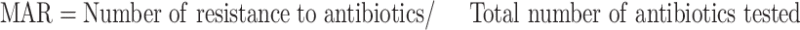Abstract
Purpose
Contamination with Salmonella on food products and poultry in particular has been linked to foodborne infections and/or death in humans. This study investigated the occurrence, genetic diversities and antibiotic resistance profiles of Salmonella strains isolated from chickens.
Patients and methods
Twenty each duplicate faecal swab samples were collected from five different poultry pens of broilers, layers and indigenous chickens in the North-West Province, South Africa. Isolates identities were confirmed through amplification and sequence analysis of 16S rRNA and the invA gene fragments after which phylogenetic tree was constructed. Salmonella enteritidis (ATCC:13076TM), Salmonella Typhimurium (ATCC:14028TM) and E. coli (ATCC:259622TM) were used as positive and negative controls, respectively. The serotypes of Salmonella isolates were determined. Antibiotic-resistant profiles of the isolates against eleven antimicrobial agents were determined.
Results
Eighty-four (84%) of representative isolates possessed the invA genes. The percent occurrence and diversity of Salmonella subspecies in chickens were 1.81–30.9% and was highest in Salmonella enterica subsp. enterica. Notably, the following serotypes Salmonella bongori (10.09%), Salmonella Pullorum (1.81%), Salmonella Typhimurium (12.72%), Salmonella Weltevreden, Salmonella Chingola, Salmonella Houten and Salmonella Bareily (1.81%). Isolates (96.6%) displayed multidrug resistance profiles and the identification of isolates with more than nine antibiotic resistance was a cause for concern.
Conclusion
This study indicates that isolates had pre-exposure histories to the antibiotics tested and may pose severe threats to food security and public health.
Keywords: Salmonella, diversity, antimicrobial resistance, phylogenetic, chickens
Introduction
Salmonella spp. are enteric pathogens that have received a lot of attention due to their ability to cause food-borne diseases and high rates of mortality amongst humans and thus were declared as agents of public health significance.1,2 Salmonellosis is the most common food-borne disease caused by Salmonella species in humans with symptoms ranging from headache, vomiting, fatigue, nausea, bloody diarrhea, gastroenteritis, and abdominal cramps and self-limiting for which often no antimicrobials are prescribed for its control.3–5 Hence, the pathogen is capable of causing socio-economic and public health implications to humans. Salmonella enterica serotype Typhimurium (S. Typhimurium) and Salmonella enterica serotype Enteritidis (S. Enteritidis) are considered of high health importance due to their ability to cause salmonellosis in humans and veterinary animals in both developed and developing countries of the world.
Salmonella has been highlighted as economically important zoonotic pathogens by the World Health Organisation (WHO) and the Food Agriculture Organisation (FAO) dated back to 1950s.1 Salmonella spp. have been enteric pathogens co-existing with pathogens such as Escherichia coli, Klebsiella spp., and Proteus spp.6 According to Kagambèga et al,5 ruminants such as cattle and sheep, non-ruminants - pigs, dogs, rodents, poultry, birds, and cold-blooded animals such as fish and lizards, and humans have been implicated as reservoirs of the typhoidal and the non-typhoidal Salmonella species. However, Poultry and its products are the major sources of Salmonella-borne infection in the food chain. The ability of Salmonella to be transmitted from reservoirs to other animals and humans calls for concern. Thus, making its survey and control among suspected reservoirs such as chickens is necessary. The influx of many antibiotic resistance strains within the environment calls for a concern. Antibiotic resistance is currently a global problem that poses a threat to public health. Therefore, a study to investigate the occurrence and antibiotic resistance profiles of Salmonella among chicken whose carcasses forms a major of the South African cuisines germane.
Materials And Methods
Sample Collection
This study was conducted within farms located at Ngaka Molema Modiri District of Mafikeng, North West Province, South Africa. The study site's (Mafikeng) geographical coordinates are 25° 52ʹ 0” South, 25° 39ʹ 0” East. Twenty samples each were collected in duplicates from five different poultry pens, housing broiler, layer and indigenous chickens in the study area. The broilers and layers belonged to the White Leghorn breed while the indigenous belonged to Potchefstroom koekoek breed (Gallus gallus domesticus). Swabs from the gut were aseptically collected in duplicates from test animals and transported on ice to the laboratory for analysis within 24 hrs of collection. Ethical clearance for the study was obtained from the Mafikeng Animal Research Ethics Committee of the North West University prior to the commencement of sampling. Samples were also collected under the supervision of trained Veterinarians and Animal Health Technicians from the Centre for Animal Health Studies, North West University, South Africa.
Isolation Of Microbial Isolates
Isolation of Salmonella spp. from chickens was done using ISO-6579:2002 procedure.8 Sample pre-enrichment and enrichment were achieved in buffered peptone water and tetrathionate broth, respectively, prior to enrichment in Rappaport vassiliadis Soy (RVS) broth and incubated at 42 °C for 24 hrs. About 1 mL of the inoculated RVS broth was plated on sterile Salmonella - Shigella Agar (SSA) and was incubated aerobically at 37 °C for 18 hrs. Colonies having creamy with or without black centre on SSA were regarded as presumptive Salmonella isolates and were further studied. Sub-culturing was done until pure colonies were obtained.
Morphological And Biochemical Characterization Of Isolates
The morphological and biochemical tests (Gram staining, catalase, Simmons citrate test, urease and Triple sugar iron (TSI) agar) were determined as described by Ateba and Mochaiwa.9 Gram-negative rods and catalase-positive colonies were kept on double-strength slants and kept under −20°C for further use.
Molecular Characterisation Of Isolates
The amplification of 16S rRNA region of the bacteria was employed for the discrimination of presumptive Salmonella isolates. The DNA was extracted using a Fungal/Bacterial DNA extraction kit (Zymo Research Corporation, Southern California, USA) following the manufacturer's specification. The pure eluted DNA was stored at −80 °C for further analysis. The pure DNA was quantified using a Nanodrop Lite spectrophotometer (Model 1558) obtained from Thermo Scientific, USA, and the genomic DNA was quantified on a 1% agarose gel. The presence of fluorescence band when viewed under the UV Transilluminator (Biorad Gel DocTM XR+) confirmed the presence of DNA of presumptive Salmonella isolates.
PCR Amplification Of 16S rRNA
The 16S ribosomal RNA (16S rRNA) PCR was employed in the identification of Salmonella isolates.10 The amplification was done using a Biorad C1000 TouchTM Thermal Cycler. The 27F (51-AGAGTTTGATCCTGGCTCAG-31) and 1492R (51- GGTTACCTTGTTACGACTT – 31) primers used were synthesized at Inqaba Biotechnical Industries (Pty) Ltd, South Africa, having an expected amplicon size of 1450 bp. For the polymerase chain reaction (PCR), a 25 µL reaction mix composed of 12 µL of master mix (Thermo Scientific PCR Master Mix 2X), oligonucleotides (1 µL), DNA template (4 µL) and nuclease-free water (7 µL) was used.10 The negative controls used include Aspergillus flavus and water as a template in the PCR assays while the positive was Salmonella Typhimurium ATCC 14028TM.
Amplification Of The invA Gene In Presumptive Salmonella Isolates
The invA gene fragment was amplified using the set of primers invA F (51–GTGAAATTATCGCCACGTTCGGGCAA-31) and invA R 51-TCATCGCACCGTCAAAGGAACC −31) with expected amplicon size of 284 bp. Slight modifications in the annealing temperature previously reported by Ateba and Mochaiwa9 were used: initial denaturation (95 °C for 2 mins), denaturation (95 °C for 15 s), annealing (47.8 °C for 1 min), elongation (72 °C for 45 s) and final elongation (72 °C for 7 mins). A 25 µL reaction mix was used in the amplification, and this is composed of 12.5 µL of master mix (Thermo Scientific PCR Master Mix 2X), oligonucleotides (1 µL), DNA template (4 µL) and nuclease-free water (6.5 µL). A positive control (Salmonella Typhimurium ATCC 14028) and a negative control (Escherichia coli ATCC 25922 and non-template water) were used.
Gel Electrophoresis Of Amplicons
The molecular weight of PCR amplicons was determined by gel electrophoresis.11 A DNA marker (Fermentas Life Science, Lithuania) of 1 kb was used and the gel was allowed to run at 60 volts, 400 amperes for 60 mins in 1% tris acetate ethylenediamineacetate (TAE) buffer before photographing under the UV transilluminator light (Biorad Gel DocTM XR+).
Gene Sequencing And Identification Of Isolates
The amplified product was sequenced using an automated DNA sequencer (SpectruMedix model SCE 2410) at Inqaba Biotechnical Industries (Pty) Ltd Pretoria. Resulting sequences were cleaned using the FinchTV software version 1.4.0 (Geospiza Inc.) and blasted against on the National Centre for Biotechnology Information12 database using the Nucleotide Basic Local Alignment Search Tool (BLAST) program (http://www.ncbi.nlm.nih.gov/BLAST). Isolates were identified based on the highest percentage of similarity and sequences were deposited in NCBI gene bank and accession numbers were obtained. The serotypes of presumptive Salmonella isolates were determined using the Salmonella antisera agglutination kits. Isolates were then classified into serotypes as described in the Kauffman–White Salmonella classification.
Phylogenetic Tree Construction
Cleaned sequences were aligned by CLUSTALW sequence alignment tool and de-gapped using Bio-Edit software package.13,14 To identify putative close phylogenetic relatives, multiple sequence alignments were obtained using Clustal-W against corresponding nucleotide sequences retrieved from the Gene bank. The evolutionary distance matrices were generated.15 Phylogenetic analysis was done using the neighbour joining method16 in MEGA program version 5.10.17 The bootstrap analysis was done using 1000 replications for neighbour joining. The sequences were checked for putative chimeric artefacts using the Chimera-Buster program and then manipulation and tree editing was done using the Tree View option.18 Salmonella enterica was used as the root to the tree.
Determination Of Antibiotics Resistance Profile Of Salmonella Isolates
Antibiotic sensitivity of Salmonella isolates was investigated against eleven antibiotics belonging to eight different classes using the disc diffusion method.19 Antibiotics used include; ampicillin (10 µg), oxy-tetracycline (30 µg), ciprofloxacin (5 µg), streptomycin (10 µg), gentamicin (10 µg), sulphamethoxazole/trimethoprim (300 µg), chloramphenicol (30 µg), erythromycin (15 µg), norfloxacin (10 µg), cephalothin (30 µg), and nalidixic acid (30 µg). Antibiotics discs were placed at an equilateral distance to each other on Muller–Hinton agar (MHA) plates and were incubated at 37 °C for 18 hrs. After incubation, zones of inhibition around the antibiotics disc were measured using a meter rule graduated in millimetres. The test was made in triplicate and the mean diameter of the inhibitory zones (IZD) were calculated. The mean IZD was determined as either susceptible, intermediate, or resistant using the Clinical and Laboratory Standards Institute20 criteria. The multiple antibiotics resistance (MAR) phenotypes were recorded for isolates showing resistance to more than two antibiotics21 and the MAR index was calculated as shown in Equation 1.22
Clustering Of Antibiotic-Resistant Patterns Of Salmonella Isolates
To determine the similarities and differences between Salmonella isolates from different sources based on their antibiotic resistance patterns, cluster analysis was done. The IZDs of Salmonella strains were clustered using a cluster analysis on the Statistica software package (Statsoft, USA) and a dendrogram was generated. Ward’s method and the Euclidean distance method were used to generate the clusters.
Statistical Analysis
The statistical analysis of data generated was evaluated using Statistical package for Social Sciences (SPSS, version 21.0 IBM Corp., USA). The frequency and percentage of occurrence of isolates and correlations between isolates antibiotics resistance and sources were determined using Pearson’s product. The cluster analysis of antibiotics sensitivity patterns of Salmonella isolates was evaluated through the Ward’s algorithm and Euclidean distances on the Statistica software version 7.0 (Statsoft, USA). Significance and goodness of fit were evaluated at 95% confidence interval while sequence algorithms were cleaned and processed using FinchTv, Bioedit and the phylogenetic tree was constructed using the MEGA6 Software’s.
Results
The morphological and biochemical characteristics of presumptive Salmonella isolates from chickens in Mafikeng community, South Africa, is as presented in Supplementary material S1. Colonies pigment morphology ranged from pink to colourless with/without black centre, on Salmonella Shigella Agar. As shown in Supplementary material A1, 96 percent of colonies had a circular shape while the opacity ranged from 83.63% (opaque) and translucent (16.36%). All selected isolates were gram-negative rods having the ability to hydrolyze hydrogen peroxide in the production of catalase enzyme. Isolates showed alkalinity by a red colour pigment on slants, yellow butt with or without gas, thus signifying acid production while black pigment in butt showed hydrogen sulphide production which is typical of Salmonella. About 81.81% of the isolates were positive to alkalinity, 16.36% were negative while 3.63% had weak alkalinity reaction while 96.36% were able to produce acid. A number of 96.36% of the presumptive isolates had the ability to utilize citrate as a sole source of carbon and energy, while 98.18% tested negative to urease and indole production.
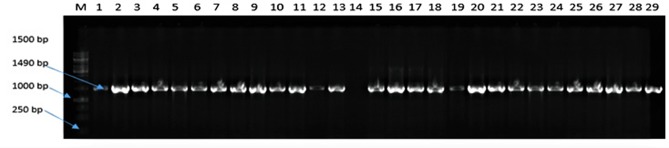
Figure 1 presents the gel picture of 16 S rRNA amplification of representative presumptive Salmonella isolates from chickens. The 16 S rRNA amplification was performed twice to ensure reliability of obtained results. There was a 100% positive amplification at an expected band size of 1450 bp. There was a positive amplification of Salmonella Typhimurium ATCC 14028 in lane 1 while no amplification was observed in lane 14 (negative control). The positive amplification confirms the use of 27F and 1497R sets of oligonucleotides for 16 S rRNA region amplification in enteric bacteria.
Figure 1.
Agarose gel (1%) electrophoresis showing the amplification of 16 S rDNA of representative presumptive bacteria isolates colonising the gut of chickens obtained from North-west province, South Africa.
Notes: Lane 1 to 29 shows the amplification of 16 S region of presumptive Salmonella DNA isolates while lanes 14 = no template (negative control), lane 13 = Salmonella Typhimurium ATCC 14028 (positive control).
Abbreviations: M, DNA marker (1kb); bp, base pairs.
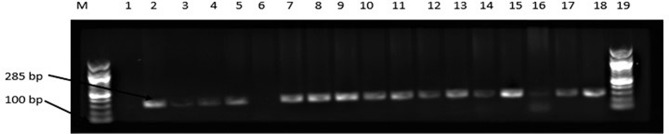
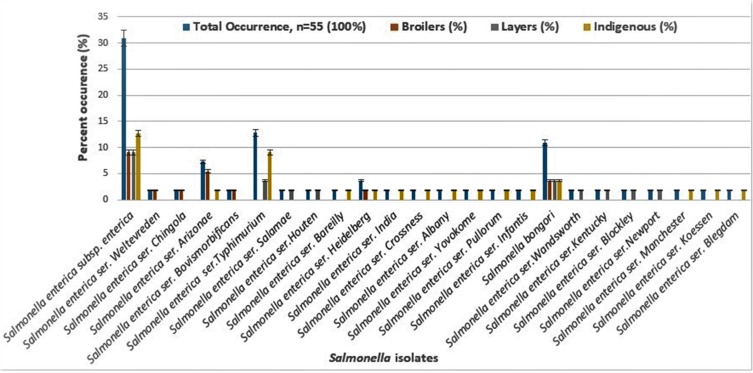
Salmonella-specific PCR was conducted using the invA genes. As shown in Figure 2, about 87.27% of the representative isolates showed positive amplification while about 12.72% were negative. As shown in Table 1, the percent similarity of isolates to data in the NCBI gene bank ranged from 85% to 99%. In percent, a portion of 26% had 99% similarity while 85% had 94% similarity to Salmonella. The percent occurrence of Salmonella in chicken is presented in Figure 3. The percent occurrence based on subspecies ranged from 2% to 61% (Supplementary material S2). The highest occurring subspecies belongs to the Salmonella enterica subsp. enterica (61%) while the least occurring serotype was Salmonella Salamae, Salmonella Weltevreden, Salmonella Chingola, Salmonella Houten, Salmonella Bareilly (2%). Based on source, Salmonella Typhimurium was highest in indigenous chickens (9.08%) followed by layers (3.63%), while in broilers, Salmonella Arizona was highest as described in Figure 3. Salmonella Salamae was not isolated in the indigenous and broiler chickens from the study site. Likewise, in layers, Salmonella Arizona and Salmonella Weltevreden were not isolated except in indigenous breeds and in broiler chickens. Autoagglutination was obtained in some Salmonella bongori and Salmonella enterica subspecies enterica isolates, hence the inability to determine the serotypes of these isolates. Non-specific agglutination as a product of loss of antigen expression could give pseudo-positive results as earlier reported by.23,24
Figure 2.
Agarose gel (1%) electrophoresis showing the amplification of invA gene in representative presumptive Salmonella isolates in chickens obtained from Mafikeng, North-west province, South Africa.
Notes: Lane 1 = no template (negative control), Lane 2 = Salmonella Typhimurium ATCC 14028TM (positive control); Lane 6 = Escherichia coli ATCC 259622TM (negative control), lane 2 to 18 = positive amplification of invA gene at 286 bp, lane 6 = no amplification.
Abbreviations: M, DNA marker (100 bp); bp, base pairs.
Table 1.
Identity Of Presumptive Salmonella Isolates From Chickens In Mafikeng, South Africa
| Isolate Number | Sample Source | Sequence | Assession Number | InvA | Serotype | Similarity (%) | Name of Organism |
|---|---|---|---|---|---|---|---|
| 1 | Broiler | Seq1 | MG663456 | +VE | AAG | 98 | Salmonella enterica subsp. enterica |
| 2 | Broiler | Seq2 | MG663457 | +VE | AAG | 92 | Salmonella enterica subsp. enterica |
| 3 | Broiler | Seq3 | MG663458 | +VE | AAG | 97 | Salmonella enterica subsp. enterica |
| 4 | Broiler | Seq4 | MG663459 | +VE | AG | 92 | Salmonella enterica ser. Weltevreden |
| 5 | Broiler | Seq5 | MG663460 | +VE | AG | 92 | Salmonella enterica ser. Chingola |
| 6 | Broiler | Seq6 | MG663461 | +VE | AG | 92 | Salmonella enterica ser. Arizonae |
| 7 | Broiler | Seq7 | MG663462 | +VE | AG | 98 | Salmonella enterica ser. Bovismorbificans |
| 8 | Layer | Seq8 | MG663463 | +VE | AAG | 99 | Salmonella enterica subsp. enterica |
| 9 | Layer | Seq9 | MG663464 | +VE | AAG | 92 | Salmonella enterica subsp. enterica |
| 10 | Layer | Seq10 | MG663465 | +VE | AG | 97 | Salmonella enterica ser. Typhimurium |
| 11 | Layer | Seq11 | MG663466 | +VE | AG | 92 | Salmonella enterica ser. Salamae |
| 12 | Layer | Seq12 | MG663467 | +VE | AG | 99 | Salmonella enterica ser. Houten |
| 13 | Layer | Seq13 | MG663468 | +VE | AAG | 99 | Salmonella enterica subsp. enterica |
| 14 | Indigenous | Seq14 | MG663469 | +VE | AAG | 98 | Salmonella enterica ser. Bareilly |
| 15 | Indigenous | Seq15 | MG663470 | +VE | AAG | 99 | Salmonella enterica subsp. enterica |
| 16 | Indigenous | Seq16 | MG663471 | +VE | AAG | 98 | Salmonella enterica subsp. enterica |
| 17 | Indigenous | Seq17 | MG663472 | +VE | AAG | 92 | Salmonella enterica subsp. enterica |
| 18 | Indigenous | Seq18 | MG663473 | +VE | AG | 92 | Salmonella enterica ser. Heidelberg |
| 19 | Indigenous | Seq19 | MG663474 | +VE | AG | 92 | Salmonella enterica ser. Arizonae |
| 20 | Indigenous | Seq20 | MG663475 | +VE | AAG | 99 | Salmonella enterica subsp. enterica |
| 21 | Indigenous | Seq21 | MG663476 | +VE | AG | 99 | Salmonella enterica ser. India |
| 22 | Indigenous | Seq22 | MG663477 | +VE | AG | 97 | Salmonella enterica ser. Crossness |
| 23 | Indigenous | Seq23 | MG663478 | +VE | AG | 97 | Salmonella enterica ser. Albany |
| 24 | Indigenous | Seq24 | MG663479 | +VE | AG | 99 | Salmonella enterica ser. Yovokome |
| 25 | Indigenous | Seq25 | MG663480 | +VE | AG | 98 | Salmonella enterica ser. Pullorum |
| 26 | Indigenous | Seq26 | MG663481 | +VE | AG | 98 | Salmonella enterica ser. Infantis |
| 27 | Broiler | Seq27 | MG663482 | +VE | AG | 92 | Salmonella enterica ser. Arizonae |
| 28 | Broiler | Seq28 | MG663483 | +VE | AG | 99 | Salmonella enterica ser. Heidelberg |
| 29 | Broiler | Seq29 | MG663484 | +VE | AAG | 92 | Salmonella enterica subsp. enterica |
| 30 | Broiler | Seq30 | MG663485 | +VE | AAG | 92 | Salmonella enterica subsp. enterica |
| 31 | Broiler | Seq31 | MG663486 | −VE | AAG | 92 | Salmonella bongori |
| 32 | Broiler | Seq32 | MG663487 | −VE | AAG | 92 | Salmonella bongori |
| 33 | Broiler | Seq33 | MG663488 | +VE | AG | 92 | Salmonella enterica ser. Arizonae |
| 34 | Layer | Seq34 | MG663489 | +VE | AAG | 92 | Salmonella enterica subsp. enterica |
| 35 | Layer | Seq35 | MG663490 | +VE | AG | 92 | Salmonella enterica ser. Wandsworth |
| 36 | Layer | Seq36 | MG663491 | +VE | AAG | 92 | Salmonella enterica subsp. enterica |
| 37 | Layer | Seq37 | MG663492 | −VE | AAG | 92 | Salmonella bongori |
| 38 | Layer | Seq38 | MG663493 | +VE | AG | 92 | Salmonella enterica ser. Kentucky |
| 39 | Layer | Seq39 | MG663494 | −VE | AAG | 92 | Salmonella bongori |
| 40 | Layer | Seq40 | MG663495 | +VE | AG | 94 | Salmonella enterica ser. Blockley |
| 41 | Layer | Seq41 | MG663496 | +VE | AG | 98 | Salmonella enterica ser. Newport |
| 42 | Layer | Seq42 | MG663497 | +VE | AG | 98 | Salmonella enterica ser. Typhimurium |
| 43 | Indigenous | Seq43 | MG663498 | −VE | AAG | 98 | Salmonella bongori |
| 44 | Indigenous | Seq44 | MG663499 | +VE | AAG | 99 | Salmonella enterica ser. Manchester |
| 46 | Indigenous | Seq46 | MG663500 | +VE | AAG | 98 | Salmonella enterica subsp. enterica |
| 47 | Indigenous | Seq47 | MG663501 | +VE | AG | 99 | Salmonella enterica ser. Typhimurium |
| 48 | Indigenous | Seq48 | MG663502 | +VE | AAG | 92 | Salmonella enterica subsp. enterica |
| 49 | Indigenous | Seq49 | MG663503 | +VE | AG | 99 | Salmonella enterica ser. Typhimurium |
| 50 | Indigenous | Seq50 | MG663504 | +VE | AG | 85 | Salmonella enterica ser. Typhimurium |
| 51 | Indigenous | Seq51 | MG663505 | +VE | AG | 97 | Salmonella enterica ser. Typhimurium |
| 52 | Indigenous | Seq52 | MG663506 | +VE | AAG | 99 | Salmonella enterica ser. Koessen |
| 53 | Indigenous | Seq53 | MG663507 | +VE | AAG | 98 | Salmonella bongori |
| 54 | Indigenous | Seq54 | MG663508 | −VE | AG | 99 | Salmonella enterica ser. Blegdam |
| 55 | Indigenous | Seq55 | MG663509 | +VE | AAG | 92 | Salmonella enterica subsp. enterica |
| Control 1 | Control 1 | ATCC 14028TM | +VE | AG | Salmonella enterica ser. Typhimurium | ||
| Control 2 | Control 2 | MG663511 | +VE | 99 | Escherichia coli O157:H7 |
Notes: Lane 1 = control 1 = Salmonella Typhimurium (positive control); control 2 = Escherichia coli (negative control) was an environmental strain.
Abbreviations: +VE, positive amplification; −VE, negative amplification; AAG, auto-agglutination against antisera; AG, positive agglutination.
Figure 3.
Percentage occurrence of Salmonella isolates based on its source.
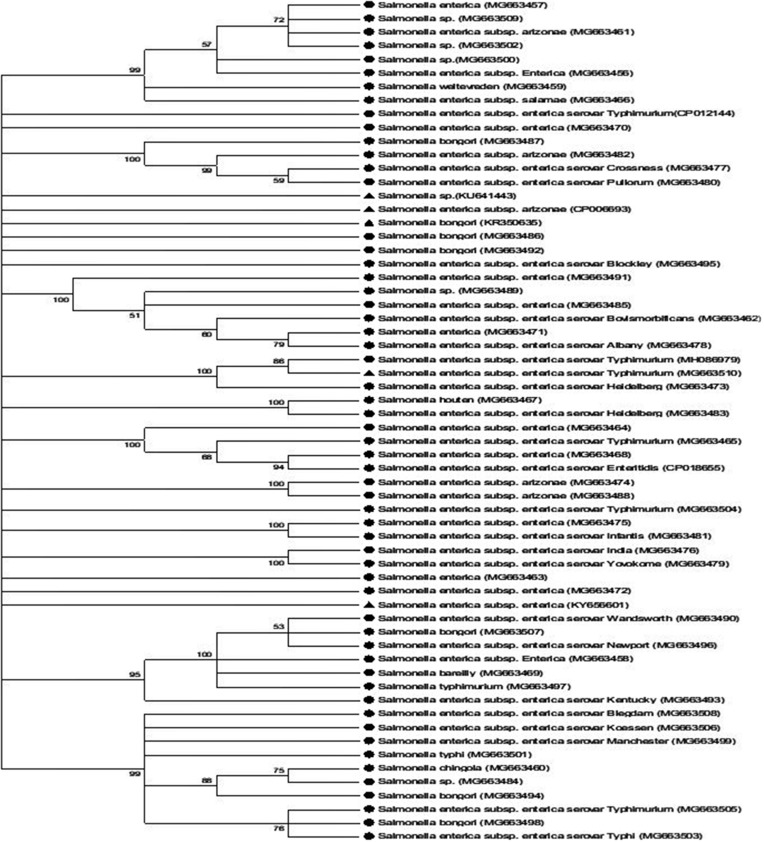
Based on the cluster algorithm of the Neighbour Joining method used, the percent evolutionary relatedness of Salmonella isolates is presented in Figure 4. The evolutionary distance of Salmonella isolates was 35.13655429. Most of the Salmonella isolates were found to evolve from the same ancestral origin with similarities higher than 70% and comparable to strains sourced from the gene bank. Salmonella enterica subsp. enterica (MG663457, MG663509, MG663461 and MG663502) were found to evolve from the same ancestor which we presumed to be Salmonella spp. However, a genetic evolution was observed in the isolates with a 72% homology compared to MG663500, MG663459, and MG663456 having 99% homology to the genetic sequences of the parent’s genome.
Figure 4.
Neighbor joining method of phylogenetic tree based on partial 16S rDNA gene sequence, showing the phylogenetic relationships between Salmonella species and the most closely related strains from the genebank.
Notes: Numbers at the nodes indicate the ranks of bootstrap based on 1000 resampled data sets, and the cut-off points were placed at 70% for condensed tree. The scale bar indicates 0.5 base substitution per site. Salmonella enterica were set as the out-group. Sequences obtained in this study are denoted with a circle shape.
Also, Salmonella bongori (MG663487) exhibited a 100% concatenated homology with other Salmonella isolates. All the comparable sequences from gene bank, Salmonella spp. (KU641443), Salmonella Arizonae (CP006693) and Salmonella bongori (KR350635), showed relatedness and were comparable to sequences identified as Salmonella bongori (MG663486), MG663492 and Salmonella Blockley (MGG3495). Isolates Salmonella enterica subsp. enterica (MG663485, MG663489, MG663485 and MG663462) had 100% evolutionary relation to the parental genus. Salmonella Houten (MG663464) and Salmonella Heidelberg (MG663483) clustered together on the same cladograph. Salmonella enterica subsp. enterica (MG663464) had 100% homology to an out-group (Salmonella enterica subsp. enterica KY656601). However, Salmonella enterica subsp. enterica (MG663468) was similar to Salmonella Enteritidis (CP018655) and had a 94% homology to Salmonella Typhimurium (MG663465). Salmonella Typhimurium (MG663510, MH086979 and MG663473) had the same node showing that they both evolved from the same ancestor.
Table 2 presents the antibiotic sensitivity profile of Salmonella isolates from chickens in Mafikeng. About 56% of the total Salmonella isolates were resistant to ampicillin treatment, 18% had intermediate resistance while 26% were susceptible (Supplementary material S3). The ampicillin-resistant strains were found more in the indigenous chickens (81%) followed by broilers (36%) and lowest in the layers (27%). About 69% of all Salmonella isolates were resistant to oxy-tetracycline with 9% being intermediate-resistant. Higher occurrence of oxy-tetracycline resistance was obtained in indigenous chickens (65%). More than 30% of the isolates were resistant to ciprofloxacin while about 20% had intermediate resistance with a distribution largest (46%) in the indigenous chickens. Ninety-five percent resistance to streptomycin was obtained in this study and was dominant in the layers. The percent resistance to trimethoprim/sulphamethoxazole ranged from 64% to 84% in the different samples investigated. When exposed to chloramphenicol, only a small proportion (8–20%) of isolates were resistant to this drug. Against erythromycin, an 100% resistance was observed and was found not to depend on sample source. Hence, the use of erythromycin in the treatment of Salmonella-borne infection should be avoided.
Table 2.
Prevalence Of Antibiotic Resistance And Multi-Drug Resistance Of Salmonella Isolates From Chicken Sourced From Mafikeng, South Africa
| Antibiotics | Resistance | Intermediate Resistance | ||||||
|---|---|---|---|---|---|---|---|---|
| Total n= 55 (%) |
Broilers n =14 (%) |
Layers n =15 (%) |
Indigenousn = 26 (%) | Total n = 55 (%) |
Broilers n = 14 (%) |
Layers n = 15 (%) |
Indigenousn = 26 (%) | |
| AMP | 31 (56) | 5 (36) | 4 (27) | 21 (81) | 10 (18) | 4 (29) | 5 (33) | 1 (4) |
| OXT | 38 (69) | 12 (86) | 9 (60) | 17 (65) | 9 (16) | 1 (7) | 4 (27) | 4 (15) |
| CIP | 17 (31) | 3 (21) | 5 (33) | 10 (39) | 11 (20) | 5 (36) | 3 (20) | 2 (8) |
| STR | 52 (95) | 13 (93) | 15 (100) | 25 (96) | 2 (4) | 1 (7) | 0 (0) | 1 (4) |
| GCN | 2 (4) | 0 (0) | 1 (7) | 1 (4) | 1 (2) | 1 (7) | 0 (0) | 0 (0) |
| SXT | 43 (78) | 9 (64) | 12 (80) | 22 (85) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| C30 | 7 (13) | 2 (14) | 3 (20) | 2 (8) | 8 (15) | 4 (29) | 2 (13) | 2 (8) |
| ERY | 55 (100) | 14 (100) | 15 (100) | 26 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| NOR | 27 (49) | 7 (50) | 6 (40) | 14 (54) | 5 (9) | 1 (7) | 0 (0) | 4 (15) |
| KF | 19 (36) | 8 (57) | 3 (20) | 8 (31) | 15 (27) | 0 (0) | 7 (47) | 8 (31) |
| NAL | 26 (47) | 8 (57) | 9 (60) | 9 (35) | 5 (9) | 0 (0) | 1 (7) | 4 (15) |
| Number of MAR phenotypes | ||||||||
| 0 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||||
| 1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||||
| 2 | 1 (2) | 1 (7) | 0 (0) | 0 (0) | ||||
| 3 | 4 (7) | 0 (0) | 0 (0) | 4 (15) | ||||
| 4 | 13 (24) | 4 (29) | 4 (27) | 5 (19) | ||||
| 5 | 13 (24) | 3 (21) | 1 (4) | 9 (35) | ||||
| 6 | 4 (7) | 2 (14) | 0 (0) | 2 (8) | ||||
| 7 | 8 (15) | 3 (21) | 0 (0) | 5 (19) | ||||
| 8 | 9 (17) | 1 (7) | 0 (0) | 8 (31) | ||||
| 9 | 3 (6) | 1 (7) | 0 (0) | 2 (8) | ||||
Notes: Ampicillin (AMP) 10 μg; chloramphenicol (C30) 30 μg; nalidixic acid (NAL) 30 μg; streptomycin (STR) 10 μg; oxy-tetracycline (OXT) 30 μg; cephalothin (KF) 30 μg; erythromycin (ERY) 15 μg; sulphamethoxazole/trimethoprim (SXT) 22 μg; gentamycin (GCN) 10μg; ciprofloxacin (CIP) 10 μg; norfloxacin (NOR) 10 μg.
Abbreviation: MAR, multiple antibiotic resistance.
The multiple antibiotic resistance index and antibiotic-resistant phenotypes of Salmonella isolate are presented in Table 3. The MAR index ranged from 0.27 to 0.81 and was highest in Salmonella Weltevreden (AMP-OXT-STR-SXT-C30-ERY-NOR-KF-NAL), Salmonella Pullorum (AMP-OXT-CIP-STR-SXT-ERY-NOR-KF-NAL) and Salmonella Typhimurium (AMP-OXT-CIP-STR-SXT-ERY-NOR-KF-NAL) with resistance against nine different groups of antibiotics investigated. However, the MAR index of isolates was lowest in Salmonella enterica subsp. enterica, Salmonella Arizonae, Salmonella Albany and Salmonella Heidelberg strains having resistance to only three groups of the antibiotics studied.
Table 3.
Multiple Antibiotic Resistance Index And Phenotype Pattern Of Salmonella Isolates From Chickens In Mafikeng, South Africa
| Salmonella Strains | Sample Source | No. Of Strains | Antibiotics Resistance Profiles | MARIndex |
|---|---|---|---|---|
| Salmonella bongori | Indigenous | 1 | AMP-CIP-STR-SXT-ERY | 0.45 |
| Indigenous | 1 | AMP-OXT-CIP-STR-SXT-ERY-NOR-KF | 0.72 | |
| Broilers, layers | 2 | OXT-CIP-STR-ERY-NOR-NAL | 0.54 | |
| Indigenous | 1 | OXT-CIP-STR-SXT-ERY-NOR-NAL | 0.63 | |
| Broiler | 1 | OXT-STR-ERY-NAL | 0.36 | |
| Salmonella enterica subsp. enterica | Indigenous | 1 | OXT-STR-ERY-NOR | 0.36 |
| Broiler | 1 | OXT-STR-SXT-ERY | 0.36 | |
| Layer | 1 | OXT-STR-SXT-ERY-NAL | 0.45 | |
| Indigenous | 1 | OXT-STR-SXT-ERY-NOR | 0.45 | |
| Indigenous | 1 | AMP-OXT-CIP-STR-SXT-ERY-NOR | 0.63 | |
| Indigenous | 1 | AMP-OXT-CIP-STR-SXT-C30-ERY | 0.63 | |
| Broiler | 1 | AMP-SXT- ERY-KF | 0.36 | |
| Indigenous | 1 | AMP-SXT-ERY | 0.27 | |
| Indigenous, layer | 2 | AMP-STR-SXT-ERY | 0.36 | |
| Broiler | 1 | AMP-STR-SXT-ERY-KF | 0.45 | |
| Indigenous | 1 | AMP-STR-SXT-ERY-NOR | 0.45 | |
| Broiler | 1 | OXT-CIP-STR-SXT-ERY-NOR-NAL | 0.63 | |
| Broiler | 1 | OXT-STR-ERY-KF | 0.36 | |
| Indigenous | 1 | OXT-STR-ERY-NAL | 0.36 | |
| Layer | 1 | OXT-STR-ERY-NOR | 0.36 | |
| Indigenous | 1 | STR-SXT-C30-ERY-KF-NAL | 0.54 | |
| Salmonella enterica ser. Arizonae | Indigenous | 1 | AMP-STR-ERY | 0.27 |
| Broiler | 1 | OXT-CIP-STR-SXT-ERY-NOR-NAL | 0.63 | |
| Broiler | 1 | OXT-STR-ERY-NOR-KF-NAL | 0.54 | |
| Broiler | 1 | OXT-STR-SXT-ERY-KF | 0.45 | |
| Salmonella enterica ser. Typhimurium | Indigenous | 1 | AMP-OXT-CIP-STR-SXT-ERY-NOR-KF | 0.72 |
| Indigenous | 1 | AMP-OXT-CIP-STR-SXT-ERY-NOR-NAL | 0.72 | |
| Indigenous | 1 | AMP-OXT-STR-SXT-ERY-NOR-KF-NAL | 0.72 | |
| Indigenous | 1 | AMP-STR-GCN-SXT-C30-ERY-NOR-NAL | 0.72 | |
| Indigenous | 1 | AMP-OXT-CIP-STR-SXT-ERY-NOR-KF-NAL | 0.81 | |
| Indigenous | 1 | STR-SXT-ERY-KF | 0.36 | |
| Indigenous | 1 | STR-SXT-ERY-KF-NAL | 0.45 | |
| Salmonella enterica ser. Heidelberg | Broiler | 1 | AMP-OXT-STR-SXT-C30-ERY-KF | 0.63 |
| Indigenous | 1 | OXT-STR-ERY | 0.27 | |
| Salmonella enterica ser. Manchester | Indigenous | 1 | AMP-CIP-STR-SXT-ERY-NOR-NAL | 0.63 |
| Salmonella enterica ser. Weltevreden | Broiler | 1 | AMP-OXT-STR-SXT-C30-ERY-NOR-KF-NAL | 0.81 |
| Salmonella enterica ser. Salamae | Indigenous | 1 | OXT-STR-SXT-ERY-NOR-NAL | 0.54 |
| Salmonella enterica ser. Blegdam | Indigenous | 1 | AMP-OXT-CIP-STR-SXT-ERY-NOR-KF | 0.72 |
| Salmonella enterica ser. Pullorum | Indigenous | 1 | AMP-OXT-CIP-STR-SXT-ERY-NOR-KF-NAL | 0.81 |
| Salmonella enterica ser. Koessen | Indigenous | 1 | AMP-OXT-STR-SXT-ERY | 0.45 |
| Salmonella enterica ser. India | Indigenous | 1 | AMP-OXT-STR-SXT-ERY | 0.45 |
| Salmonella enterica ser. Crossness | Indigenous | 1 | AMP-OXT-STR-SXT-ERY | 0.45 |
| Salmonella enterica ser. Bovismorbificans | Broiler | 1 | AMP-OXT-STR-SXT-ERY-NOR-KF-NAL | 0.72 |
| Salmonella enterica ser. Infantis | Indigenous | 1 | AMP-OXT-STR-SXT-ERY-NOR-NAL | 0.63 |
| Salmonella enterica ser. Albany | Indigenous | 1 | AMP-STR-ERY | 0.27 |
| Salmonella enterica ser. Newport | Layer | 1 | AMP-STR-SXT-ERY | 0.36 |
| Salmonella enterica ser. Yovokome | Indigenous | 1 | AMP-STR-SXT-ERY-KF | 0.45 |
| Salmonella enterica ser. Wandsworth | Indigenous | 1 | OXT-CIP-STR-SXT-ERY-NOR-NAL | 0.63 |
| Salmonella enterica ser. Kentucky | Layer | 1 | OXT-STR-ERY-NAL | 0.36 |
| Salmonella enterica ser. Blockley | Indigenous | 1 | STR-SXT-ERY-KF | 0.36 |
| Salmonella enterica ser. Chingola | Broiler | 1 | OXT-STR-ERY-NOR-NAL | 0.45 |
| Salmonella enterica ser. Bareilly | Indigenous | 1 | OXT-STR-SXT-ERY-NAL | 0.45 |
| Salmonella enterica ser. Houten | Indigenous | 1 | AMP-OXT-CIP-STR-S300-ERY-NOR-NAL | 0.72 |
Note: Ampicillin53 10 μg; chloramphenicol (C30) 30 μg; nalidixic acid (NAL) 30 μg; streptomycin (STR) 10 μg; oxy-tetracycline (OXT) 30 μg; cephalothin (KF) 30 μg; erythromycin (ERY) 15 μg; sulphamethoxazole/trimethoprim (SXT) 22 μg; gentamycin (GCN) 10μg; ciprofloxacin (CIP) 10 μg; norfloxacin (NOR) 10 μg.
Abbreviation: MAR, multi-antibiotic resistance index.
Salmonella bongori strains from the indigenous chickens had the highest multiple resistance index of 0.72 with a wide range of resistant phenotypes (AMP-OXT-CIP-STR-SXT-ERY-NOR-KF) while similar strains from broilers (0.3) had the lowest MAR index with resistance phenotype patterns (OXT-STR-ERY-NAL). Also, a large portion of the Salmonella spp. and Salmonella enterica subsp. enterica had a hepta-multi-antibiotic-resistant patterns (AMP-OXT-CIP-STR-SXT-ERY-NOR) with MAR of 0.63 majorly from the indigenous chickens. Salmonella Koessen, Salmonella India, Salmonella Crossness, Salmonella Yovokome (AMP-STR-SXT-ERY-KF) had penta-resistant phenotype patterns.
An octa-antibiotics resistance was obtained in Salmonella Houten, Salmonella Bovismorbificans, Salmonella Blegdam, Salmonella Typhimurium and Salmonella bongori. Salmonella Typhimurium isolated in this study were found to belong to the indigenous chickens only, with a MAR index ranging from 0.72 to 0.81. This is indicative of a high multi-antibiotic resistance profiles against the main streams of antibiotics often prescribed in the treatment of Salmonella infections in both humans and animals. All octa-antibiotic-resistant strains had resistance to nalidixic acid except Salmonella Blegdam and Salmonella bongori.
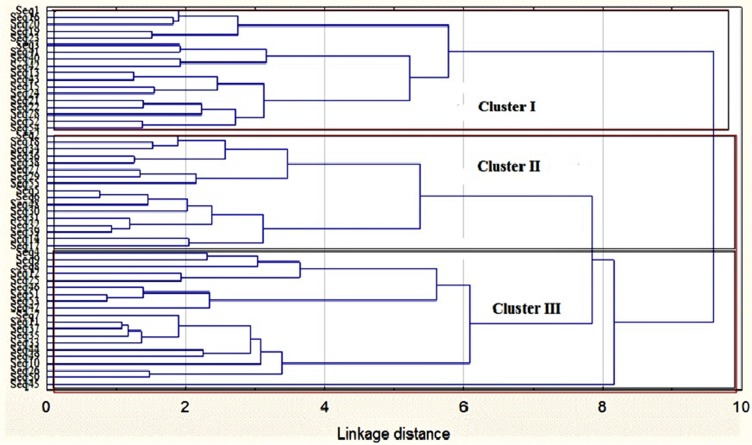
The relatedness and differences between the MAR-resistant strains of Salmonella are as shown in Figure 5. Three (3) major clusters (Clusters I, II and III) were observed and were traced to the source of isolates as described in Table 5. A total of 18 isolates clustered in cluster I, while in cluster II (17) and cluster III (20).7 Cluster III recorded the highest distribution of Salmonella isolates. Table 4 presents the percentage distribution of Salmonella based on sample source and antibiotics resistance clustering patterns. The percentage distribution of Salmonella isolates based on antibiotics resistance clusters ranged from 15% to 73.3%. The distribution of resistant strains in cluster I ranged from 16.6% to 73.3% and the largest proportion were from indigenous chickens (11; 73.3%). In clusters I and III, respectively, isolates from layers (22.2%; 35%) and indigenous chickens (73.3%; 50%) had the highest relatedness in terms of antibiotic resistance patterns as opposed to isolates from broilers. However, there was no significant difference between isolates from layers, clustered in clusters II (4; 23%) and I (4; 22.2%) at P≥0.05. The percent distribution of Salmonella isolates from indigenous chickens ranged from 29.4% to 73.3%. As shown in Table 5, there exists a positive correlation in the antibiotic-resistance patterns of Salmonella isolates from layers and broilers while a negative correlation was obtained against indigenous chickens.
Figure 5.
Dendrogram of antibiotic resistance profiles of Salmonella strains isolated from chickens in Mafikeng, South Africa, using cluster analysis.
Notes: Seq represents sequence numbering of Salmonella isolates from different types of chickens in Mafikeng. The tree was constructed using the Ward’s method and Euclidean distances in the Statistica version 7 software (Statsoft US).
Abbreviation: Seq, sequences of Salmonella isolates from chickens.
Table 5.
Pearson Correlations Between Percent Antibiotic Resistance Of Salmonella Isolates And Source
| Antibiotic Resistance | Total Resistance (%) | Broilers (%) | Layers (%) | Indigenous (%) |
|---|---|---|---|---|
| Total resistance (%) | 1 | |||
| Broilers (%) | 0.925** | 1 | ||
| Layers (%) | 0.931** | 0.874** | 1 | |
| Indigenous (%) | 0.959** | 0.800** | 0.825** | 1 |
Note: **Correlation is significant at the 0.01 level (2-tailed).
Table 4.
The Percent Distribution Of Resistant Salmonella Isolates Based On Sample Source And Antibiotic Resistant Clusters
| Type of Chicken/Source | Cluster I N = 18 |
Cluster II N = 17 |
Cluster III N = 20 |
|---|---|---|---|
| Broiler | 3 (16.6%) | 8 (47.1%) | 3 (15%) |
| Layers | 4 (22.2%) | 4 (23.5%) | 7 (35%) |
| Indigenous | 11 (73.3%) | 5 (29.4%) | 10 (50%) |
Discussion
The morphological characteristics observed in this study support the previous observation contained in the WHO Global Salm-Surv as described by Hendriksen et al.25 The triple sugar iron test of the presumptive Salmonella isolates depicts their ability to utilize lactose, saccharose and dextrose sugars. A positive to indole test depicts the ability of isolates to utilize amino acid in the form of tryptophan to produce the enzyme indole. However, some isolates showed a positive reaction to urease and indole which contradicts the expected results stipulated in the Bergey’s Manual of Determinative Bacteriology. Nevertheless, a similar variation has been reported by Shan et al.26 However, the observation in this study might be due to a shift in the nutrient utilisation pattern of Salmonella as a result of ecological stress emanating from competition for food and other stress inducers. Hence, biochemical characteristics might not be adequate to effectively discriminate a microbial community, hence the need for the use of more reliable approaches such as the molecular techniques. The PCR discrimination method was effective in the discrimination of Salmonella spp. as opposed to the use of biochemical and morphological characteristics as obtained from this study. Therefore, the polymerase chain reaction could present a rapid, sensitive and reliable method for pathogen detection.
The positive amplification of the invA genes in Salmonella isolates supports the findings of previous authors on the presence of invasive genes in Salmonella.27 Demonstrated the presence and functionality of invA, B and C genes which are regions of high similarity in diverse Salmonella serovars except in Salmonella arizona in which invD gene regions were detected. The presence of invA genes has been reported in Salmonella isolates from broiler chickens in Iran,28 poultry, pigs, humans and other food commodities in Brazil.29 The use of invA genes has been regarded as the most reliable in Salmonella discrimination since many possess the invA gene within their genomes. Therefore, it is pertinent in the tracking of the pathogenesis of Salmonella-borne infections in animals and humans. The virulence of Salmonella in hosts has been linked to their ability to invade the epithelial tissues. On ingestion, Salmonella attaches itself to the intestinal mucosa lining contributing to a decrease in the pH of the gastrointestinal tract, thus causing an irritation. Invasive Salmonella species could deplete the mucosa layer by penetrating through the M cells overlying the Peyer’s patches.30 In some patients, this situation may progress to a systemic infection resulting from the invasion of the intestinal lymphoid follicles by Salmonella strains which presents clinical signs associated with drained mesenteric lymph nodes.
From the study, some isolates were found not to possess the invA gene, thus implying their inability to cause infections in hosts. However, the occurrence of the invasive Salmonella isolates among the chicken samples within the Mafikeng Community suggests that consumers and other stakeholders within the food and value chain might be at a risk of Salmonella-borne infections. This can hamper the safety and health of both veterinary and humans and the socioeconomic status of the people living in Mafikeng community, South Africa.
Salmonella species such as Salmonella Bovismorbificans was isolated from this study as opposed to the previous report of it been found only in humans.31 Many diverse Salmonella strains identified in this study have been implicated to possess the extended spectrum of the β-lactamases (ESBLs) enzymes coding for antibiotic-resistant genes and have been linked to salmonellosis in humans.31–34 The isolation of these strains from chickens may have resulted from human-to-animal interaction along the available interfaces such as contaminated feed, water, handling, infected hosts (rodents) and animal care personnel on the farms.
The dominance of Salmonella enterica species in chickens supports the findings of35 who reported a high occurrence of Salmonella Typhimurium compared to the serotype Salmonella Enteritidis in a study conducted in the Democratic Republic of Congo. Also, the isolation of Salmonella Heidelberg, Salmonella Koessen, Salmonella Pullorum and Salmonella Gallinarum36 has previously been isolated from poultry justifying that aves (poultry) are reservoirs of Salmonella.34 Salmonella Koessens, Salmonella Pullorum and Salmonella Gallinarum have been linked to salmonellosis originating from eggs. These Salmonella serovars constitute a threat to food safety and are capable of causing human sicknesses. Variations in Salmonella serovars occurrences have been reported in different countries and said to be a function of geographical location.36–38 The typhoidal groups have been implicated as the most cause of ill health in the developing countries.39,40 However, the non-typhoidal Salmonella serovars (Salmonella Typhimurium and Salmonella Enteritidis) are regarded as the major causes of salmonellosis outbreak in developing countries like India, Iran and many sub-saharan Africa.41 Hence, the isolation of virulent strains of Salmonella in chickens which happens to form a major part of South African cuisines brings a concern to food security and safety of consumers.
The bootstrap values within the evolutionary trend were higher than 70%, which supports the previous report of Wayne et al42 on decision of a close relatedness between organisms. However, the disparity in homology between CP012144 and other isolates could be due to mutation and development of new traits as evolution proceeds. However, CP012144 was comparable to Salmonella enterica subsp. enterica (MG663470) isolated in this study confirming the sharing of the same ancestral origin. The development of new traits having a 99% homology in isolates MG663482, MG66347, and MG663480 was observed showing that they all evolved from the same ancestor. This corroborates the previous report that Salmonella strains evolved from two broad genus Salmonella enterica and Salmonella bongori.43 The bootstrap clustering of MG663495 with S. bongori could be due to evolutionary traits that are not pronounced in the isolates. However, the high bootstrap values within the Salmonella enterica group are indicative of high genetic relatedness and reliability of traits developed which cannot easily disappear or wiped out overnight.44 The bootstrap values of Salmonella isolates and control strains in this study were higher than 50% which showed a high level of repetitive clustering within the isolates. This supports the findings of Soltis and Soltis45 on the acceptable bootstrap value (100–70%) in the construction of phylogenies. The observation of 100% bootstrap values of isolates as shown in the phylogenetic tree showed that 100% level of repetition exists in the genome compared.
Ampicillin belongs to the group of aminopenicillins which are often administered in the treatment of diseases caused by the gram-negative pathogenic enteric bacteria. Gentamycin belongs to the group of aminoglycosides alongside streptomycin, but in this study, gentamycin showed effectiveness in the control of Salmonella. Gentamycin has been reported to have a higher sensitivity on Salmonella strains compared to other antibiotics used in previous studies.46 This might be due to the fact that gentamycin does not fall within the most common antibiotics administered in the treatment of Salmonella caused infection. However, it must be noted that an uncontrolled use of these antibiotics could also lead to a build-up of resistance to these antibiotics. All isolated Salmonella bongori strains had resistance to ciprofloxacin and nalidixic acid which belong to the fluoroquinolones often regarded as the last resort in Salmonella infection treatment. Similar reports have been made on the isolation of fluoroquinolone-resistant Salmonella in Taiwan.47 Nalidixic acid is a new generation of the fluoroquinolones often prescribed in the treatment of Salmonella-caused infections. This finding is concurrent with48 reports on multiple antibiotics resistance of Salmonella isolates from poultry in India and Egypt, respectively.
Higher antibiotic resistance phenotype profile was observed in Salmonella Typhimurium as opposed to the previous report of its penta-antibiotic resistance phenotype profiles. The increased antibiotic resistance obtained in this study could be due to misuse of antibiotics, thus resulting in adaptation and change in the antibiotic sensitivity behaviour of this pathogen with the aim to survive stress condition within the eco-system. Isolation of multiple antibiotics resistance strains in indigenous chickens calls for concern as it is believed that this breed of chickens is not often administered antibiotics during ill health. Albeit, the occurrence of multi-drug resistance could be due to the effect of a possible lateral gene transfer within the ecological niche. With regard to sample source, the occurrence of Salmonella multiple-antibiotics resistance followed the order (layers ≤ broilers ≤ indigenous chickens).
The resort to the prolonged use of fluoroquinolones in the treatment of Salmonella-borne infections has led to many cases of antimicrobial resistance globally.49 This resistance has been explained to be caused by mutations of the gyrase DNA gene and change in the efflux pump which is a target for the fluoroquinolones.50,51 However, Lauderdale et al47 have suggested the use of the extended spectrum of cephalosporins as the last resort in the treatment of Salmonella infections. Hence, there is the need to develop an effective therapeutic approach in the control of these evolving virulent Salmonella strains. Furthermore, a negative correlation exists between the antibiotic-resistant profiles of Salmonella isolates from broilers, indigenous and layer chickens. The Pearson partial correlation was significant at p≤0.01. A positive correlation shows closer similarities in the antibiotic-resistant patterns of different Salmonella isolates in the study area, while a negative correlation within the sample source is implicative of non-source-dependent profiles.
The high occurrence of the antibiotic-resistant Salmonella strains from indigenous chickens could be due to a pick-up of virulence determinants from the environment or through interaction hosts such as rodents and livestock whom they share feeding and drinking troughs. Also, the high percent distribution of antibiotic-resistant strains in the broilers as shown in the clustering profiles indicates that isolates do share the same antibiotic resistance histories. According to Forshell et al,52 the abuse and misuse of antibiotics is a major cause of increasing antibiotic resistance among microorganisms of public health significance such as Salmonella. During processing or dressing operations, care for poultry, situations of gastrointestinal content shedding could arise which could lead to spillages of gut content into the environment. Also, unhygienic practices on farms and processing industries could aid in the transport of these virulent strains to the public either through the release of untreated effluents into river channels or other water bodies. These water bodies form a major resource for livelihood among the rural dwellers.
Conclusion
From this study, it is reported that the similarities in the antibiotic-resistant patterns among isolates from broilers, layers and indigenous chickens reveal similarities in antibiotic exposure histories. It is therefore suggested that there is need to sensitize farmers to adhere to prescribed guidelines on the use of antibiotics. In addition, the implementation of good sanitation among farm workers as well as standard operating procedures in farms where animals are housed should be encouraged to curb the spread of multi-drug-resistant strains of Salmonella since the latter may pose a threat to public health. The detection of large proportions of diverse multi-antibiotic-resistant Salmonella strains in chickens within Mafikeng community, especially in indigenous breed, indicates that these animals may pose a threat on food security and safety. Further studies on the antibiotic-resistant genes harboured by this pathogen could advance knowledge in the development of suitable antibiotics and other prophylaxis to curb Salmonella-caused infections.
Acknowledgments
Authors acknowledge the assistance received from Dr M. Ogunrinu and Dr L. Ngoma during this study. This work was supported by The World Academy of Science in collaboration with the Directorate of Science and Technology, National Research Foundation (TWAS DST-NRF) of South Africa (grant number UID: 110854, 2018).
Disclosure
Mr Stephen Abiola Akinola reports grants from TWAS DST-NRF, outside the submitted work. The authors report no other conflicts of interest in this work.
References
- 1.Ulaya WD. Determination of Virulence Factors in Salmonella Isolates of Human, Poultry and Dog Origin in Lusaka District, Zambia, in Department of Paraclinical Studies Lusaka. The University of Zambia; 2013:98. [Google Scholar]
- 2.Acheson D, Hohmann EL. Nontyphoidal salmonellosis. Clin Infect Dis. 2001;32(2):263–269. doi: 10.1086/318457 [DOI] [PubMed] [Google Scholar]
- 3.Gill CJ, Hamer DH. Foodborne illnesses. Curr Treat Options Gastroenterol. 2001;4(1):23. doi: 10.1007/s11938-001-0044-0 [DOI] [PubMed] [Google Scholar]
- 4.Kemal J. A review on the public health importance of bovine salmonellosis. Vet Sci Technol. 2014;5(2):1. [Google Scholar]
- 5.Kagambèga A, Lienemann T, Aulu L, et al. Prevalence and characterization of Salmonella enterica from the feces of cattle, poultry, swine and hedgehogs in Burkina Faso and their comparison to human Salmonella isolates. BMC Microbiol. 2013;13(1):253. doi: 10.1186/1471-2180-13-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drzewiecka D. Significance and roles of Proteus spp. bacteria in natural environments. Microb Ecol. 2016;72(4):741–758. doi: 10.1007/s00248-015-0720-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO. World Health Statistics 2016: Monitoring Health for the SDGs Sustainable Development Goals. WHO Press. 2016. [Google Scholar]
- 8.Zouboulis A, Loukidou M, Matis K. Biosorption of toxic metals from aqueous solutions by bacteria strains isolated from metal-polluted soils. Process Biochem. 2004;39(8):909–916. doi: 10.1016/S0032-9592(03)00200-0 [DOI] [Google Scholar]
- 9.Ateba CN, Mochaiwa B. Use of invA gene specific PCR analysis for the detection of virulent Salmonella species in beef products in the north west province, South Africa. J Food Nutr Res. 2014;2(p):294–300. doi: 10.12691/jfnr-2-6-5 [DOI] [Google Scholar]
- 10.Muyzer G, Teske A, Wirsen CO, Jannasch HW. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbial. 1995;164:165–172. doi: 10.1007/BF02529967 [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J, Russel DW. Molecular Cloning. A Laboratory Manual. 3. ed. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 12.National Center for Biotechnology Information, U.S. National Library of Medicine, 8600 Rockville Pike, BethesdaMD, 20894, USA. [Google Scholar]
- 13.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall TA. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. In Nucleic Acids Symposium Series. London: Information Retrieval Ltd.; 1999:c1979–c2000. [Google Scholar]
- 15.Jukes TH, Cantor CR. Evolution of protein molecules. Mamm Protein Metab. 1969;3(21):132. [Google Scholar]
- 16.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- 17.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493. doi: 10.1093/ajcp/45.6_ts.764 [DOI] [PubMed] [Google Scholar]
- 20.Wayne PA. Clinical and laboratory standards institute. Perform Stand Antimicrob Susceptibility Test. 2007;M100-S17. [Google Scholar]
- 21.Rota C, Yangüela J, Blanco D, Carramiñana JJ, Ariño A, Herrera A. High prevalence of multiple resistance to antibiotics in 144 Listeria isolates from Spanish dairy and meat products. J Food Prot. 1996;59(9):938–943. doi: 10.4315/0362-028X-59.9.938 [DOI] [PubMed] [Google Scholar]
- 22.Singh S, Yadav AS, Singh SM, Bharti P. Prevalence of Salmonella in chicken eggs collected from poultry farms and marketing channels and their antimicrobial resistance. Food Res Int. 2010;43:2027–2030. doi: 10.1016/j.foodres.2010.06.001 [DOI] [Google Scholar]
- 23.Wattiau P, Weijers T, Andreoli P, et al. Evaluation of the Premi®Test Salmonella, a commercial low-density DNA microarray system intended for routine identification and typing of Salmonella enterica. Int J Food Microbiol. 2008;123(3):293–298. doi: 10.1016/j.ijfoodmicro.2008.01.006 [DOI] [PubMed] [Google Scholar]
- 24.Schrader KN, Fernandez-Castro A, Cheung WKW, Crandall CM, Abbott SL. Evaluation of commercial antisera for Salmonella serotyping. J Clin Microbiol. 2008;46(2):685–688. doi: 10.1128/JCM.01808-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendriksen R, Jaap W, Van Bergen M. Global Salm-Surv. A Global Salmonella Surveillance and Laboratory Support Project of the World Health Organization. Identification of Thermotolerant Campylobacter. WHO; 2003. [Google Scholar]
- 26.Shan Y, Lai Y, Yan A. Metabolic reprogramming under microaerobic and anaerobic conditions in bacteria Subcell. Biochem. 2012:64,159–179. [DOI] [PubMed] [Google Scholar]
- 27.Galán JE, Curtiss R. Distribution of the invA,-B,-C, and-D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect Immun. 1991;59(9):2901–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salehi TZ, Mahzounieh M, Saeedzadeh A. Detection of invA gene in isolated Salmonella from broilers by PCR method. Int J Poult Sci. 2005;4(8):557–559. doi: 10.3923/ijps.2005.557.559 [DOI] [Google Scholar]
- 29.Oliveira SDD, Rodenbusch CR, Michael GB, Cardoso MIR, Canal CW, Brandelli A. Detection of virulence genes in Salmonella enteritidis isolated from different sources. Braz J Microbiol. 2003;34:123–124. doi: 10.1590/S1517-83822003000500042 [DOI] [Google Scholar]
- 30.Ohl ME, Miller SI. Salmonella: a model for bacterial pathogenesis. Annu Rev Med. 2001;52(1):259–274. doi: 10.1146/annurev.med.52.1.259 [DOI] [PubMed] [Google Scholar]
- 31.Cavaco L, Hasman H, Xia S, Aarestrup FM. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob Agents Chemother. 2009;53(2):603–608. doi: 10.1128/AAC.00997-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harakeh S, Yassine H, El-Fadel M. Antimicrobial-resistant patterns of Escherichia coli and Salmonella strains in the aquatic Lebanese environments. Environ Pollut. 2006;143(2):269–277. doi: 10.1016/j.envpol.2005.11.027 [DOI] [PubMed] [Google Scholar]
- 33.Morpeth SC, Ramadhani HO, Crump JA. Invasive non-typhi Salmonella disease in Africa. Clin Infect Dis. 2009;49(4):606–611. doi: 10.1086/603553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiao J, Zhang Q, Alali WQ, et al. Characterization of extended-spectrum β-lactamases (ESBLs)-producing Salmonella in retail raw chicken carcasses. Int J Food Microbiol. 2017;248:72–81. doi: 10.1016/j.ijfoodmicro.2017.02.016 [DOI] [PubMed] [Google Scholar]
- 35.Phoba M-F, Boeck H, Ifeka BB, et al. Epidemic increase in Salmonella bloodstream infection in children, Bwamanda, the Democratic Republic of Congo. Eur J Clin Microb Infect Dis. 2014;33(1):79–87. doi: 10.1007/s10096-013-1931-8 [DOI] [PubMed] [Google Scholar]
- 36.Shivaprasad H. Fowl typhoid and pullorum disease. Revue Scientifique Et Technique-Office Int Des Epizooties. 2000;19(2):405–416. doi: 10.20506/rst.19.2.1222 [DOI] [PubMed] [Google Scholar]
- 37.Shilangale RP, Giannatale ED, Chimwamurombe PM, Kaaya GP. Prevalence and antimicrobial resistance pattern of Salmonella in animal feed produce in Namibia. Vet. Ital. 2012;48:125–132. [PubMed] [Google Scholar]
- 38.Foley S, Lynne A, Nayak R. Salmonella challenges: prevalence in swine and poultry and potential pathogenicity of such isolates 1 2. J Anim Sci. 2008;86(14_suppl):E149–E162. doi: 10.2527/jas.2007-0464 [DOI] [PubMed] [Google Scholar]
- 39.Foley S, Lynne A. Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance 1. J Anim Sci. 2008;86(14_suppl):E173–E187. doi: 10.2527/jas.2007-0447 [DOI] [PubMed] [Google Scholar]
- 40.Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Bäumler AJ. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 2001;3(14–15):1335–1344. [DOI] [PubMed] [Google Scholar]
- 41.Gilks CF. Acute bacterial infections and HIV disease. Br Med Bull. 1998;54(2):383–393. doi: 10.1093/oxfordjournals.bmb.a011695 [DOI] [PubMed] [Google Scholar]
- 42.Wayne L, Stackebrandt E, Kandler O, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol. 1987;37(4):463–464. doi: 10.1099/00207713-37-4-463 [DOI] [Google Scholar]
- 43.Tuchili L, Ulaya W, Kato Y, Kaneuchi C. Recent characterisation of Salmonella strains isolated from chickens in Zambia. J Vet Med Sci. 1996;58(1):77–78. doi: 10.1292/jvms.58.77 [DOI] [PubMed] [Google Scholar]
- 44.Konstantinidis KT, Stackebrandt E. Defining Taxonomic Ranks, in the Prokaryotes. Springer; 2013:229–254. [Google Scholar]
- 45.Soltis PS, Soltis DE. Applying the bootstrap in phylogeny reconstruction. Stat Sci. 2003;256–267. doi: 10.1214/ss/1063994980 [DOI] [Google Scholar]
- 46.Arora D,Kumar S, Singh D, Jindal N, Mahajan N. Isolation, characterization and antibiogram pattern of Salmonella from poultry in parts of Haryana, India. Adv Anim Vet Sci. 2013;1(5):161–163. [Google Scholar]
- 47.Lauderdale T-L, Aarestrup FM, Chen P-C, et al. Multidrug resistance among different serotypes of clinical Salmonella isolates in Taiwan. Diagn Microbiol Infect Dis. 2006;55(2):149–155. doi: 10.1016/j.diagmicrobio.2006.01.002 [DOI] [PubMed] [Google Scholar]
- 48.Mahmoud MS, Esmail M, Khairy R, Mazher O. Detection of multi-drug resistant non-typhoid Salmonella isolates in cases of gastroenteritis in Egypt. Br Microb Res J. 2015;6(3):167. doi: 10.9734/BMRJ [DOI] [Google Scholar]
- 49.Threlfall EJ. Antimicrobial drug resistance in Salmonella: problems and perspectives in food‐and water‐borne infections. FEMS Microbiol Rev. 2002;26(2):141–148. doi: 10.1111/j.1574-6976.2002.tb00606.x [DOI] [PubMed] [Google Scholar]
- 50.Piddock LJ. Fluoroquinolone resistance in Salmonella serovars isolated from humans and food animals. FEMS Microbiol Rev. 2002;26(1):3–16. doi: 10.1111/j.1574-6976.2002.tb00596.x [DOI] [PubMed] [Google Scholar]
- 51.Yang B, Xi M, Cui S, et al. Mutations in gyrase and topoisomerase genes associated with fluoroquinolone resistance in Salmonella serovars from retail meats. Food Res Int. 2012;45(2):935–939. doi: 10.1016/j.foodres.2011.01.031 [DOI] [Google Scholar]
- 52.Forshell LP, Wierup M. Salmonella contamination: a significant challenge to the global marketing of animal food products. Rev Sci Tech off Int Epiz. 2006;25(2):541–554. [PubMed] [Google Scholar]
- 53.Guariguata L, Whiting DR, Hambleton I, Beagley J, Linnenkamp U, Shaw JE. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res Clin Pract. 2014;103(2):137–149. doi: 10.1016/j.diabres.2013.11.002 [DOI] [PubMed] [Google Scholar]