffer (Upstate Biotechnology) and were immunoprecipitated with rabbit anti-H2BK56ac (2 µg, ab195494, Abcam, Cambridge, UK) overnight at 4°C. The normal anti-IgG antibody (2 µg, ab171870, Abcam) was used as a control of immunoprecipitation. The beads were washed in low-salt for 5 mins at 4°C, and were washed twice in 1 × TE (Upstate Biotechnology) for 2 mins at room temperature. The DNA was eluted from the beads were treated as Liu et al described.19 The serial dilutions of the 10% input DNA (1:4, 1:20, 1:100 and 1:500) were analyzed by using RT-qPCR.
Western Blot Analysis
MG-63 cells were collected after transfection for 48 hrs, and these collected cells were washed once with PBS. The whole-cell lysates were prepared by using RIPA lysis buffer (Beyotime Biotechnology). The concentration of the protein samples was quantified by using a BCA protein assay kit (Pierce, Appleton, WI, USA). The total protein was resolved by SDS-PAGE and transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, NH). The membranes were blocked in BSA solution at room temperature for 1 hr, and then incubated with the appropriate primary antibodies as anti-Histone H2B (acetyl K12) antibody (ab61228), anti-Histone H2B antibody (ab1790), anti-p-ERK1/2 antibody (ab223500), anti-ERK1/2 antibody (ab115799), anti-HA antibody (ab71113), anti-GFP antibody (ab6556), anti-CBP antibody (ab119488), anti-His antibody (ab83831), anti-MDM2 antibody (ab38618) and anti-β-actin antibody (ab8227, Abcam) overnight at 4°C. Afterward, the membranes were incubated with HRP-conjugated secondary antibody (ab205718, Abcam) for 1 hr at room temperature. The Western blots were developed by using Western Blotting Luminol Reagent (Santa Cruz Biotechnology, California, USA). The intensity of these bands was quantified by using ImageJ software (Bio-Rad).
Statistical Analysis
These data from the current study are shown as the mean ± standard deviation (SD). Statistical analyses were performed using SPSS 19.0 statistical software (SPSS, Inc., Chicago, IL, USA). One-way ANOVA followed by the Duncan’s post hoc test was used to analyze the significant differences among different groups. A P-value of < 0.05 represented a statistically significant result.
Results
Ras-ERK1/2 Activation Down-Regulated H2BK12ac Expression Level
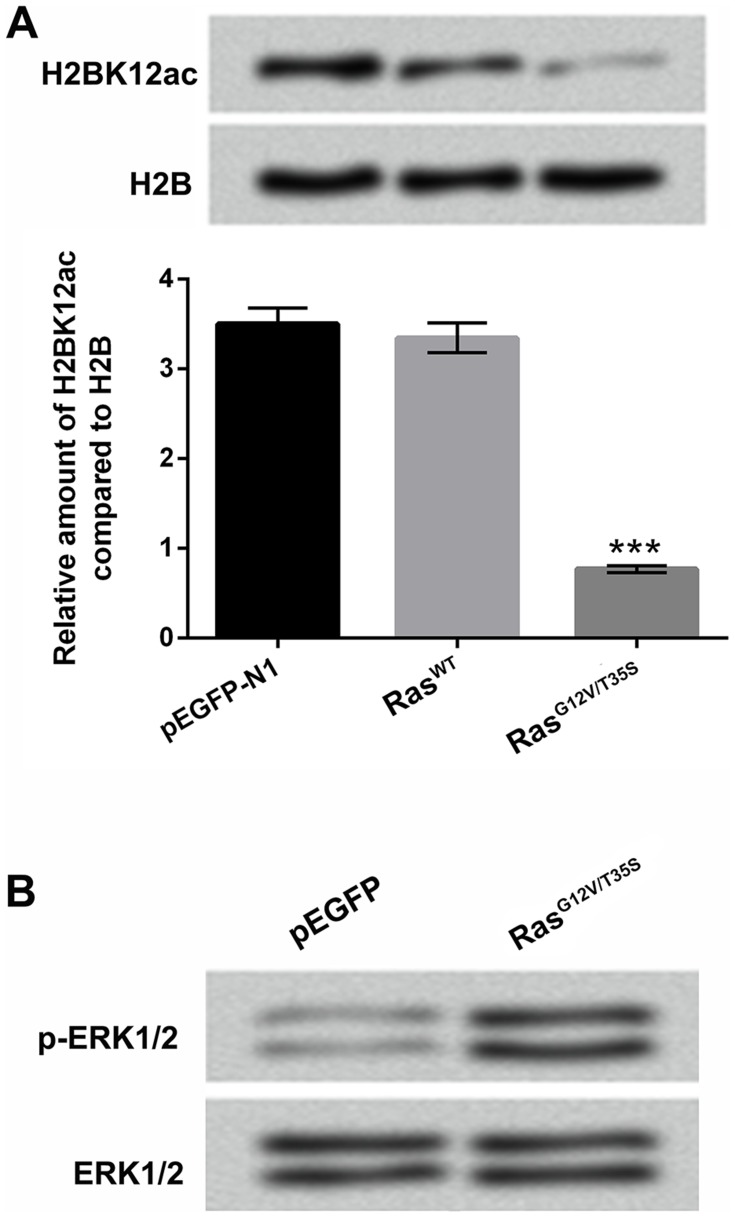
We constructed the plasmids of pEGFP-RasWT (RasWT) and pEGFP-K-RasG12V/T35S (RasG12V/T35S), and the empty plasmid of pEGFP-N1 served as a negative control. These plasmids were transfected into MG-63 cells, and the H2BK12ac levels were then assessed by using Western blot assay in these transfected cells. Results in Figure 1A revealed that the H2BK12ac level was significantly declined in MG-63 cells transfected with RasG12V/T35S plasmid compared with in MG-63 cells transfected with pEGFP-N1 (P < 0.001). There was no significant difference of the H2BK12ac level in RasWT-transfected cells and in pEGFP-N1-transfected cells (Figure 1A). By Western blot assay verification, we observed that the specific mutant plasmid of RasG12V/T35S significantly up-regulated p-ERK1/2 protein level, indicating that it indeed activated ERK1/2 signaling pathway (Figure 1B). These data demonstrated that H2BK12ac was specifically down-regulated by ERK1/2 signaling pathway.
Figure 1.
H2BK12ac is specifically regulated by the ERK1/2 pathway. (A) Expression vectors of pEGFP-N1, RasWT and RasG12V/T35S were transfected into MG-63 cells. The relative H2BK12ac levels were determined in the transfected cells by using Western blot assay. (B) The protein levels of p-ERK1/2 and ERK1/2 were detected by using Western blot to verify the pathway was indeed activated after transfection. ***P < 0.001 represents significant difference.
Abbreviations: H2BK12ac, histone H2B acetylated on lysine 12; ERK, extracellular signal-regulated kinase.
H2BK12ac Was Involved In Regulating Cell Proliferation And Migration In MG-63 Cells
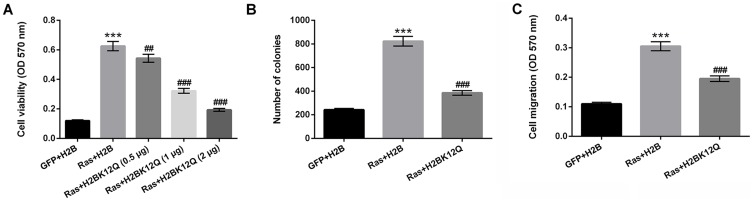
To explore the effect of H2BK12ac on MG-63 cells' proliferation and migration, the H2BK12Q plasmid was constructed to mimic the acetylation state of H2BK12ac. Different amounts of the H2BK12Q (0.5, 1 and 2 µg) expression plasmids and RasG12V/T35S were co-transfected into MG-63 cells, and viability of these cells was subsequently analyzed. As shown in Figure 2A, results displayed that cell viability was significantly increased in RasG12V/T35S-transfected MG-63 cells (P < 0.001). When cells were co-transfected with H2BK12Q, the increased cell viability induced by RasG12V/T35S was obviously inhibited at the amounts of 0.5 (P < 0.01), 1 and 2 µg (P < 0.001). Next, colony formation assay demonstrated that the number of colonies was significantly increased in RasG12V/T35S-transfected MG-63 cells (P < 0.001). But, the inhibition was exhibited in RasG12V/T35S and H2BK12Q co-transfected MG-63 cells (P < 0.001, Figure 2B). Additionally, cell migration was also increased in RasG12V/T35S-transfected cells (P < 0.001); however, in RasG12V/T35S and H2BK12Q co-transfected MG-63 cells, the promoting effect of cell migration was obviously decreased (P < 0.001, Figure 2C). Collectively, these data indicated that H2BK12Q could regulate Ras-ERK1/2 activation-induced cell proliferation and migration in MG-63 cells.
Figure 2.
Ras-ERK1/2 activation-induced H2BK12ac participates in regulating cell proliferation and migration. The expression plasmids of pEGFP-N1, pEGFP-H2B, pEGFP-K-RasG12V/T35S and pEGFP-H2BK12Q were constructed (H2BK12Q was constructed to mimic the acetylation state of H2BK12ac, the plasmids were indicated as GFP, H2B, Ras and H2BK12Q, respectively). MG-63 cells were transfected with H2B, or 0.5, 1, and 2 µg of H2BK12Q and 0.6 µg Ras plasmids. (A) Cell viability, (B) colony formation, and (C) cell migration were assessed by MTT, soft-agar colony formation and Transwell migration assays. ***P < 0.001: Ras+H2B vs GFP+H2B; ##P < 0.01, ###P < 0.001: Ras+H2BK12Q vs Ras+H2B.
Abbreviations: H2BK12ac, histone H2B acetylated on lysine 12; ERK, extracellular signal-regulated kinase; MTT, 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide.
H2BK12ac Was Involved In Regulating The Transcription Of ERK1/2-Downstream Genes
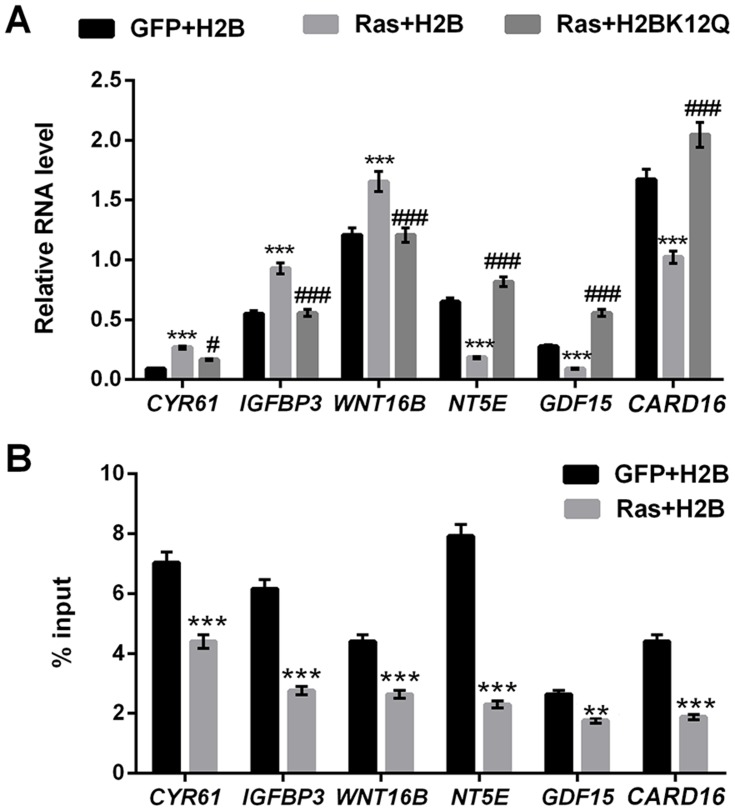
We next investigated the effect of H2BK12Q on the expression of the downstream target genes of ERK (CYR61, IGFBP3, WNT16B, NT5E, GDF15 and CARD16) by using RT-qPCR. Results in Figure 3A revealed that the expression levels of CYR61, IGFBP3 and WNT16B were significantly increased in Ras and H2B co-transfected MG-63 cells (P < 0.001). On the contrary, the expression levels of NT5E, GDF15 and CARD16 were significantly decreased in Ras and H2B co-transfected MG-63 cells (P < 0.001). However, these results were obviously reversed in Ras and H2BK12Q co-transfected MG-63 cells (P < 0.05 or P < 0.001, Figure 3A). To further verify the relationship between H2BK12ac and these transcription genes, the ChIP assay was performed. Results in Figure 3B revealed the reduction input levels of H2BK12ac were presented in these gene promoter regions (P < 0.01 or P < 0.001). The data hinted that H2BK12ac could be directly incorporated into the promoter region of these genes for regulation of the transcription.
Figure 3.
H2BK12ac is involved in regulating the transcription of ERK1/2-targeted genes. (A) The relative RNA level of CYR61, IGFBP3, WNT16B, NT5E, GDF15 and CARD16 were analyzed by using RT-qPCR. (B) The relationship between H2BK12ac and these transcription genes was assessed by using ChIP assay. ***P < 0.001: Ras+H2B vs GFP+H2B; #P < 0.05, ###P < 0.001: Ras+H2BK12Q vs Ras+H2B.
Abbreviations: H2BK12ac, histone H2B acetylated on lysine 12; RT-qPCR, reverse transcription-quantitative PCR; ChIP, chromatin immunoprecipitation.
HDAC1 Knockdown Regulates H2BK12ac Expression And Inhibited The Carcinogenic Role Of Ras-ERK1/2 In MG-63 Cells
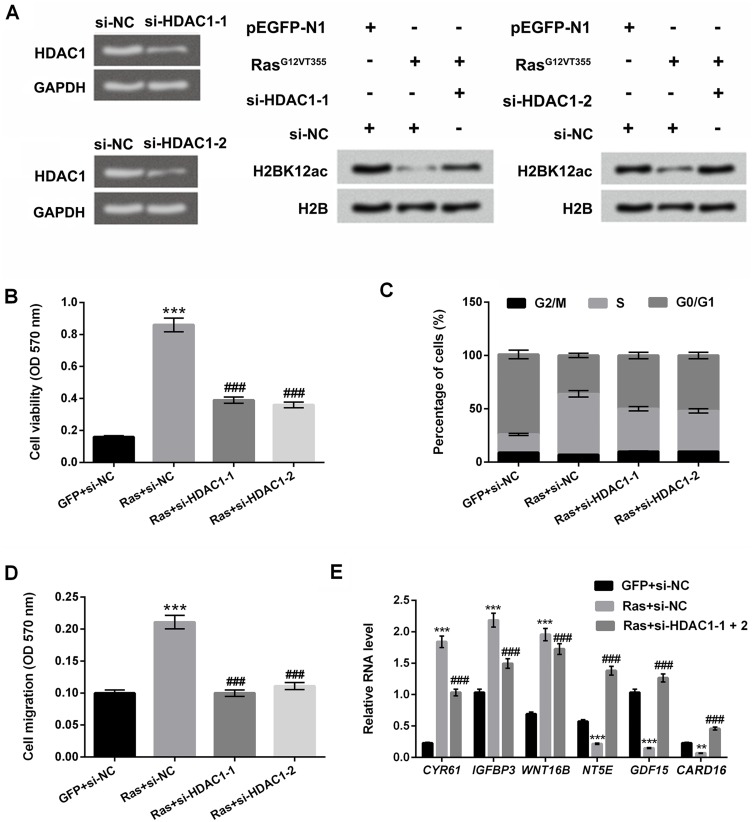
To confirm the relationship between HDAC1 and H2BK12ac, the vectors of si-HDAC1-1 and si-HDAC1-2 were synthesized and transfected into MG-63 cells to knockdown HDAC1 expression. In Figure 4A, RT-qPCR assay results showed that the mRNA expression level of HDAC1 was notably down-regulated in si-HDAC1-1- and si-HDAC1-2-transfected cells. Additionally, we observed that HDAC1 knockdown reversed Ras-ERK activation-induced reduction in H2BK12ac. In the subsequent experiments, we further confirm the effect of HDAC1 knockdown on cell viability, migration, cell cycle. The results showed that HDAC1 knockdown significantly declined the promoting effect of Ras on cell viability, migration and S phase arrest of cell cycle (P < 0.001, Figure 4B–D). Finally, RT-qPCR assay revealed that HDAC1 knockdown significantly reversed the regulatory effect of Ras on the downstream genes of ERK1/2 pathway (CYR61, IGFBP3, WNT16B, NT5E, GDF15 and CARD16) (P < 0.001, Figure 4E). Data indicated that HDAC1 knockdown up-regulated H2BK12ac expression and suppressed the carcinogenic role of Ras-ERK1/2 in MG-63 cells.
Figure 4.
HDAC1 knockdown prevents ERK1/2 activation-induced decrease in H2BK12ac. (A) MG-63 cells were transfected with si-NC, si-HDAC1-1 and si-HDAC1-2. The mRNA expression level of HDAC1 was examined by using RT-qPCR assay. Western blot assay was performed to reveal the HDAC1 knockdown on ERK1/2 activation-induced decrease in the H2BK12ac. MG-63 cells were co-transfected with Ras, si-HDAC1-1, si-HDAC1-2 and the control plasmids (GFP and si-NC), (B) Cell viability, (C) migration and (D) cell cycle were determined by using MTT, Transwell migration, and flow cytometry assays. (E) The relative RNA level of CYR61, IGFBP3, WNT16B, NT5E, GDF15 and CARD16 were analyzed by RT-qPCR. **P < 0.01, ***P < 0.001: Ras+si-NC vs GFP+si-NC; ###P < 0.001: Ras+si-HDAC1-1 or si-HDAC1-2 vs Ras+si-NC.
Abbreviations: HDAC1, histone deacetylases 1; H2BK12ac, histone H2B acetylated on lysine 12; ERK, extracellular signal-regulated kinase; MTT, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide; RT-qPCR, reverse transcription-quantitative PCR.
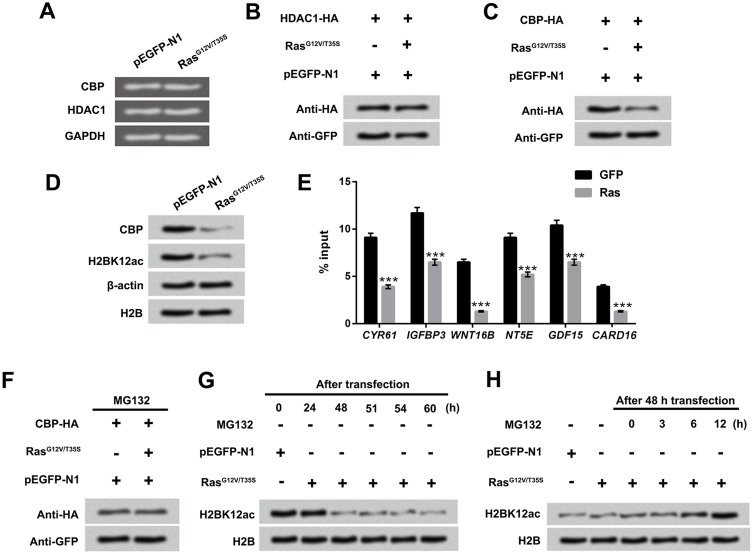
H2BK12ac Was Down-Regulated By Ras-ERK1/2 Activation Through Degradation Of CBP
RT-qRCR and Western blot assay displayed the mRNA and protein levels of CBP and HDAC1, and results showed that the mRNA expression levels of CBP and HDAC1 were not affected by Ras-ERK activation (Figure 5A and B); however, the protein level of CBP was notably down-regulated by Ras-ERK activation (Figure 5C and D). Subsequently, ChIP was performed to investigate whether CBP physically was associated with the downstream genes of ERK1/2 pathway. Results in Figure 5E displayed that the CBP binding in the promoter regions of downstream genes of ERK1/2 (CYR61, IGFBP3, WNT16B, NT5E, GDF15 and CARD16) were reduced by Ras-ERK1/2 activation (P < 0.001). According to the results of Figure 5A–D, we speculate that CBP is likely to be regulated post-transcriptionally. To verify the hypothesis, the MG132 (a proteasome inhibitor) was used to treat these transfected MG-63 cells. Results in Figure 5F showed that the CBP degradation was increased after the activation of the Ras-ERK pathway. To further confirm this result, we performed the time gradient experiments, which showed that H2BK12ac level was gradually recovered after adding the MG132 (Figure 5G and H). These data indicated that Ras-ERK1/2 activation-induced H2BK12ac reduction might be through mediating the degradation of CBP.
Figure 5.
Down-regulated H2BK12ac triggered by Ras-ERK1/2 activation is associated with the degradation of CBP. (A) MG-63 cells were transfected with RasG12V/T35S and pEGFP-N1, the mRNA expression of CBP and HDAC1 was examined by using RT-qPCR assay. (B and C) CBP-HA or HDAC1-HA plasmids were transfected into MG-63 cells. After transfection for 48 hrs, the protein levels of CBP and HDAC1 were determined by Western blot assay. (D) MG-63 cells were transfected with RasG12V/T35S and pEGFP-N1, and the protein levels of CBP and H2BK12ac were also examined by Western blot assay. (E) The binging abilities of H2BK12ac in promoter region of CYR61, IGFBP3, WNT16B, NT5E, GDF15 and CARD16 were detected by ChIP analysis. (F) The CBP-HA, RasG12V/T35S and pEGFP-N1 transfected into MG-63 cells were treated with MG132 (a protease inhibitor), and CBP protein levels were analyzed by using Western blot assay. (G) After transfection for 24, 48, 51, 54 and 60 hrs, the protein level of H2BK12ac were determined by Western blot. (H) After transfection for 48 hrs, the transfected cells were treated with MG132 for 0, 3, 6 and 12 hrs, and the protein levels of H2BK12ac were examined by Western blot assay. ***P < 0.001.
Abbreviations: H2BK12ac, histone H2B acetylated on lysine 12; ERK, extracellular signal-regulated kinase; HDAC1, histone deacetylases 1; CBP, CREB binding protein; RT-qPCR, reverse transcription-quantitative PCR; ChIP, chromatin immunoprecipitation.
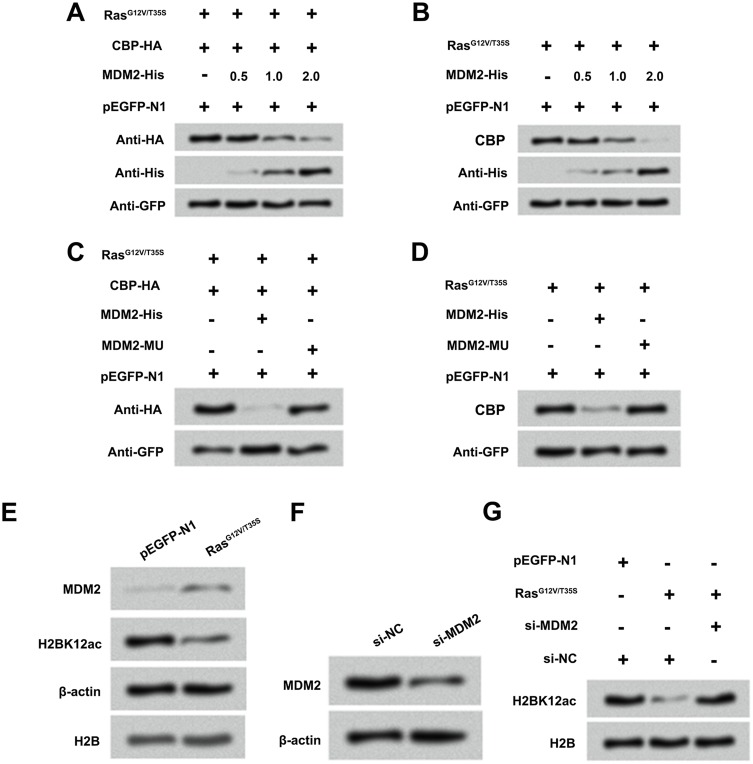
Ras-ERK Activation-Induced Degradation Of CBP Was Regulated By MDM2
MDM2 is known to be linked to the degradation of various histone acetylase molecules.20 Therefore, we want to examine whether MDM2 participates in regulating the degradation process of CBP after activation of ERK1/2 pathway. MG-63 cells were co-transfected with RasG12V/T35S and CBP-HA plasmids following with or without MDM2-His plasmids. Results in Figure 6A and B revealed that RasG12V/T35S and MDM2 significantly reduced the endogenous and exogenous expression of CBP. However, when MG-63 cells were transfected with MDM2-MU plasmids, the protein level of CBP had no significant changes (Figure 6C and D), indicating the CBP degradation required MDM2 participation. In addition, we analyzed whether the CBP degradation caused by Ras-ERK1/2 activation was through up-regulation of MDM2 level. Western blot assay displayed that the protein level of MDM2 was up-regulated after activation of ERK1/2 signaling pathway, assuredly (Figure 6E). Then, si-MDM2 and si-NC plasmids were transfected into MG-63 cells to further demonstrate the relationship between MDM2 and CBP degradation. Results showed that the protein level of MDM2 was notably down-regulated in si-MDM2-transfected cells (Figure 6F). In Figure 6G, results displayed that inhibition of MDM2 notably prevented the decrease in H2BK12ac. Data indicated that Ras-ERK1/2 activation down-regulated H2BK12ac might be via MDM2-mediated CBP degradation.
Figure 6.
Ras-ERK1/2 activation-induced degradation of CBP is mediated by MDM2. (A and B) MG-63 cells were co-transfected with CBP-HA, RasG12V/T35S, pEGFP-N1 plasmids and increasing amounts of MDM2-His plasmid (0.5, 1 and 2 µg), and the protein levels of CBP and MDM2 were assessed by Western blot assay. (C and D) A mutation in MDM2 (MDM2-MU), and the above plasmids were transfected into MG-63 cells, and the protein levels of CBP were analyzed by Western blot assay. (E) Protein levels of MDM2 and H2BK12ac in pEGFP-N1 and RasG12V/T35S transfected cells were examined by Western blot assay. (F) Protein levels of MDM2 in si-NC and si-MDM2 transfected cells were examined by Western blot assay. (G) Protein level of H2BK12ac in pEGFP-N1, RasG12V/T35S, si-NC and si-MDM2 was examined by Western blot assay.
Abbreviations: H2BK12ac, histone H2B acetylated on lysine 12; ERK, extracellular signal-regulated kinase; MDM2, murine double minute 2; CBP, CREB binding protein.
Discussion
OS is the most common type of primary tumor of human bone tissue. The existing therapeutic methods failed to satisfactorily improve the survival rate of patients with OS.21 As everyone knows, histone acetylation/de-acetylation modification is one of the key mechanisms of gene transcription regulation, which affects the occurrence and development of cancers.22,23 H2BK12ac is a pattern of histone H2B acetylated on lysine 12, and whether H2BK12ac joins in the regulation of the development of OS remains uninvestigated. In this study, we explored the effect of H2BK12ac on cell proliferation and migration in MG-63 cells. Interesting results showed that H2BK12ac was specifically down-regulated by Ras-ERK1/2 activation, which was involved in regulation of cell proliferation and migration, as well as affected the transcription of ERK1/2 downstream genes. In addition, HDAC1 silence up-regulated H2BK12ac expression level, and inhibited the promoting effect of Ras-ERK1/2 on MG-63 cells. Besides, Ras-ERK1/2 activation-induced down-regulation of H2BK12ac might be associated with MDM2-mediated CBP degradation.
Histone acetylation and deacetylation affect the interaction between chromatin and DNA, which can cause chromosomal structural changes; this effect has been confirmed in many solid cell tumors, such as leukemia and colon cancer.24,25 H2BK12ac is a crucial histone acetylation pattern of H2B, which has been reported to be a target for autoantibodies in systemic lupus erythematosus (SLE).26 Additionally, it is confirmed that H2BK12ac has a greater impact on prognosis in early non-small-cell lung cancer (NSCLC), which may help to select early NSCLC patients for adjuvant treatment.27 But, the effect of H2BK12ac on OS remains unclear. In our study, we constructed H2BK12Q to mimic H2BK12ac, and the inhibitory effect of H2BK12Q on Ras-ERK activation-induced cell viability, migration and colony formation in MG-63 cells was observed. Further, we found that H2BK12ac was involved in regulating the expression and transcription of ERK1/2-targeted genes (CYR61, IGFBP3, WNT16B, NT5E, GDF15 and CARD16). Collective data indicated that Ras-ERK1/2 signaling promoted the development of OS which was closely associated with H2BK12ac.
HDAC1 is a histone deacetylase, which is involved in regulation of the eukaryotic gene expression, and plays a crucial role in cell proliferation, apoptosis and migration in various cancers.28–30 For example, Burdelski et al found that HDAC1 was associated with rapid tumor cell proliferation and genomic instability in prostate cancer.31 Tang et al demonstrated that HDAC1 could trigger breast cancer cells' proliferation and migration.32 Based on these researches, whether HDAC1 participates in mediating the proliferation and migration of OS cells has aroused our strong interest. In the current study, si-HDAC1-1 and si-HDAC1-2 were transfected into MG-63 cells to knockdown HDAC1 expression. We observed that Ras-ERK1/2 activation-induced H2BK12ac down-regulation was alleviated in si-HDAC1-1 and si-HDAC1-2 transfected MG-63 cells. Moreover, the regulation effect of Ras-ERK1/2 on MG-63 cells' viability, migration, cell cycle and ERK1/2-targeted genes were all reversed by HDAC1 knockdown. These data hinted that HDAC1 knockdown could regulate H2BK12ac expression and suppress the carcinogenic role of Ras-ERK1/2 in OS cells.
CBP is an acetyltransferase, which acts as a co-activator for various transcription factors through directly contacting DNA-bound activators.33 It is now known to play vital roles in DNA repair, cell growth and differentiation.34,35 Emerging evidence has reported that CBP histone acetyltransferase activity could affect cell proliferation and invasion in hepatocellular carcinoma cells (HCC).35 Additionally, several studies found that CBP could mediate H2B acetylation.17,36 Based on these researches, we explored the relationship between CBP and Ras-ERK1/2-indiced H2BK12ac reduction. Results revealed that Ras-ERK1/2-activation decreased H2BK12ac expression through degradation of CBP. MDM2 is a critical negative regulator of the p53 tumor suppressor, which is closely related to tumor metastasis.37 Recent studies have demonstrated that MDM2 could directly interact with histones and degrade several histone acetyl transferases.20,38 Therefore, we speculated that MDM2 might regulate the degradation of CBP, thereby affecting Ras-ERK1/2-induced H2BK12ac down-regulation. As we expect, our experimental results confirmed this conjecture.
Taken together, this study demonstrated that Ras-ERK1/2 signaling could promote OS cells' proliferation and migration by specifically down-regulating H2BK12ac. The process is closely associated with MDM2-mediated CBP degradation. The findings provided a new sight for exploring a novel treatment method for OS. However, the research currently remains lack of the correlative clinical and in vivo validation; moreover, the potential regulatory mechanism is still lacking systematic elaboration. The further explorations will conduct to delve into these issues.
Acknowledgment
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Disclosure
Authors declare that there is no conflict of interest in this work.
References
- 1.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. Cancer. 2009;115(7):1531–1543. doi: 10.1002/cncr.24121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jimenez C, Yang Y, Kim HW, et al. Primary hyperparathyroidism and osteosarcoma: examination of a large cohort identifies three cases of fibroblastic osteosarcoma. J Bone Miner Res. 2005;20(9):1562–1568. doi: 10.1359/JBMR.050507 [DOI] [PubMed] [Google Scholar]
- 3.Ma K, Huang MY, Guo YX, Hu GQ. Matrine-induced autophagy counteracts cell apoptosis via the ERK signaling pathway in osteosarcoma cells. Oncol Lett. 2016;12(3):1854–1860. doi: 10.3892/ol.2016.4848 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Yu Y, Luk F, Yang JL, Walsh WR. Ras/Raf/MEK/ERK pathway is associated with lung metastasis of osteosarcoma in an orthotopic mouse model. Anticancer Res. 2011;31(4):1147–1152. [PubMed] [Google Scholar]
- 5.Yang R, Piperdi S, Gorlick R. Activation of the RAF/mitogen-activated protein/extracellular signal-regulated kinase kinase/extracellular signal-regulated kinase pathway mediates apoptosis induced by chelerythrine in osteosarcoma. Clin Cancer Res. 2008;14(20):6396–6404. doi: 10.1158/1078-0432.CCR-07-5113 [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, Lee JC, Kumar S, Gowen M. ERK pathway mediates the activation of Cdk2 in IGF-1-induced proliferation of human osteosarcoma MG-63 cells. J Bone Miner Res. 1999;14(4):528–535. doi: 10.1359/jbmr.1999.14.4.528 [DOI] [PubMed] [Google Scholar]
- 7.Poudel B, Kim DK, Ki HH, Kwon YB, Lee YM, Kim DK. Downregulation of ERK signaling impairs U2OS osteosarcoma cell migration in collagen matrix by suppressing MMP9 production. Oncol Lett. 2014;7(1):215–218. doi: 10.3892/ol.2013.1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhary SC, Siddiqui MS, Athar M, Alam MS. Geraniol inhibits murine skin tumorigenesis by modulating COX-2 expression, Ras-ERK1/2 signaling pathway and apoptosis. J Appl Toxicol. 2013;33(8):828–837. doi: 10.1002/jat.2739 [DOI] [PubMed] [Google Scholar]
- 9.Buhrman G, Kumar VSS, Cirit M, Haugh JM, Mattos C. Allosteric modulation of Ras-GTP is linked to signal transduction through RAF kinase. J Biol Chem. 2011;286(5):3323. doi: 10.1074/jbc.M110.193854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng ZY, Cheng C, Fu XR, Zhou S, Chang EC. Abstract 5074: identification of Ras compartment-specific interacting proteins. Cancer Res. 2011;70(8 Supplement):5074. [Google Scholar]
- 11.Liu RT, Hou CY, You HL, et al. Selective occurrence of Ras mutations in benign and malignant thyroid follicular neoplasms in Taiwan. Thyroid. 2004;14(8):616–621. doi: 10.1089/1050725041692882 [DOI] [PubMed] [Google Scholar]
- 12.Demunter A, Ahmadian MR, Libbrecht L, et al. A novel N-Ras mutation in malignant melanoma is associated with excellent prognosis. Cancer Res. 2001;61(12):4916–4922. [PubMed] [Google Scholar]
- 13.Neilsen BK, Frodyma DE, Lewis RE, Fisher KW. KSR as a therapeutic target for Ras-dependent cancers. Expert Opin Ther Targets. 2017;21(5):499–509. doi: 10.1080/14728222.2017.1311325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen M, Lin H, Zhao H. Change point analysis of histone modification reveals epigenetic blocks linking to physical domains. Ann Appl Stat. 2016;10(1):506–526. doi: 10.1214/16-AOAS905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445(7124):214–218. doi: 10.1038/nature05458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li D, Shi J, Du Y, et al. Profiling analysis of histone modifications and gene expression in Lewis lung carcinoma murine cells resistant to anti-VEGF treatment. PLoS One. 2016;11(6):e0158214. doi: 10.1371/journal.pone.0158214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Ruiz PD, Novikov L, Casill AD, Park JW, Gamble MJ. MacroH2A1.1 and PARP-1 cooperate to regulate transcription by promoting CBP-mediated H2B acetylation. Nat Struct Mol Biol. 2014;21(11):981–989. doi: 10.1038/nsmb.2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imwong M, Dondorp AM, Nosten F, et al. Exploring the contribution of candidate genes to artemisinin resistance in plasmodium falciparum. Antimicrob Agents Chemother. 2010;54(7):2886–2892. doi: 10.1128/AAC.00032-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Wang D-L, Chen S, Zhao L, Sun F-L. Oncogene Ras/Phosphatidylinositol 3-kinase (PI3K) signaling targets histone H3 acetylation at lysine K56. J Biol Chem. 2012;287(49):41469–41480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu CE, Esfandiari A, Ho YH, et al. Targeting negative regulation of p53 by MDM2 and WIP1 as a therapeutic strategy in cutaneous melanoma. Br J Cancer. 2018;118(4):495–508. doi: 10.1038/bjc.2017.433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Guo W, Shen D, et al. Surgical treatment of primary osteosarcoma of the sacrum: a case series of 26 patients. Spine. 2017;42(16):1207–1213. doi: 10.1097/BRS.0000000000002043 [DOI] [PubMed] [Google Scholar]
- 22.Anderson L, Gomes MR, daSilva LF, et al. Histone deacetylase inhibition modulates histone acetylation at gene promoter regions and affects genome-wide gene transcription in schistosoma mansoni. PLoS Negl Trop Dis. 2017;11(4):e0005539. doi: 10.1371/journal.pntd.0005539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yen CY, Huang HW, Shu CW, et al. DNA methylation, histone acetylation and methylation of epigenetic modifications as a therapeutic approach for cancers. Cancer Lett. 2016;373(2):185–192. doi: 10.1016/j.canlet.2016.01.036 [DOI] [PubMed] [Google Scholar]
- 24.Nouzova M, Holtan N, Oshiro MM, et al. Epigenomic changes during leukemia cell differentiation: analysis of histone acetylation and cytosine methylation using CpG island microarrays. J Pharmacol Exp Ther. 2004;311(3):968–981. doi: 10.1124/jpet.104.072488 [DOI] [PubMed] [Google Scholar]
- 25.Ashktorab H, Belgrave K, Hosseinkhah F, et al. Global histone H4 acetylation and HDAC2 expression in colon adenoma and carcinoma. Dig Dis Sci. 2009;54(10):2109–2117. doi: 10.1007/s10620-008-0601-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Bavel CC, Dieker J, Muller S, et al. Apoptosis-associated acetylation on histone H2B is an epitope for lupus autoantibodies. Mol Immunol. 2009;47(2–3):511–516. doi: 10.1016/j.molimm.2009.08.009 [DOI] [PubMed] [Google Scholar]
- 27.Barlesi F, Giaccone G, Gallegos-Ruiz MI, et al. Global histone modifications predict prognosis of resected non small-cell lung cancer. J Clin Oncol. 2007;25(28):4358–4364. doi: 10.1200/JCO.2007.11.2599 [DOI] [PubMed] [Google Scholar]
- 28.Cai RL, Yan-Neale Y, Cueto MA, Xu H, Cohen D. HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J Biol Chem. 2000;275(36):27909–27916. doi: 10.1074/jbc.M000168200 [DOI] [PubMed] [Google Scholar]
- 29.Hayashi A, Horiuchi A, Kikuchi N, et al. Type-specific roles of histone deacetylase (HDAC) overexpression in ovarian carcinoma: HDAC1 enhances cell proliferation and HDAC3 stimulates cell migration with downregulation of E-cadherin. Int J Cancer. 2010;127(6):1332–1346. doi: 10.1002/ijc.25151 [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Bu L, Hu J, et al. HDAC1 knockdown inhibits invasion and induces apoptosis in non-small cell lung cancer cells. Biol Chem. 2018;399(6):603–610. doi: 10.1515/hsz-2017-0306 [DOI] [PubMed] [Google Scholar]
- 31.Burdelski C, Ruge OM, Melling N, et al. HDAC1 overexpression independently predicts biochemical recurrence and is associated with rapid tumor cell proliferation and genomic instability in prostate cancer. Exp Mol Pathol. 2015;98(3):419–426. doi: 10.1016/j.yexmp.2015.03.024 [DOI] [PubMed] [Google Scholar]
- 32.Tang Z, Ding S, Huang H, et al. HDAC1 triggers the proliferation and migration of breast cancer cells via upregulation of interleukin-8. Biol Chem. 2017;398(12):1347–1356. doi: 10.1515/hsz-2017-0155 [DOI] [PubMed] [Google Scholar]
- 33.Kar S. p300/CBP proteins: a HAT trick to determine cell fate. Prajnan O Sadhona. 2015;143. [Google Scholar]
- 34.Philip P, Boija A, Vaid R, et al. CBP binding outside of promoters and enhancers in drosophila melanogaster. Epigenetics Chromatin. 2015;8:48. doi: 10.1186/s13072-015-0042-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inagaki Y, Shiraki K, Sugimoto K, et al. Epigenetic regulation of proliferation and invasion in hepatocellular carcinoma cells by CBP/p300 histone acetyltransferase activity. Int J Oncol. 2016;48(2):533–540. doi: 10.3892/ijo.2015.3288 [DOI] [PubMed] [Google Scholar]
- 36.Abell AN, Jordan NV, Huang W, et al. MAP3K4/CBP-regulated H2B acetylation controls epithelial-mesenchymal transition in trophoblast stem cells. Cell Stem Cell. 2011;8(5):525–537. doi: 10.1016/j.stem.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouska A, Eischen CM. Mdm2 affects genome stability independent of p53. Cancer Res. 2009;69(5):1697–1701. doi: 10.1158/0008-5472.CAN-08-3732 [DOI] [PubMed] [Google Scholar]
- 38.Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16(4):631–639. doi: 10.1016/j.molcel.2004.10.016 [DOI] [PubMed] [Google Scholar]