Abstract
Pathogenic enterobacteria face various oxygen (O2) levels during intestinal colonization from the O2-deprived lumen to oxygenated tissues. Using Shigella flexneri as a model, we had previously demonstrated that epithelium invasion is promoted by O2 in a Type III secretion system (T3SS)-dependent manner1. However, subsequent pathogen adaptation to tissue oxygenation modulation remained unknown. Assessing single-cell distribution, together with tissue oxygenation, we demonstrate here that the colonic mucosa O2 is actively depleted by Shigella flexneri aerobic respiration, not host neutrophils, during infection, leading to the formation of hypoxic foci of infection. This process is promoted by T3SS inactivation in infected tissues, favoring colonizers over explorers. We identify the molecular mechanisms supporting infectious hypoxia induction, and we demonstrate here how enteropathogens optimize their colonization capacity in relation to their ability to manipulate tissue oxygenation during infection.
Pathogenic enterobacteria virulence and first-line immune cells’, particularly neutrophils’, survival and function are highly modulated by oxygen (O2)1,2,3,4. Most virulent enterobacteria (Shigella spp., Listeria spp., Salmonella spp., pathogenic E. coli, or Yersinia pestis) are well adapted to these changing microenvironments5, suggesting adaptability to various O2 levels represents a selective advantage crucial for their infectious capacity. The colonization process involves three key steps: degradation of the mucus layer, invasion of the epithelium mediated by the T3SS, and formation of primary foci of infection. We previously demonstrated that S. flexneri T3SS activation requires O2, which diffuses on the epithelium surface1. However, tissue oxygenation modulation during the dissemination of enteropathogens and its impact on colonization or immune response efficiency remains largely unknown, although hypoxia has been previously reported in a TNBS colitis model of inflammation6.
In this work, we show that oxygen is depleted during tissue colonization by enteropathogens, using S. flexneri as a model, leading to the formation of hypoxic foci of infection. We designed a quantitative image analysis method allowing the assessment of tissue oxygenation at a single-cell level for both bacteria and neutrophils. We demonstrate that S. flexneri aerobic respiration is essential for O2 depletion within the colonic mucosa, not neutrophils, as previously demonstrated in a non-infectious model of inflammation7. We show that formation of hypoxic foci of infection is the primary S. flexneri colonization strategy, which is promoted by the repression of T3SS activity. Our results demonstrate that the interaction between S. flexneri and immune cells occurs mainly in the absence of O2. We anticipate our results will have a significant impact on new vaccine and antibiotic development strategies, as well as reappraisal of immune response and host-pathogen interactions in low O2 conditions.
We recently observed that hypoxia was induced within the colonic mucosa upon S. flexneri infection8 using a hypoxia reporter, EF59. Here, measuring relative hypoxia levels from the epithelial surface to the submucosa (Supplementary Table 1), we confirmed this preliminary observation (Figure 1a) and demonstrated that during infection, compared to basal conditions, the degree of hypoxia increased significantly to a depth of 130 µm (Figure 1b, p < 0.05). In basal conditions, the ‘physiological hypoxic status’ of the epithelium was confirmed, as previously reported in the mouse6,7 (Figure 1b, uninfected tissues).
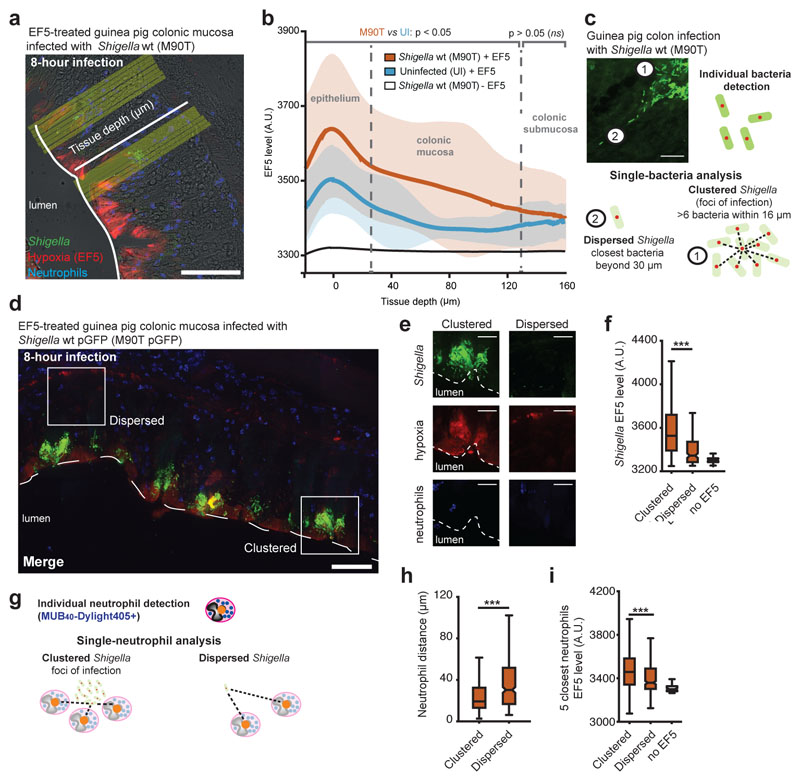
Figure 1. Hypoxia is specifically induced by Shigella within foci of infection.
a. Hypoxia was detected in guinea pig colonic mucosa infected with S. flexneri 5a pGFP strain (green) using the EF5 reporter9. EF5 was immunodetected with an a-EF5-Cy3 (red); neutrophils were labeled with the Myelotracker-Dylight40512 marker (or Myelotracker-Alexa40512, blue). Scale bar: 50 µm.
b. Hypoxia profiles through the colonic mucosa were reported. The EF5 level is reported as mean over all aligned profiles (thick lines) ± standard deviation (shaded area) against tissue depth (n = 128, 61 and 48 profiles averaged for respectively the M90T, uninfected and no-EF5 conditions). The gray segments atop the curves represent the range of depths for which the EF5 levels are significantly different between the M90T and uninfected conditions (two-sided Student t-test, p < 0.05).
c. Individual bacteria were detected by quantitative image analysis (red dot, see Supplementary Figure 1a), and two populations of S. flexneri were defined: as ‘dispersed’ if its closest neighbor was located beyond 30 µm or as ‘clustered’ if at least 6 bacteria were within a 16 µm diameter, forming a focus of infection. Scale bar: 20 µm.
d. Detection of ‘clustered’ and ‘dispersed’ bacteria populations within the colonic mucosa. Scale bar: 50 µm.
e-f. Hypoxia levels were compared at single ‘dispersed’ and ‘clustered’ bacteria levels by quantitative image analysis. Scale bar: 20 µm. Hypoxia levels were significantly higher around ‘clustered’ compared to ‘dispersed’ bacteria (ANOVA 1-way test + Tukey test, on n = 61119, 186 and 11400 bacteria, respectively for the clustered+EF5, dispersed+EF5 and no-EF5 conditions, see Supplementary Table 2). The no-EF5 signal was measured on infected tissues not stained with EF5.
g. Individual neutrophils (orange dot), stained with Myelotracker-Dylight40512, were localized in tissue by quantitative image analysis.
h. Neutrophils were found closer to ‘clustered’ bacteria compared to ‘dispersed’ bacteria (distance from n = 61119 and 186 bacteria to the closest neutrophil for respectively the ‘clustered’ and ‘dispersed’ bacteria, p < 10-10).
i. The hypoxia level detected around neutrophils (averaged over the 5 closest) was higher around ‘clustered’ bacteria compared to ‘dispersed’ bacteria (EF5 levels around the 5 closest neutrophils, measured for n = 61119, 186 and 11400 bacteria, respectively for the clustered+EF5, dispersed+EF5 and no-EF5 conditions, p ~ 10-10).
*** indicates p < 0.001. In the box-plots of panels f and h, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points not considered outliers, and the outliers are not plotted. Outliers are data points that are further away than 1.5 times the range from the 1st to the 3rd quartile, respectively above or below the 3rd and 1st quartile.
Taking into account the potential heterogeneity of the bacteria population10, we developed a quantitative imaging strategy at a single-bacteria level, as previously reported11, to localize individual bacteria and assess hypoxia levels (Figure 1c and Supplementary Figure 1a-c). First, we revealed that most bacteria (99.71%, Supplementary Figure 1a) were located within foci of infection formed in the colonic extracellular matrix (‘clustered’ bacteria, defined as having at least 6 neighboring bacteria within 16 µm) not dispersed within the mucosa (‘dispersed’ bacteria, defined as having a closest neighboring bacteria beyond 30 µm, Figure 1c and Supplementary Figure 1a). We demonstrated that S. flexneri faced hypoxic conditions specifically within foci of infection, not when dispersed elsewhere in the mucosa (Figure 1d-f, p < 10-10). This result implies that S. flexneri adaptation to low-O2 levels is crucial for tissue colonization. We demonstrated that a S. flexneri ∆fnr mutant strain (FNR, which mediates the adaptation of S. flexneri to anaerobiosis1) did not propagate within foci of infection and was avirulent (Supplementary Figure 2a), as previously demonstrated1; also, the neutrophil population was reduced, a sign of limited inflammation (Supplementary Figure 2a). In a rabbit ileal loop model, we confirmed that FNR was specifically required for S. flexneri propagation in an inflammatory environment, but not mandatory for its fitness within the intestinal lumen (Supplementary Figure 2b).
Neutrophils represent the most abundant immune cell population recruited upon Shigella infection, and their activation has been previously identified as responsible for hypoxia induction in a TNBS colitis model of inflammation7. Neutrophils were specifically labeled with MUB40, (or Myelotracker, a neutrophil lactoferrin marker12), allowing single-cell analyses (Figure 1g). We observed that neutrophils were in closest proximity to ‘clustered’ bacteria compared to ‘dispersed’ bacteria (Figure 1h) and that the level of hypoxia quantified at a single-neutrophil level was significantly higher when located in the vicinity of ‘clustered’ bacteria (Figure 1i). Hypoxia levels measured at a single-bacteria level around ‘clustered’ and ‘dispersed’ S. flexneri remained similar in conventional and neutropenic animals (Figure 2a-b), suggesting that neutrophil contribution is not an essential factor for hypoxia induction. As expected, a significantly reduced amount of neutrophils were detected in the infected colonic mucosa of neutropenic animals; the S. flexneri population was only mildly reduced (Supplementary Figure 2c). Neutrophil hypoxia levels were compared to the closest bacteria hypoxia profiles; for more than 95% of neutrophils, hypoxia levels were not found to be significantly higher than the hypoxia generated by bacteria in their vicinity (Supplementary Figure 2d).
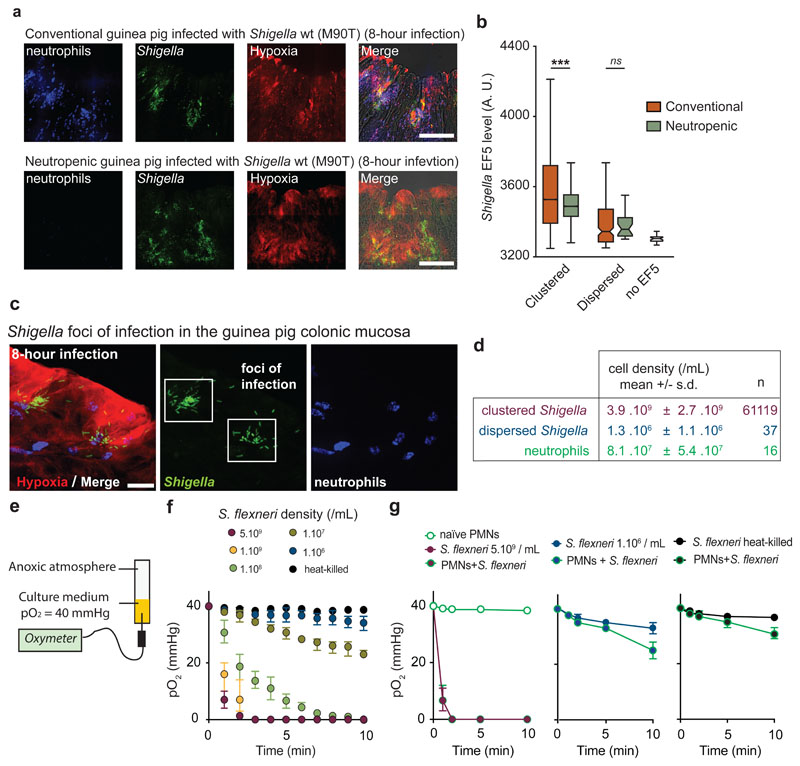
Figure 2. Neutrophils are not essential for O2 depletion in infected tissues, which is mainly caused by Shigella aerobic respiration.
a. S. flexneri pGFP (green) foci of infection were detected in conventional (8 animals, 28 acquisitions) and neutropenic (3 animals, 7 acquisitions) guinea pig colonic mucosa. Neutrophils were labeled with Myelotracker-Dylight40512 (blue) and hypoxia with an α-EF5-Cy3 (red). Scale bar: 50 µm.
b. Hypoxia levels around ‘clustered’ bacteria were comparable between conventional vs neutropenic animals (p < 0.0001, see Supplementary Table 3).
c-d. Cell density of ‘clustered’ (in foci of infection) and ‘dispersed’ S. flexneri pGFP (green) populations were calculated in vivo (expressed as mean ± S.D). Neutrophils were labeled with Myelotracker-Dylight40512 (blue) and hypoxic areas with an α-EF5-Cy3 (red). Scale bar: 2 µm. For clustered bacteria, the density is calculated as the number of bacteria around each clustered bacterium within 16 µm for 61119 bacteria. For dispersed bacteria and neutrophils, the density is calculated as the number of dispersed bacteria (n = 37 loci imaged), respectively neutrophils (n = 16 loci imaged), divided by the imaged tissue volume.
e. O2 consumption rates were assessed in vitro with an oximeter in a RPMI 1640 + 10 mM Hepes culture medium stabilized at 40 mmHg (hypoxic chamber) in tubes sealed under anoxic conditions (see Methods). Exponentially grown bacterial cultures were inoculated at the indicated concentration (t = 0).
f. O2 depletion rate was correlated with S. flexneri bacterial density. O2 tensions (expressed as mean ± S.D.) were assessed at indicated cell densities surrounding ‘clustered’ and ‘dispersed’ population densities calculated in vivo (see d.) or in the presence of 5.109 heat-killed S. flexneri (n = 3 independent experiments).
g. O2 depletion rate (O2 tensions expressed as mean ± S.D., n = 3 independent experiments) in the presence of S. flexneri and neutrophils (PMNs, 8.107 neutrophils/mL). In the presence of 1.106 S. flexneri/mL or 5.109 heat-killed S. flexneri/mL, anoxia was not reached over the measurement period, although neutrophil O2-consumption was observed (10 min, see Supplementary Table 4).
Since neutrophils’ contribution seemed limited, O2 consumption mediated by S. flexneri was investigated as a potential cause of local O2 depletion observed within foci of infection. S. flexneri population density in foci was estimated from confocal images to be 3.9.109 ± 2.7.109 cell/mL for ‘clustered’ bacteria, compared to 1.3.106 ± 1.1.106 cell/mL for bacteria ‘dispersed’ in the tissue (Figure 2c-d). The impact of bacterial density on S. flexneri O2 consumption kinetics was then assessed in vitro using a dedicated device allowing the inoculation of bacteria in a sealed anoxic tube containing culture medium stabilized at 40 mmHg O2, reflecting estimated physiological O2 levels13 (Figure 2e). O2 consumption rate was correlated to the S. flexneri population cell density (Figure 2f). At a cell density measured within foci of infection (between 1.109 and 5.109 cell/mL), complete O2 depletion by S. flexneri occurred rapidly (tanoxia = 2.3 ± 1.1 min, Figure 2f); similar results were obtained with other pathogenic enterobacteria, including E. coli, S. typhimurium, L. monocytogenes, and Y. pestis, though not with Lactobacillus casei, an aerotolerant anaerobe (Supplementary Figure 3). When a similar experiment was conducted at a density corresponding to the ‘dispersed’ population, complete O2 depletion was not achieved over the measurement period (Figure 2f). The co-incubation of neutrophils with S. flexneri at a pathophysiological cell density (8.1.107 ± 5.4.107 cell/mL, Figure 2d) had no impact on the timing of complete O2 depletion compared to S. flexneri alone (clustered, tanoxia = 2.4 ± 1.0 min, p > 0.05, Figure 2g). A mild increase of O2 consumption was observed when co-incubating neutrophils with S. flexneri at a cell density corresponding to the ‘dispersed’ population or heat-killed bacteria (t = 10 min, p < 0.01 and p < 0.05, respectively), although complete O2 depletion was not achieved over the measurement period (Figure 2g). We demonstrate here that O2 depletion during infection seems driven by S. flexneri, while neutrophils contribute comparatively much less.
We reasoned that S. flexneri aerobic respiration is responsible for the O2 depletion observed in vivo. At the end of the aerobic respiratory chain, O2 is reduced to H2O by terminal cytochrome oxidases14: S. flexneri expresses two cytochrome oxidases named bd-I (CydAB) and bd-II (AppCB), similarly to E. coli, (S. flexneri cyoAB genes are truncated) (Figure 3a). Therefore, we engineered S. flexneri ∆cydAB and ∆appCB mutants and noted that their cellular morphologies were indiscernible from the wild-type (Supplementary Figure 4a).
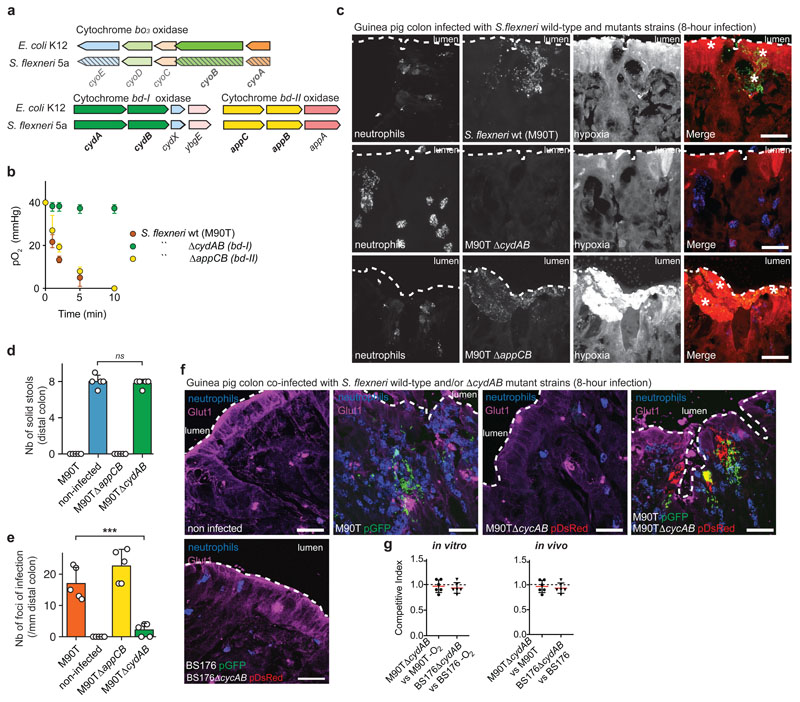
Figure 3. Aerobic respiration is required for hypoxia induction and efficient colonic mucosa colonization by Shigella in vivo.
a. Genetic organization of cytochrome oxidases (bo3, bd-I and bd-II) encoding genes in Escherischia coli K12 and Shigella flexneri 5a. cyoA and cyoB genes are not functional in S. flexneri due to genetic insertions.
b. S. flexneri 5a ∆cydAB (bd-I) was unable to consume O2 in vitro in a RPMI 1640 + 10 mM Hepes medium stabilized at a pO2 = 40 mmHg and had a growth defect in the presence of O2 (n = 3 independent experiments, p < 10-8, 2-way ANOVA test, see Supplementary Figure 4c and Supplementary Table 5). O2 tensions expressed as mean ± S.D.
c. S. flexneri 5a ∆cydAB (bd-I) pGFP (green) was avirulent in vivo compared to wild-type and ∆appCB (bd II) mutant strains. Hypoxia was detected with a-EF5-Cy3 (red), neutrophils with Myelotracker-Dylight40512 (blue). Scale bar: 50 µm. This experiment was repeated independently three times with similar results.
d. Tissues infected with the ∆cydAB strain were not inflamed as compared to the wild-type strain, as indicated by the presence of solid stools in the colon lumen (mean ± S.D., p > 0.8, see Supplementary Table 6).
e. The number of foci of infection was significantly reduced in tissues infected by the ∆cydAB strain compared to the wild-type strain (mean ± S.D., p < 10-4, see Supplementary Table 7).
f. GLUT-1 was detected with a monoclonal antibody (magenta) within S. flexneri 5a pGFP (green) hypoxic foci of infection. S. flexneri 5a ∆cydAB pDsRed (red) was able to colonize the colonic mucosa upon co-infection with the wild-type strain (S. flexneri 5a pGFP). Plasmid-cured BS176 and BS176∆cydAB remain in the luminal compartment. Neutrophils were labeled with Myelotracker-Dylight40512 (blue). Scale bar: 70 µm. See Supplementary Figure 5 for extended imaging. This experiment was repeated independently three times with similar results.
g. M90T/M90T∆cydAB and BS176/BS176∆cydAB Competitive Indexes were determined in co-cultures in vitro (-O2 conditions) and upon guinea pig co-infection in vivo (lumen and colonic mucosa). CI=1 means no growth difference (mean ± S.D., n=5 biologically independent animal samples). *** indicates p < 0.001.
We showed that the bd-I complex (CydAB), but not the bd-II complex (AppCB), was essential for S. flexneri’s ability to consume O2 (Figure 3b). Consistently, cytochromes b and d expressed by S. flexneri belong mainly to the bd-I complex (Supplementary Figure 4b), and S. flexneri ∆cydAB displayed a growth defect in the presence of O2 compared to the wild-type strain, confirming that aerobic respiration is defective in this mutant (Supplementary Figure 4c). The S. flexneri ∆cydAB (bd-I) mutant was not virulent in vivo, O2 depletion was not observed, and foci of infection were not formed in the colonic mucosa (Figure 3c-d); although it remained invasive and was phagocytized by neutrophils as per the wild-type strain in vitro (Supplementary Figure 4d). Consistent with previous results (Figure 2f-g), the co-incubation of neutrophils (with or without DPI, a neutrophil NADPH oxidase inhibitor) with S. flexneri ∆appCB (bd-II) had no impact on timing of anoxia induction (Supplementary Figure 4e). When similar experiments were conducted with S. flexneri ∆cydAB or upon activation with N-Formyl-Met-Leu-Phe (FmlF), a mild increase of oxygen consumption was observed and abolished by DPI (Supplementary Figure 4e, p < 0.05); although anoxia was not reached in both conditions over the measurement period.
We confirmed that GLUT-1, a hypoxia-induced glucose transporter7, was overexpressed in non-infected epithelial cells (physiological hypoxia), but also within S. flexneri foci of infection (Figure 3f and Supplementary Figure 5a) though not upon S. flexneri ∆cydAB mutant infection (Supplementary Figure 5a). To confirm that S. flexneri ∆cydAB avirulence was caused by its defect in O2 consumption, we co-infected guinea pigs with S. flexneri pGFP wild-type and ∆cydAB pDsRed mutant strains. We confirmed that hypoxia was induced within foci of infection (GLUT-1 stabilization, Figure 3f), mediated by S. flexneri wild-type strain, and we demonstrated that, in this context, the S. flexneri ∆cydAB mutant was able to colonize the colonic mucosa together with the wild-type strain (Figure 3f and Supplementary Figure 5b). The relative amount of S. flexneri wild-type and ∆cydAB mutant strains in the lumen (comparing plasmid-cured BS176 and BS176∆cydAB mutant strains, Figure 3f and Supplementary Figure 5b) or within the colonic mucosa (comparing M90T and M90T∆cydAB mutant strains, Figure 3f and Supplementary Figure 5b) was similar (Competitive Index value around 1), as observed in vitro under anoxic conditions (Figure 3g). We therefore confirmed our hypothesis that S. flexneri aerobic respiration was the primary cause of hypoxia induction in vivo, explaining its essential role for tissue colonization15. The ability of S. flexneri to grow in the absence of O2 supports the expansion of foci, whose formation appears to be the preferential strategy for tissue colonization (composed of 99.71% total bacteria, Supplementary Figure 1a).
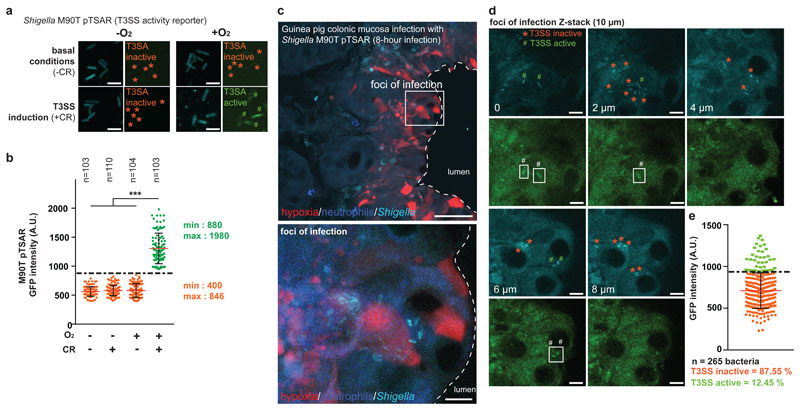
We hypothesized that the repression of S. flexneri’s dissemination capacity may promote foci of infection extension by favoring colonizing over exploring bacteria. S. flexneri’s ability to invade host cells relies on the T3SS, whose activity is significantly reduced in hypoxic conditions1. To assess the level of T3SS activity of individual bacterium within colonic tissues, we measured the fluorescence signal produced by a S. flexneri strain harboring the pTSAR plasmid, which has a fast-maturating eGFP under the control of a T3SS activity-dependent promoter16.
The positive correlation between O2 availability and the T3SS-activity in the presence of the secretion inducer Congo red (congo red, +CR) was confirmed in vitro (fast-maturating eGFP fluorescent signal and IpaB secretion, Figure 4a-b, p<0.001). We also previously confirmed oxygen-dependent type III secretion of effectors in vitro by western blot1. The activity of S. flexneri T3SS was also assessed in vivo within foci of infection (Figure 4c). Performing a single-bacteria analysis, we demonstrated that S. flexneri T3SS was mostly inactive in infected tissues (87.55% total bacteria, Figure 4d-e, n = 265 bacteria). These results confirm that hypoxic conditions encountered in foci of infection blocks T3SS activation, hence limiting S. flexneri dissemination to adjacent host cells and promoting tissue colonization through foci expansion.
Figure 4. S. flexneri T3SS is inactive in the colonic mucosa, supporting foci of infection extension.
a-b. O2-dependent modulation of S. flexneri Type III secretion system (T3SS)1 was confirmed in vitro in the presence of congo red (0.01%) using the activity reporter pTSAR16 (bacteria are constitutively cyan (CFP); bacteria with an active T3SS have a high pTSAR signal (eGFP, green, #), not bacteria with an inactive T3SS (*)); T3SS was active at a GFP signal intensity above 900 AU, as indicated by a 1-way ANOVA test (see Supplementary Table 8, *** indicates p < 0.001. Red lines and black bars: mean ± standard deviation.)
c. T3SS activity was assessed at a single-bacteria level in vivo, in hypoxic foci of infection formed by S. flexneri pTSAR This experiment was repeated independently three times with similar results.
d-e. Bacteria with an active T3SS are indicated in the green channel (#) in Z-stack image series, and bacteria with an inactive T3SS are indicated in the cyan channel (*). Most of S. flexneri pTSAR individual bacteria detected within hypoxic foci of infection have an inactive T3SS (*, 87.66% total population with eGFP signal intensity above 900 AU, n = 286 bacteria. Red and black bars are mean ± S.D. n = 3 biologically independent animal samples).
Here we show that intestinal mucosa colonization by S. flexneri occurs through the formation and expansion of hypoxic foci of infection. In summary, while O2 is essential for S. flexneri T3SS-dependent epithelium invasion1, its depletion is conversely mandatory for subsequent tissue colonization. The ability of S. flexneri to survive this low-oxygenated environment, as well as the associated blockade of the T3SS, contributes to the expansion of foci of infection.
Our results reveal that the adaptation of invasive pathogens to various O2 levels is essential for tissue colonization. In fact, to our knowledge, pathogenic microorganisms that are not facultative anaerobes are not invasive. We demonstrate that O2 depletion by pathogenic enterobacteria may be considered a community behavior17, querying previous concepts such as the O2 availability in colonic inflammation models and bacteria adaptation to these pathophysiological environments18. In particular, improved methods will be required to study the population heterogeneity within foci in regards to O2 levels. For example, more than 40% of individual bacteria were neither ‘clustered’ nor ‘dispersed’ in neutropenic animals (Supplementary Table 1), highlighting the importance of foci formation in response to neutrophil recruitment and antimicrobial activity.
O2 depletion by pathogenic enterobacteria is expected to modulate the survival and function of immune cells19,20, particularly neutrophils2,3,4; important O2-dependent antimicrobial functions, such as ROS production, are likely to be altered. The contribution of alternative electron transfer machineries for oxygen-depletion and enteropathogen virulence, such as the recently described Listeria monocytogenes extracellular electron transport (EET)21, requires further investigation. The respective contributions of enteropathogens, commensal flora, and host cells (i.e. neutrophil activation, epithelial cell death, etc.) in oxygen depletion may be further investigated in other acute or chronic models of inflammation. In conclusion, this “battle to breathe” is anticipated to be crucial for the outcome of infection and will have to be taken into account in the development of truly effective antibiotics or vaccines targeting pathogens in low-O2 environments for efficient clearance or protection.
Material and Methods
Bacterial strains, growth conditions
Shigella flexneri 5a (Shigella) strains (wild-type M90T, plasmid-cured BS176 M90TΔfnr mutant, and BS176Δfnr mutant) and Staphylococcus aureus were propagated in Trypticase soy media (TCS, Oxoid) or on TCS plates, supplemented with Congo Red (0.01%, Sigma) for Shigella strains. Escherischia coli, Salmonella typhimurium, and Listeria monocytogenes were cultured in Luria Bertani (LB) medium. Lactobacillus casei was grown in deMan, Rogosa and Sharpe (MRS) broth (Sigma-Aldrich). S. flexneri M90T was used as a wild-type Shigella strain. Targeted deletion of appCB and cydAB genes was performed using λ red recombination using the following primers appCB1 (5’-ATGTGGGATGTCATTGATTTATCGCGCTGGCAGTTTGCTCCATATGAATATCCTCCTTAGTTCC-3’) and appCB2 (5’-TTAGTACAACTCGTTTTTGTTACGGCGGAGAGTTTCTGTTGTGTAGGCTGGAGCTGCTTC-3’) with plasmid pKD3 to generate a PCR product containing a chloramphenicol resistance cassette flanked on each end with 50bp corresponding to the S. flexneri appCB genes. Primers cydAB1 (5’-TTAGTACAGAGAGTGGGTGTTACGTTCAATATCTTCTTTGCATATGAATATCCTCCTTAGTTCC-3’) and cydAB2 (5’-ATGTTAGATATAGTCGAACTGTCGCGCTTACAGTTTGCCTGTGTAGGCTGGAGCTGCTTC-3’) were used with plasmid pKD4 to generate a PCR product containing a kanamycin resistance cassette flanked on each end with 50bp corresponding to the S. flexneri cydAB genes. KO generation was performed under anoxic conditions to limit potential toxicity from inactivating the respiratory pathway. Isolated mutants were passaged on TSA plates containing 0.01% congo red to confirm virulence plasmid maintenance.
Shigella flexneri 5a fluorescent strains (wild-type M90T, M90TΔcydAB, plasmid-cured BS176, BS176ΔcydAB, M90TΔfnr mutant, and BS176Δfnr mutant) were generated by transforming the pFPV25.1 (AmpR) or pDsRed (AmpR) plasmids, as indicated. Antibiotics were added as necessary at the following concentrations: chloramphenicol, 50 µg/mL; ampicillin, 100 µg/mL; kanamycin 50 µg/mL. Experiments under different oxygen (O2) concentrations were performed in an anaerobic cabinet (Don Whitley DG250) or in a cabinet in which O2 tensions can be controlled (Don Whitley H35 Hypoxystation).
To assess the level of the T3SS activity, S. flexneri 5a was transformed with pTSAR16, allowing the constitutive expression of CFP (cyan) and inducible expression of the fast maturating eGFP (dependent of the T3SS activity).
Cytochrome quantification
The cellular content of cytochromes b and d hemes were calculated as described by Dejean et al.22, taking into account the respective molar extinction coefficient values (18.0 kM-1cm-1 and 27.9 kM-1cm-1, respectively) and the reduced minus oxidized spectra recorded using a dual beam spectrophotometer (Aminco DW2000). Briefly, S. flexneri wt, ∆cydAB, and ∆appCB mutants strains were grown until OD600 = 2 was reached. For each measurement, 100 OD units were centrifuged for 10 minutes at 5000 rpm, bacterial pellets were resuspended in distilled water (dH2O) up to 2 ml; 1 mL for the oxidized condition and 1 mL for the reduced condition (dithionite addition). The cytochrome maximum and minimum absorbance values were measured at 560 and 575, and 630 and 655 nm for cytochromes b and d, respectively, and expressed in mol/OD.
Cell culture
Blood collection, Neutrophils purification
All participants gave written, informed consent in accordance with the Declaration of Helsinki principles. Peripheral Human blood was collected from healthy patients at the ICAReB service of the Institut Pasteur (authorization DC No.2008-68). Human blood was collected from the antecubital vein into tubes containing sodium citrate (3,8% final) as anticoagulant molecules. Guinea pig blood samples were collected on anesthetized animals by cardiac puncture.
Human or guinea pig neutrophils were purified as described previously23. Briefly, whole blood samples were centrifuged at 450x g for 15 min. Platelet rich plasma (PRP) was collected and centrifuged at 2500x g for 20 min to form platelet poor plasma (PPP). Blood cells were resuspended in NaCl 0.9% and dextran sulfate (0.72%). After 30 min sedimentation, the neutrophil-containing upper layer was centrifuged at 240x g for 10 min. The resuspended pellet was separated on a 2-layers Percoll (GE Healthcare) gradient (51%/42%) by centrifugation at 240x g for 20 min. Neutrophils were collected, and remaining red blood cells were removed using CD235a (glycophorin) microbeads (negative selection, Miltenyi Biotec).
For functional assays, human or guinea pig purified neutrophils were cultured in RMPI 1640 (Life Technologies) supplemented with 10 mM Hepes (Life Technologies).
Epithelial cell culture
The Hep2 cell line was purchased at the ATCC and was tested for mycoplasma contamination. Hep2 epithelial cells were cultured in a DMEM medium (ThermoFischer) supplemented with 10 % Fetal Calf Serum (ThermoFischer). For functional assays, Hep2 cells were incubated in DMEM medium supplemented with 10 mM Hepes, without FCS.
Antibodies and fluorescent dyes
Hypoxia detection in guinea pigs was achieved by 2-nitroimidazole derivative EF5 molecule (University of Pennsylvania, USA) accumulation in tissues. An intracardiac injection (1.5 mL for 150 g guinea pig) of a 10 mM EF5 solution was performed one hour prior to Shigella challenge.
EF5 was immunodetected on fixed cells or tissues using an α-EF5 antibody (ELK3-51) conjugated with a Cy3 fluorophore (ready-to-use solution, University of Pennsylvania, USA). Dapi (Invitrogen) was used at 1 µg/mL. Neutrophils were labeled with the MUB40-Dylight405 marker12, binding to lactoferrin stored in specific and tertiary granules. GLUT-1 was detected with a mouse monoclonal antibody (Abcam, ab40084), used at 1:40 dilution and a secondary antibody conjugated with an Alexa647 fluorophore (Thermofisher Scientific).
Bacterial cell infection in vitro
Cell entry assays, Hep2 epithelial cells were seeded (5. 105 cells/well) and grown overnight in 24-well tissue culture plates in indicated conditions (0, 5% and 21% O2). Neutrophils were directly seeded (5. 105 cells/well) in 24-well tissue culture plates. Cells were challenged at an MOI of 100 (Hep2) or 20 (neutrophils) with bacteria grown until the mid-log phase was reached in liquid culture under the same conditions. Bacteria were spun briefly onto cells by centrifugation at 2000x g for 10 min.
To measure bacterial entry, cells were challenged as above, incubated for 30 min at 37ºC, after which gentamicin (50 µg/ml) was added for 30 min to kill extracellular bacteria. Cells were then washed three times with PBS, lysed with 1 mL 1% saponin in PBS, and bacteria recovered by plating to solid media. Results are expressed as the number of CFU/ml of cell lysate and are the average of at least three independent experiments performed in triplicate. Statistical significance was calculated using the Student’s T-test.
In vitro O2 quantification
Gas proof glass tubes were first left in an anaerobic cabinet to remove all traces of exogenous oxygen and generate an anoxic atmosphere. The tubes were then filled with RPMI 1640 culture medium supplemented with 10 mM Hepes pre-stabilized at 37ºC and pO2 = 40 mmHg. Bacteria and neutrophils were prepared in similar conditions in adequate culture media and resuspended in the prepared glass tubes. Direct oxygen quantification was performed with a Clark-type sensor mounted with a needle (Unisense oximeter) over time and initiated immediately after bacteria or cell inoculation. Results are averaged from at least three independent experiments.
Animal models of shigellosis
Rabbit ligated ileal loop model
Experiments on rabbits were approved by the Institut Pasteur local ethic committee (CETEA).
For evaluation of competitive indices, experiments were performed on independent occasions in at least two ileal loops in four animals in total. For competitive indices calculations, bacteria were grown in liquid media for three hours at 37°C, and a total 107 CFU of bacteria consisting of equal numbers of two or three strains in 500 μl of physiological water were injected into loops. After an 18-hour infection, animals were sacrificed, and bacteria were recovered from the loops. Series of dilutions of the outputs were plated onto TCS plates containing antibiotics and congo red (CR), allowing the selection and counting of M90T (CR+ (red colony), CmR, KmS), M90TΔfnr (CR+ (red colony), CmR, KmR), BS176 (CR- (white colony), CmR, KmS), BS176Δfnr (CR- (white colony), CmR, KmR). The competitive index was calculated as the ratio of mutant to wild-type bacteria recovered from animals divided by the ratio in the inoculum; the results are the average of at least four samples originating from four different rabbits. For single strain infections (M90T, BS176, Δfnr, and BS176Δfnr), 107 CFU were inoculated in ligated loops. After an 18-hour infection, animals were sacrificed and infected ileal loops collected. For immunohistochemistry, ileal loops were fixed in 4% buffered formalin, embedded in paraffin, and 5 µm sections obtained and stained with haematoxylin-eosin. Shigella strains were detected using a primary mouse anti-LPS antibody and an HRP-conjugated anti-mouse antibody (Dako).
Guinea pig intrarectal infection
Experiments on guinea pigs were approved by the Institut Pasteur local ethic committee (CETEA).
Young conventional guinea pigs (3 weeks, female, Dunkin-Hartley, < 150g) were used for Shigella infection and hypoxia detection studies23. When indicated, neutropenia was generated in guinea pigs by two intraperitoneal injections of cyclophosphamide (100 mg/kg on day 7 and 50 mg/kg on day 1). Shigella infection was achieved by an intrarectal challenge of animals with 1010 CFU exponentially grown. Infection proceeded for 8 hours before animals were sacrificed and the distal 10 cm of colon harvested and then flushed with 4% paraformaldehyde (PFA) in 1xPBS and inverted on wooden skewers. Tissues were kept in 4% PFA-1xPBS for 1- to 2-hours to complete fixation and then incubated in 1xPBS-glycine (100 mM) for 30 minutes to quench the PFA. Tissues were immersed successively in 15% and 30% sucrose at 4°C overnight and then released from the skewers by a longitudinal incision and prepared as swissrolls7. Swissrolls were embedded in Tissue-Tek OCT compound (Sakura) using a flash-freeze protocol and frozen at -80°C. For standard immunofluorescence staining, 10 µm sections were obtained using a cryostat CM-3050 (Leica). For hypoxia quantitative detection, 30 µm sections were generated.
Competitive Indexes were assessed and calculated in vivo on homogenized luminal and colonic mucosa bacteria content (M90T vs M90TΔcydAB mutant and BS176 vs BS176ΔcydAB mutant).
Colonic tissue processing for Quantitative analysis
Immunostaining
For IF samples used for quantitative analyses, transversal colon sections of 30 μm thickness were stained using a modified protocol from Arena and colleagues24. Briefly, tissues on slides were fixed with 4% PFA for 10 minutes at room temperature. They were then washed with 1xPBS and stained using an α-EF5-Cy3 antibody solution (supplied by University of Pennsylvania) supplemented with 0.1% Triton, 1% BSA, and MUB40-Dylight405 (1 µg/mL) overnight at 4°C. The following day, the slides were washed with 1xPBS and mounted with ProLongGold® (Invitrogen).
Imaging
For quantification, fluorescence 3D images were acquired with an automated spinning-disk microscope (CellVoyager 1000, Yokogawa Electrics, Japan). The swissrolls were first scanned with a 10x air objective (Olympus, UPLSAPO 10x 0.4NA air) in bright-field to locate the tissue, followed by a scan to locate GFP-expressing S. flexneri. Infection foci were imaged with a 40x oil objective (Olympus, UPLSAPO 40x 1.3NA oil) over 3 fluorescence channels (405, GFP, AF568) and bright field, capturing a 40 µm thick section to encompass the whole tissue. Full fields made of 1x2 to 7x5 3D tiles of 200 µm × 200 µm × 40 µm each were acquired depending on the size of the given site.
Image Quantitative analysis
Hypoxia profiles through colonic mucosa
The lamina propria was delineated using Fiji25. Intensity profiles were generated from 10 µm thick lines on maximum-intensity projections of the 3D images in the EF5 channel averaged with a Gaussian filter with σ = 10 µm (Figure 1a). The profiles ran into the tissue, orthogonally to the lamina propria, and their location was selected to traverse an infectious focus for infected tissues and randomly arranged for non-infected tissues.
The hypoxia curves of Figure 1b are generated by measuring several profiles that run through the LP from the apical surface to the basal surface (Figure 1a). Within one condition (infected or non-infected), all profiles are aligned with respect to the first local maximum (that crosses the LP surface). After alignment, we have the EF5 signal distribution - one value per profile - as a function of the tissue depth. We then compute the mean EF5 signal (thick line on Figure 1b) and its standard deviation (shaded area on Figure 1b) and report them as function of the tissue depth.
We then perform a Student t-test at each position along the tissue depth, comparing the distribution of EF5 signal at a given depth between profiles through tissues of infected (red) and non-infected (blue) animals, for all depths. We then report the range of depths for which this test reveals significant differences as black horizontal line over the plot. Figure 1b displays one of three repetitions. The infected and not-infected curves are made respectively of averaging 128 and 61 profiles. Sample size was adjusted to reach statistical differences between compared conditions. A blind analysis was used for image acquisition.
Hypoxia measurements at single-cell level
Automatic segmentation of individual S. flexneri and neutrophils was carried out using TrackMate26, yielding their X, Y, and Z coordinates and their local hypoxia levels measured from the EF5 channel averaged with a 3D Gaussian filter with σ = 2 µm. Together, bacterial and neutrophil positions and the manual annotations were processed using MATLAB (The MathWorks) for further analysis (Figure 2 and Figure 3).
We inspected infection foci manually and estimated their radius to be approximately 16 µm. Clustered bacteria were therefore defined as bacteria that have at least 6 other bacteria in a neighborhood of 16 µm in radius from their position (Supplementary Figure 1a). Isolated bacteria were defined as those having no other bacteria in a neighborhood of a 30 µm radius from their position. Bacteria that did not fall into these two categories were not included in the analysis (Supplementary Table 1).
Mean hypoxia spatial profile generated by bacteria
The hypoxia profile around single-bacteria was calculated by averaging the EF5 signal (filtered with σ = 2 µm) of all pixels at a fixed distance from the closest bacterium (Supplementary Figure 1b). Pixel distance to the closest bacterium was calculated thanks to the image distance transform, taking into account the non-isotropy of the pixel size (0.2 µm/pixel in XY and 1 µm/pixel in Z). Locations outside of the tissue were not included in the profile. Pixels were then pooled in 1-µm bins from 0 to 50 µm. All the EF5 intensity of pixels in a single bin contribute to the mean and standard deviation at the bin distance and build the profiles of Supplementary Figure 2d. Single bins incorporated between 600 and 3. 106 measurements for a single image. This procedure was repeated on 16 images, treating separately clustered and dispersed bacteria. These hypoxia profiles average EF5 levels over all pixels at a specified distance, hereby canceling contributions from other potential sparse hypoxia sources such as neutrophils.
To investigate whether or not neutrophils contribute a significant addition to this hypoxia, we sought for each neutrophil the closest bacterium and categorized them as dispersed or clustered. We then reported on the hypoxia profile, the EF5 level, of the neutrophil at the measured distance to the closest bacterium, respectively, whether the closest bacterium was dispersed or clustered. A one-sided z-test was used to determine whether or not the neutrophil hypoxia could be explained by the bacteria hypoxia profile at this distance (dots) or if it was significantly larger (crosses, significance level at 5%). Less than 5% of neutrophils had a significantly higher hypoxia levels than what was generated by bacteria over 16 repetitions of this analysis.
Bacteria and neutrophil density
Whole tissue densities (Supplementary Figure 2a,c-d) were calculated by counting the number of neutrophils or bacteria, regardless of their clustering status, in the total tissue volume imaged for each experimental condition.
Clustered Shigella density (Figure 2d) was calculated by counting the number of neighbors in a sphere of radius 16 µm around the 61119 clustered bacteria detected in the conventional animals, the M90T + EF5 condition. Dispersed Shigella density was calculated by counting the number of dispersed bacteria in the volume of the imaged tissue averaged over the 37 tissue sections of the same condition. Neutrophil density was calculated by counting the number of neutrophils in the volume of the imaged tissue averaged over a subset of 16 tissue sections of the same condition.
T3SS activity reporter signal analysis
We expressed the TSAR T3SS activity reporter2 in S. flexneri wild-type strain; EF5 injected guinea pigs were infected with this strain as described above. The reporter emits in the Cyan channel (CFP) for all bacteria and in the Green channel for bacteria that are positive for TS3 activity (fast-maturating eGFP, Rep+). GFP level corresponding to an active T3SS (Rep+) was determined in vitro by culturing S. flexneri 5a pTSAR in the presence or absence of oxygen, with or without congo red (0.01%). More than 100 individual bacteria were analyzed in each condition in vitro. We then imaged tissue sections, focusing on bacteria foci and proceeded to a similar analysis. We quantified signals in 2488 WT bacteria located in foci of infection.
Statistical analysis
All statistical analyses have been performed with MATLAB software (The MathWorks). Comparisons with two groups (Figures 1b, 1h, 1i, 2b) were supported with Student unpaired-t tests. Comparisons with three groups or more were supported by 1-way ANOVA test (Figures 1f, 3d, 3e, 4b and Supplementary Figures 4b,d-e) and 2-way ANOVA test (Figures 2g, 3b and Supplementary Figure 4c), followed by a post hoc Tukey test.
When p-values for the significance of pairwise comparisons were calculated (either from a Student-t test or a post hoc Tukey test after an ANOVA test) and displayed on figures, we used the following convention:
ns not significant; p ≥ 0.05;
*p < 0.05 & p ≥ 0.01;
** p < 0.01 & p ≥ 0.001;
*** p <0.001.
Box-plots
When results are presented in the shape of a box-plot (Figures 1f, 1h, 1i, 2b), the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points not considered outliers, and the outliers are not plotted. Outliers are data points that are further away than 1.5 times the range from the 1st to the 3rd quartile, respectively above or below the 3rd and 1st quartile.
Direct oxygen consumption quantification in vitro
Direct oxygen quantifications were performed in gas-proof glass punchable tubes (BD Vacutainer Z, reference 368430) using an oximeter (OXY-Meter, Unisense) combined with a needle sensor for piercing (OX-NP). Data was recorded using the SensorTrace logger software (Unisense). Glass tubes were first opened in an anaerobic cabinet (MiniMac DG250, Don Withley) for 30 min to ensure that all traces of exogenous oxygen were removed from the tube (anoxic atmosphere). Such conditioned tubes were subsequently filled with 2 mL of a culture medium (RPMI 1640 + 10 mM Hepes for neutrophils), pre-stabilized at 37°C and at pO2 = 40 mmHg (5% O2) in a hypoxic chamber (In vivo 500, Don withley) (Figure 2e). Prior to their inoculation, bacteria (S. flexneri 5a wild-type and mutant strains, E. coli, S. typhimurium, L. monocytogenes, S. aureus or L. casei) were grown in similar conditions until OD600 = 0.3 was reached. Bacteria alone or with neutrophils were resuspended at the indicated concentrations in RPMI 1640 + 10 mM Hepes. When indicated Diphenyleneiodonium chloride (DPI, Sigma-Aldrich) was added at a 25 μM final concentration, N-Formyl-Met-Leu-Phe (FmlF, Sigma-Aldrich) at 1 μM. Direct oxygen quantification were performed over time and initiated immediately after bacteria or cell inoculation. Results were averaged from at least three independent measurements.
Supplementary Material
Acknowledgements
We acknowledge France-BioImaging infrastructure supported by the French National Research Agency (ANR-10-INBS-04, Imagopole, JYT), ANR JCJC 2017-17-CE15-0012 (BSM), and the European Research Council (ERC Grant n°232798: 2009-AdG HOMEOEPITH, PJS). E.T.A. was a Pasteur Foundation and Pasteur-Roux Fellow.
Footnotes
Competing interest statement
The authors declare no competing interests
Author contributions
BSM, JYT and ETA designed the experiments, interpreted the data, and wrote the paper. MA designed Shigella mutants. GN, LI, AA, MF and FXCV contributed to their study in vitro and in vivo. JYT conducted quantitative analysis of the data. AD, SLS and PSJ contributed to data interpretation.
Data availability.
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1.Marteyn B, et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature. 2010;465:355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cramer T, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peyssonnaux C, et al. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J Clin Invest. 2005;115:1806–1815. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walmsley SR, et al. Hypoxia-induced neutrophil survival is mediated by HIF-1alpha-dependent NF-kappaB activity. J Exp Med. 2005;201:105–115. doi: 10.1084/jem.20040624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huether SE, McCance KL. Understanding Pathophysiology. Elsevier Health Sciences; 2015. [Google Scholar]
- 6.Karhausen J, et al. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J Clin Invest. 2004;114:1098–1106. doi: 10.1172/JCI21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell EL, et al. Transmigrating Neutrophils Shape the Mucosal Microenvironment through Localized Oxygen Depletion to Influence Resolution of Inflammation. Immunity. 2014 doi: 10.1016/j.immuni.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arena ET, Tinevez J-Y, Nigro G, Sansonetti PJ, Marteyn BS. The infectious hypoxia: Occurrence and causes during Shigella infection. Microbes Infect. 2017 Mar;19(3):157–165. doi: 10.1016/j.micinf.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Ziemer LS, et al. Noninvasive imaging of tumor hypoxia in rats using the 2-nitroimidazole 18F-EF5. Eur J Nucl Med Mol Imaging. 2003;30:259–266. doi: 10.1007/s00259-002-1037-5. [DOI] [PubMed] [Google Scholar]
- 10.Bumann D. Heterogeneous host-pathogen encounters: act locally, think globally. Cell Host Microbe. 2015;17:13–19. doi: 10.1016/j.chom.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Davis KM, Isberg RR. Defining heterogeneity within bacterial populations via single cell approaches. Bioessays. 2016;38:782–790. doi: 10.1002/bies.201500121. [DOI] [PubMed] [Google Scholar]
- 12.Anderson MC, et al. MUB40 Binds to Lactoferrin and Stands as a Specific Neutrophil Marker. Cell Chem Biol. 2018 Apr 19;25(4):483–493. doi: 10.1016/j.chembiol.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Sheridan WG, Lowndes RH, Young HL. Intraoperative tissue oximetry in the human gastrointestinal tract. Am J Surg. 1990;159:314–319. doi: 10.1016/s0002-9610(05)81226-7. [DOI] [PubMed] [Google Scholar]
- 14.Unden G, Trageser M. Oxygen regulated gene expression in Escherichia coli: control of anaerobic respiration by the FNR protein. Antonie Van Leeuwenhoek. 1991;59:65–76. doi: 10.1007/BF00445650. [DOI] [PubMed] [Google Scholar]
- 15.Way SS, Sallustio S, Magliozzo RS, Goldberg MB. Impact of either elevated or decreased levels of cytochrome bd expression on Shigella flexneri virulence. J Bacteriol. 1999;181:1229–1237. doi: 10.1128/jb.181.4.1229-1237.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell-Valois F-X, et al. A fluorescent reporter reveals on/off regulation of the Shigella type III secretion apparatus during entry and cell-to-cell spread. Cell Host Microbe. 2014;15:177–189. doi: 10.1016/j.chom.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Davis KM, Mohammadi S, Isberg RR. Community behavior and spatial regulation within a bacterial microcolony in deep tissue sites serves to protect against host attack. Cell Host Microbe. 2015;17:21–31. doi: 10.1016/j.chom.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes ER, et al. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe. 2017;21:208–219. doi: 10.1016/j.chom.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nizet V, Johnson RS. Interdependence of hypoxic and innate immune responses. Nat Rev Immunol. 2009;9:609–617. doi: 10.1038/nri2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor CT, Colgan SP. Regulation of immunity and inflammation by hypoxia in immunological niches. Nat Rev Immunol. 2017;17:774–785. doi: 10.1038/nri.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Light SH, et al. A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature. 2018;562:140–144. doi: 10.1038/s41586-018-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dejean L, Beauvoit B, Guérin B, Rigoulet M. Growth of the yeast Saccharomyces cerevisiae on a non-fermentable substrate: control of energetic yield by the amount of mitochondria. Biochim Biophys Acta. 2000;1457:45–56. doi: 10.1016/s0005-2728(00)00053-0. [DOI] [PubMed] [Google Scholar]
- 23.Monceaux V, et al. Anoxia and glucose supplementation preserve neutrophil viability and function. Blood. 2016;128:993–1002. doi: 10.1182/blood-2015-11-680918. [DOI] [PubMed] [Google Scholar]
- 24.Arena ET, et al. Bioimage analysis of Shigella infection reveals targeting of colonic crypts. Proc Natl Acad Sci USA. 2015;112:E3282–90. doi: 10.1073/pnas.1509091112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tinevez JY, et al. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.