Abstract
Genetic interactions identify combinations of genetic variants that impinge on phenotype. With whole-genome sequence information available for thousands of individuals within a species, a major outstanding issue concerns the interpretation of allelic combinations of genes underlying inherited traits. In this review, we discuss how large-scale analyses in model systems have illuminated the general principles and phenotypic impact of genetic interactions. We focus on studies in budding yeast, including the mapping of a global genetic network. We emphasize how information gained from work in yeast translates to other systems, and how a global genetic network not only annotates gene function, but also provides new insights into the genotype to phenotype relationship.
[A]. Context
Geneticists have long appreciated the major impact that genetic interactions (GIs) can have on the phenotypic landscape of a population. In 1909, William Bateson ascribed ‘epistasis’ to a specific type of GI, whereby one mutation masks the effects of another mutation, such that a double mutant resembles the more extreme single mutant rather than the expected combined effect of both mutations, a concept useful for inferring biological pathways. Ronald Fisher generalized this term to include any multi-locus mutant effect that deviates from the expected phenotype associated with combining of the corresponding individual loci (Fisher, 1918). While the term epistasis has since evolved many different meanings, Fisher’s generalized concept of epistasis informs how we define a GI today.
GIs are thought to underlie diverse aspects of biology, including the evolution of sex, speciation, and complex disease (Phillips et al., 2000). Despite their central importance, and the ability for comprehensive genotyping through whole genome sequencing, the mapping of critical GIs associated with the natural variation within an individual’s genome remains incredibly difficult (Zuk et al., 2012). However, inbred model systems, such as yeast and worms, as well as cell cultures derived from fruit flies and mammals, provide an experimental format for mapping GIs systematically through the utilization of genome-wide collections of either defined mutants or gene perturbation systems (e.g., libraries of double-stranded RNAs, shRNAi, CRISPR-based approaches)(Bassik et al., 2013; Blomen et al., 2015; Byrne et al., 2007; Costanzo et al., 2016; Dixon et al., 2008; Du et al., 2017; Fischer et al., 2015; Han et al., 2017; Horlbeck et al., 2018; Lehner et al., 2006; Najm et al., 2018; Roguev et al., 2008; Shen et al., 2017; Vizeacoumar et al., 2013). Importantly, with prior knowledge of the general principles of genetic networks derived from model systems, it may be possible to map GIs and networks based upon natural variation of individuals in outbred populations.
The concept of GIs is simple, but the physiological repercussions can be profound. A GI between two genes occurs when an allele of one gene combines with an allele of another gene to generate a double mutant with an unexpected phenotype. The phenotype may be the exacerbation of the expected combined single mutant phenotypes, a so-called negative GI. The most extreme negative GI phenotype is ‘synthetic lethality’, which occurs when the combination of two mutations, neither by itself lethal, causes lethality. Geneticist Theodosius Dobzhansky first coined the term ‘synthetic lethality’, when he observed that a cross of outbred flies, isolated from natural populations, revealed a lethal developmental defect that could be traced back to two different genes, one from each parent (Dobzhansky, 1946).
Decades later, the yeast community began to embrace the potential of synthetic lethal GIs to reveal functional relationships. Pioneering genetic screens from David Botstein’s group were aimed at revealing the genetic network controlling cytoskeletal dynamics in yeast. Some of the first examples of synthetic lethality were observed for a combination of partial loss-of-function mutant alleles in TUB1 and TUB3 (Stearns and Botstein, 1988), both of which encode α-tubulin, the major component of the microtubule cytoskeleton, highlighting the essential role of this paralog pair. Another type of GI was demonstrated by a study that was designed to identify mutants that carry a suppressor of conditional temperature-sensitive (TS) alleles in ACT1, yeast’s single actin gene (Novick et al., 1989). This screen identified an allele in the SAC1 gene, sac1–6, as a suppressor of the act1–1 TS allele, enabling act1–1 sac1–6 double mutant cells to grow at the restrictive temperature. This is an extreme example of a positive GI, where the double mutant grows better than expected based on the phenotype predicted for the combined single mutants. Interestingly, when sac1–6 was crossed to a different actin TS allele, act1–2, the resulting act1–2 sac1–6 double mutant was synthetically lethal. Subsequent work showed that SAC1 encodes a phosphatidylinositol phosphate phosphatase, and thus a role for phosphatidylinositol signaling and actin dynamics was revealed by both genetic suppression and synthetic lethal GIs between ACT1 and SAC1 (Novick et al., 1989).
Panels of defined mutants can be crossed directly to one another to generate double mutants and identify novel GIs. For example, the analysis of TS alleles of genes required for secretory pathway activity revealed that specific subsets of genes involved in either vesicle budding or vesicle fusion tend to be synthetic lethal with one another (Kaiser and Schekman, 1990). This study clearly demonstrates that synthetic lethal GIs are driven by functional specificity, a powerful notion that inspired the development and implementation of the first yeast synthetic lethal screen based on an elegant colony-sectoring assay (Bender and Pringle, 1991). In this assay, a query gene is deleted and the same gene is placed on a plasmid whose requirement for growth can be monitored using a simple visual assessment of red/white sectoring in yeast colonies. A colony sectoring screen using a strain mutated for MSB1, a poorly characterized gene implicated in bud emergence, identified two genes, BEM1 and BEM2, as synthetic lethal interacting partners. Subsequent work revealed that Bem1 is a scaffold for major signaling molecules involved in cell polarity, while Bem2 is a Rho GTPase activation protein involved in budding (Bi and Park, 2012). Thus, a synthetic lethal screen involving a relatively uncharacterized gene identified two significant regulators of cell polarity and bud emergence, developmental programs controlled by conserved signaling pathways.
Types of genetic interactions
GIs can be quantified by measuring single and double mutant phenotypes and calculating an interaction factor that reflects any deviation from the expected combined effect of the single mutants. For yeast cell fitness or growth phenotypes, the most widely adopted model combines the phenotypes, associated with mutations in independent genes, using a multiplicative equation (Mani et al., 2008; Phillips et al., 2000). Thus, the phenotype expected for a double mutant is predicted to be equivalent to the product of the phenotypes associated with the corresponding single mutants. Consequently, as illustrated in Figure 1, GIs are scored by detecting double mutants whose phenotype differs from the expected value.
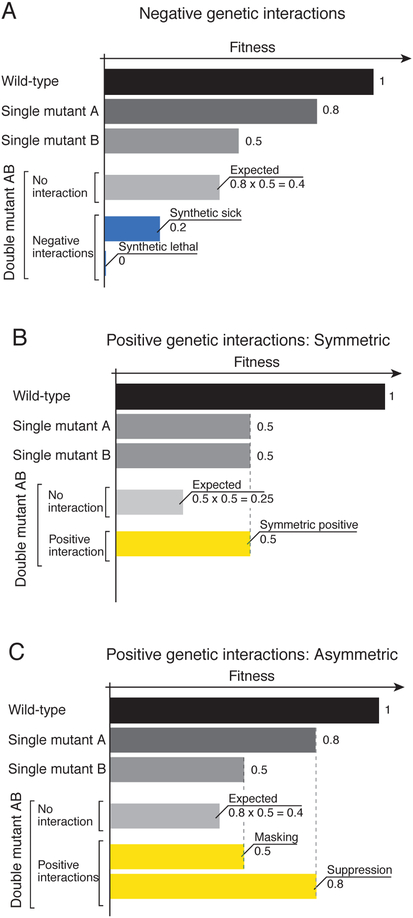
Figure 1. A graphical representation of quantitative genetic interactions.
Wild type fitness is defined as 1.0 and each single mutant (A and B) exhibits a fitness defect relative to wild-type. (A) Negative genetic interactions. A negative genetic interaction (e.g. synthetic lethal or synthetic sick interaction) occurs if the observed fitness of the double mutant is less than the double mutant fitness expected from a multiplicative model. (B) Symmetric positive interactions. In this specific case, each single mutant (A and B) exhibits a two-fold fitness defect (0.5) relative to wild type (1.0). The fitness of the resultant AB double mutant is greater than expected (0.25) and identical to the fitness of the two single mutants (0.5). Symmetric positive interactions are enriched among members of the same nonessential protein complex. (C) Asymmetric positive interactions. In this case, single mutants and double mutants differ in fitness. Positive interactions that deviate from expectation allow classification into masking or suppression subcategories.
In principle, any phenotype could be used to detect GIs, but a cell growth phenotype, which can be readily measured and quantified, integrates cellular physiology and has proven functionally informative for exploring GIs in both yeast and animal cells. For fitness phenotypes, negative GIs are scored when double mutants display a more severe growth phenotype than expected, including synthetic sickness and synthetic lethality (Fig. 1)(Dobzhansky, 1946; Mani et al., 2008; Phillips et al., 2000). In contrast, positive GIs are scored in double mutants that show a less severe phenotype than expected based on the multiplicative model (Mani et al., 2008; St Onge et al., 2007). Positive GIs can be classified into categories associated with a different mechanistic basis (Fig. 1) (Drees et al., 2005; Mani et al., 2008; St Onge et al., 2007). For example, when deletion of any gene encoding a member of the same nonessential protein complex eliminates complex activity, the phenotypes of single and double mutants involving protein complex members are expected to be quantitatively indistinguishable (Fig. 1)(Drees et al., 2005; Mani et al., 2008; St Onge et al., 2007). This means the double mutant does not conform to the expected multiplicative model and but rather shows a symmetrical positive GI. Positive GIs can also be asymmetric, in which the strength of the phenotypic effect varies between single and double mutants. The asymmetric subclass includes so-called masking interactions, where the fitness of the double mutant is better than expected, resembling that of the sickest single mutant (Fig. 1)(Drees et al., 2005; Mani et al., 2008; St Onge et al., 2007). Genetic suppression represents a mechanistically important but relatively rare asymmetric positive GI, such that the double mutant phenotype fitness is greater than that of the sickest single mutant (Fig. 1) (Baryshnikova et al., 2010; Drees et al., 2005; St Onge et al., 2007). Genetic suppression can result from the combination of two loss-of-function mutations but often involves specialized allelic combinations, and is thus a relatively small subset of the positive interactions observed in large-scale surveys of mutants with general defects in gene function (Costanzo et al., 2016; van Leeuwen et al., 2016).
[B]. Mapping a reference genetic interaction network
As mentioned above, systematic mutant collections and gene editing tools have been developed and applied to explore genetic interactions on a large-scale in several model microbial systems, including Escherichia coli (Butland et al., 2008; Typas et al., 2008) and the fission yeast Schizosaccharomyces pombe (Dixon et al., 2008; Roguev et al., 2008), as well as in the Caenorhabditis elegans metazoan model system (Byrne et al., 2007; Lehner et al., 2006) and cultured Drosophila melanogaster cell lines (Fischer et al., 2015). However, by far the most comprehensive analyses of GI mapping efforts have involved the budding yeast, Saccharomyces cerevisiae model system, which we review in detail below.
Genome-wide yeast mutant collections
With its elegant and facile genetics, the budding yeast Saccharomyces cerevisiae has been the eukaryotic model of choice for exploring the fundamental biology of eukaryotic cells at a molecular level and yeast has served as a primary test bed for development of most functional genomic methodologies. Systematic genetic and phenotypic analyses have been enabled by collaborative community efforts to create genome-wide mutant collections, within the context of reference strain background. A consortium of groups developed a comprehensive collection of deletion mutants where each of the ~6000 yeast open reading frames was replaced with a dominant, drug-resistance marker flanked by unique synthetic ‘barcode’ sequences. This systematic endeavour defined the set of ~1,000 genes essential for viability in laboratory growth conditions and generated a set of ~5,000 viable haploid deletion mutants (Winzeler et al., 1999). More recently, complementary strain collections have also been constructed in which subsets of the ~1000 essential yeast genes are individually altered to produce conditional or hypomorphic alleles [see (Costanzo et al., 2016)] that can be assayed at a semi-permissive state, enabling systematic scrutiny of this highly conserved set of genes. As described below, these arrayed mutant collections have catalyzed the development and application of high-throughput methodologies for systematic GI analysis and mapping of the yeast GI network.
Yeast genetic interaction mapping technologies
Synthetic Genetic Array (SGA) analysis is an automated method that combines arrays of either non-essential gene deletion mutants, or arrays of yeast strains carrying conditional alleles of essential genes, with robotic manipulation for high throughput construction of combinations of mutant alleles and identification of GIs (Tong et al., 2004). In its first large-scale application, SGA methodology was used to cross ~130 gene-specific query mutant strains to the complete array of ~5,000 viable haploid deletion mutants resulting in a network consisting of ~1,000 genes and ~4,000 synthetic lethal/sick digenic interactions (Tong et al., 2004). Complementary methods have been developed to map S. cerevisiae GIs in pooled cultures, including dSLAM (Pan et al., 2004), GI Mapping (GIM) (Decourty et al., 2008), Barcode Fusion Genetics (BFG-GI) (Diaz-Mejia et al., 2018) and iSeq (Jaffe et al., 2017). These various methods have been applied to identify GIs associated with subsets of genes annotated to specific bioprocesses, including DNA integrity (Pan et al., 2006), mRNA processing (Decourty et al., 2008), as well GIs among selected genes in response to DNA damaging agents (Diaz-Mejia et al., 2018). However, comprehensive SGA analysis has been the most extensively used method for GI studies [for example (Collins et al., 2007; Schuldiner et al., 2005; Tong et al., 2004)] and its large-scale application enabled mapping of a global GI network for budding yeast (Costanzo et al., 2010; Costanzo et al., 2016).
Quantitative analysis of genetic interactions
As described above, negative or positive GIs between a pair of genes can be experimentally defined based on the comparison of three properties: the single mutant phenotypes, an estimate of the expected double mutant phenotype, and a measurement of the observed double mutant phenotype. In theory, a GI can be identified using any quantitative phenotypic assay. Indeed, GIs in yeast have been revealed by accurately monitoring gene expression (van Wageningen et al., 2010), filamentous growth (Drees et al., 2005), receptor endocytosis (Burston et al., 2009), and the unfolded protein response (Jonikas et al., 2009). Flux balance analysis has also been used to detect GIs in silico using predicted changes in yeast biomass production as a phenotypic readout (Segre et al., 2005). However, most large-scale efforts to map quantitative genetic networks in yeast have focused on colony size as a proxy for cell growth or fitness, a phenotype that is easily quantified, and arguably integrates the general ‘state’ of the cell (Baryshnikova et al., 2010; Collins et al., 2006). Alternative methods that have been used to quantify GIs include: [1] profiling of growth in liquid culture to assess GIs between a subset of genes involved in DNA replication and repair (St Onge et al., 2007); [2] the use of fluorescence-labeled populations of wild-type cells co-cultured with either single or double mutant yeast strains to map a quantitative GI network for genes encoding components of the 26S proteasome (Breslow et al., 2008) -of note, this map was based on hypomorphic proteasome alleles designed to reduce mRNA stability, a type of mutation that may be associated with different GIs, both in terms of type and strength, than those associated with TS alleles (Breslow et al., 2008); and [3] a competitive growth assay to quantify GIs between duplicated genes (DeLuna et al., 2008). Although these methods provided considerable new insights into conserved biological processes and features, they have not yet been applied to genome-scale studies.
A functional map of a cell: genetic interaction profiles identify functional relationships between genes
Early studies using SGA (Tong et al., 2004) demonstrated that the set of synthetic lethal GIs for a given gene, termed a GI profile, provides a rich phenotypic signature indicative of gene function. As a result, grouping genes according to their GI profiles, using standard clustering algorithms, is an effective and powerful way to precisely predict gene function (Tong et al., 2004). As described above, the use of quantitative assays of colony size (Baryshnikova et al., 2010; Collins et al., 2006), generates rich GI profiles for each query gene. These enriched profiles further demonstrate that genes belonging to similar biological processes share overlapping subsets of both negative and positive GIs, clearly highlighting that genes encoding proteins that function together in the same pathway or protein complex often display highly similar GI profiles (Bandyopadhyay et al., 2008; Collins et al., 2007; Costanzo et al., 2010; Costanzo et al., 2016; Kelley and Ideker, 2005; Tong et al., 2004).
The combination of SGA with a genome-scale colony size-scoring methodology enabled assessment of growth defects associated with the majority of all possible yeast gene pairs (~18 million). These large-scale studies measured single and double mutant fitness to identify nearly one million GIs (~550,000 negative and ~350,000 positive) enabling assembly of the first complete GI network for any organism (Costanzo et al., 2010; Costanzo et al., 2016). Importantly, genome-wide GI analysis required the analysis of essential genes, which was achieved through the construction of a comprehensive set of mutants carrying conditional temperature-sensitive (TS) alleles. Essential genes participate in ~5-fold more negative and positive interactions than nonessential genes, showing that essential genes represent highly connected network hubs. Moreover, the GI profiles of essential genes provide more accurate gene function predictions across diverse biological processes, indicating that essential genes form the basic scaffold of the global GI network (Costanzo et al., 2016). Since essential genes are more conserved than non-essential genes and participate in a large fraction of the interactions on the global yeast genetic network, they may define a GI network that is generally conserved.
Quantitative GI profiles, composed of both essential and non-essential genes, enabled construction of a global network in which genes with similar interactions patterns are located close to one another (Fig. 2)(Costanzo et al., 2010; Costanzo et al., 2016). Consequently, the resultant network groups genes into obvious clusters and, when scrutinized for varying levels of genetic profile similarity, provides a hierarchical view of the functional organization within a cell (Fig. 2)(Costanzo et al., 2010; Costanzo et al., 2016; Dutkowski et al., 2013; Ma et al., 2018). For example, at the most stringent level of network resolution, where genes share many common GIs, the clusters are composed of relatively small, densely connected modules corresponding to known protein complexes and biological pathways (Fig. 2). At an intermediate level of network resolution, functionally-related pathway and protein complex modules, such as ‘kinetochore’ or ‘GPI-anchor’, are grouped together to highlight distinct biological processes, including large subsets of genes annotated with general roles in different bioprocesses, such as ‘DNA replication and repair’ or ‘Cell polarity and morphogenesis’ (Fig. 2). At the most general level of network resolution, bioprocess gene clusters group together into larger modules corresponding to specific cellular compartments, such as ‘Nucleus’ or ‘ER’ (Fig. 2)(Costanzo et al., 2016). In other words, computational visualization of the results of systematic quantitative analysis of a simple growth phenotype provides a remarkably nuanced view of cell function, ranging from known subcellular organelles to protein complexes. Importantly, the functional organization embedded within the global genetic network is not completely captured by available functional standards, including the Gene Ontology (GO), emphasizing the need for unbiased, genome-scale studies to accurately describe cell function (Costanzo et al., 2016; Dutkowski et al., 2013; Ma et al., 2018). The global genetic profile similarity network also provides a powerful resource for precise functional predictions because the network position and connectivity of a particular gene reflects its cellular role (Costanzo et al., 2016). A gene whose GI profile includes subsets of genes that cluster into several bioprocesses may reflect a highly pleiotropic function, such as that associated with molecular chaperones like Hsp90, which controls many signaling pathways.
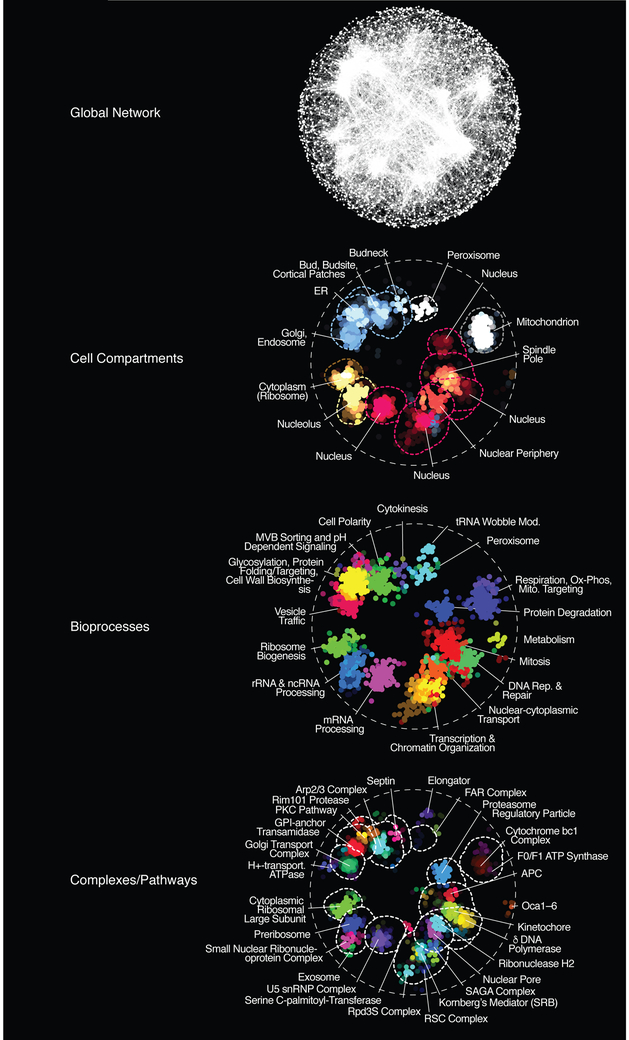
Figure 2. A functional map of a yeast cell.
(A) A global genetic profile similarity network encompassing most nonessential and essential genes was constructed by computing Pearson correlation coefficients (PCCs) for genetic interaction profiles of all pairs of genes (nodes). Gene pairs whose profile similarity exceeded a PCC > 0.2 were connected and graphed using a spring-embedded layout algorithm (Smoot et al., 2011). Genes sharing similar genetic interactions profiles map proximal to each other, whereas genes with less similar genetic interaction profiles are positioned further apart. (B-D) The global genetic interaction profile similarity network is organized as a hierarchy of functional modules enriched for specific (B) cellular compartments, (C) biological processes or (D) protein complexes and pathways. Functional annotation of the networks was done using Spatial Analysis of Functional Enrichment (SAFE) (Baryshnikova, 2016). Adapted from (Costanzo et al., 2016).
Functional information associated with negative or positive genetic interactions
Negative GIs, for both nonessential and essential genes, overlap significantly with other types of molecular interactions (Costanzo et al., 2016). For example, a negative GI can be highly predictive of a physical interaction because ~50% (~10-fold enrichment) of essential gene pairs that encode physically interacting proteins are also connected by a negative GI (Costanzo et al., 2016). Moreover, the likelihood of observing a negative GI is correlated with the functional overlap shared between a given pair of genes. This means that the closer two genes are within the hierarchical model of cell function the more likely they are to show a negative GI (Fig. 3A–B)(Costanzo et al., 2016). Likewise, the strength of the GI is also associated with the extent of functional similarity, such that stronger GIs tend to connect genes with closer functional relationships. This suggests that the functional relationship between two genes can be quantified based on the strength of their negative interaction (Costanzo et al., 2016).
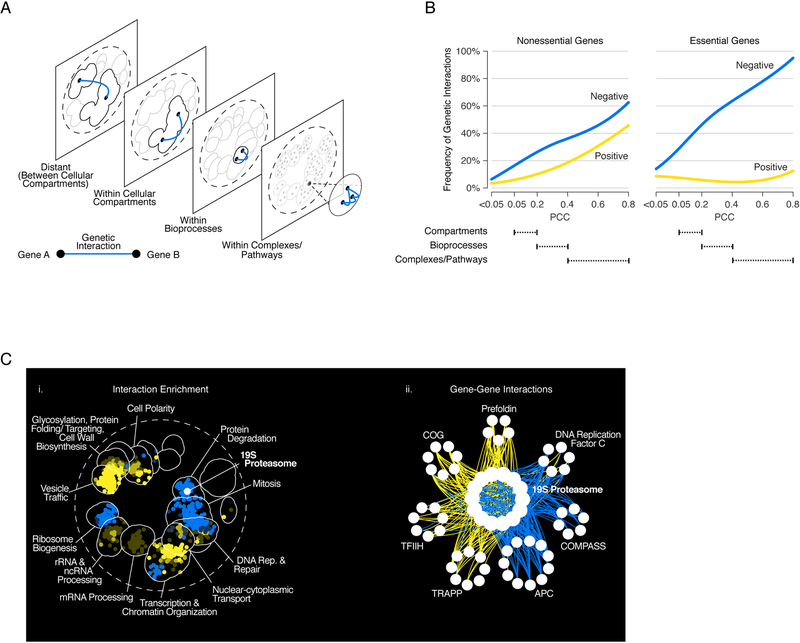
Figure 3. Mapping negative and positive interactions across the genetic network–based functional hierarchy.
(A) Schematic representation of the genetic network–based functional hierarchy illustrating functionally-defined clusters of interactions between genes within the same complex/pathway, bioprocess, or cellular compartment, as well as distant interactions that span two different cellular compartments. (B) The frequency of genetic interactions between genes in the same functional cluster (as defined in (A)), at a given level of profile similarity (PCC) in the genetic network hierarchy for negative (blue) or positive (yellow) genetic interactions. Dashed lines indicate the PCC range within which clusters in the genetic network hierarchy were enriched for cell compartments, bioprocesses, and protein complexes. (C) Functional wiring diagram from the 19S proteasome identifies within pathway modules (WPM) and between pathway models (BPM) of genetic interactions. (i) Regions of the yeast global similarity network significantly enriched for genes exhibiting negative (blue) or positive (yellow) genetic interactions with 19S proteasome genes are shown using SAFE (Baryshnikova, 2016). (ii) Genes belonging to a subset of protein complexes that showed coherent negative (blue) or positive (yellow) genetic interactions with genes encoding the 19S proteasome. Adapted from (Costanzo et al., 2016).
Positive GIs can also capture functional relationships between non-essential gene pairs since, in certain situations, they can overlap with protein-protein interactions (Bandyopadhyay et al., 2008; Baryshnikova et al., 2010; Costanzo et al., 2010; Costanzo et al., 2016; Mani et al., 2008; St Onge et al., 2007). Specifically, if any defect in a single complex component produces a null phenotype, then the simultaneous perturbation of two genes encoding members of the same nonessential protein complex will show a fitness defect equivalent to that of the single mutants (Costanzo et al., 2016; Mani et al., 2008; Segre et al., 2005; St Onge et al., 2007). However, this subclass of positive GIs accounts for only a small fraction of those observed for nonessential gene pairs (Costanzo et al., 2010; Costanzo et al., 2016). The vast majority of all positive interactions, especially those involving essential genes, do not share any direct functional relationship (Fig. 3A–B). Instead, positive interactions tend to connect genes whose products function in different cell compartments and capture more general regulatory connections, associated with cell cycle progression or mRNA and protein turnover (Costanzo et al., 2016). For example, TS alleles, which may lower protein levels or reduce protein activity, often show positive interactions with genes involved in protein degradation. In contrast, mutant alleles of essential genes that may lower mRNA levels (Breslow et al., 2008; Schuldiner et al., 2005), tend to share numerous positive GIs with genes involved in mRNA turnover (Costanzo et al., 2016). Thus, while negative GIs identify clear functional relationships between genes, positive GIs reflect more general regulatory connections, related to protein homeostasis and cell cycle kinetics. While positive GIs associated with two loss-of-function alleles do not generally connect functionally related genes, suppression interactions involving specific gene pairs, an important subclass of the strongest positive GIs, are highly predictive of shared function and are described in more detail below (Section D).
Genetic network structure and topology
Consistent with a previous theoretical analysis (Segre et al., 2005), negative and positive GIs tend to be highly organized, often occurring in coherent sets such that genes within the same protein complex or pathway are connected by a single type of interaction, either negative or positive, forming a local genetic network structure, referred to as a Within Pathway Module (WPM) (Bandyopadhyay et al., 2008; Baryshnikova et al., 2010; Bellay et al., 2011; Costanzo et al., 2010; Costanzo et al., 2016; Kelley and Ideker, 2005). The type of interaction depends on essentiality because WPMs composed of nonessential genes are enriched for positive GIs, whereas essential gene WPMs are enriched for negative GIs (Bandyopadhyay et al., 2008; Baryshnikova et al., 2010; Bellay et al., 2011; Costanzo et al., 2016). Indeed, ~80% of WPMs corresponding to essential protein complexes are enriched for negative GIs among their members (Costanzo et al., 2016). This remarkable overlap of protein interactions and GIs highlights the reduced ability of a cell to tolerate multiple mutations in the same essential complex and provides a roadmap for both discovering and predicting GIs in other organisms, including humans. In addition to WPM structures, GIs occurring between functional modules are also highly organized, whereby pairs of complexes or pathways tend to be connected exclusively by negative GIs or only by positive GIs, forming a network structure referred to as a Between Pathway Module (BPM) (Bandyopadhyay et al., 2008; Bellay et al., 2011; Costanzo et al., 2010; Costanzo et al., 2016; Kelley and Ideker, 2005).
Previous studies highlighted the prevalence of these types of genetic network structures (Bandyopadhyay et al., 2008; Bellay et al., 2011; Kelley and Ideker, 2005) and a survey of the yeast genetic network revealed that most negative GIs appear in WPMs and/or BPMs (Bellay et al., 2011). Indeed, WPMs and BPMs derived from the yeast global genetic network map a highly organized functional wiring diagram that defines specific modules and reveals functional relationships between modules (Costanzo et al., 2016). For example, negative GIs occur among genes encoding members of the 19S proteasome, reflecting the essential role of this complex (Fig. 3C). Negative GIs also connect the 19S proteasome and the APC (anaphase promoting complex), which controls mitotic exit, reflecting that their combined roles are critical for cell cycle progression. In contrast, positive GIs connect the 19S proteasome and the Prefoldin cochaperone complex, which may indicate that these complexes work in tandem to monitor protein folding and degradation (Fig. 3C)(Costanzo et al., 2016). The prevalence of coherent module structures in the yeast reference GI network has practical implications that may facilitate discovery of disease-relevant GIs in human genotyping datasets (Wang et al., 2017b), as discussed further below (Sections F,G)
[C]. Condition-specific genetic interaction networks
While a global genetic network reveals the fundamental organization of a cell (Costanzo et al., 2016), a complete analysis should include an understanding of network dynamics in response to developmental programs, environmental changes, as well as different genetic backgrounds (Section E). Indeed, cataloging the global genetic network showed that ~20% (~1000) of yeast genes are refractory to SGA GI profiling in standard laboratory growth conditions because they exhibit relatively few double mutant interactions precluding accurate assessment of their functional connections (Costanzo et al., 2016). A comprehensive understanding of gene function and GI networks therefore requires further analysis in a variety of different settings that depend on the activity of otherwise dispensable genes.
Previous studies measured condition-specific or differential GIs in the absence and presence of DNA damaging agents (Bandyopadhyay et al., 2010; Diaz-Mejia et al., 2018; Guenole et al., 2013; St Onge et al., 2007) as well as stress conditions known to trigger mitogen-activated protein kinase signaling pathways (Martin et al., 2015) or the autophagy program (Kramer et al., 2017). The resultant differential GI networks recapitulated known condition-specific functional interactions and identified novel connections between gene pairs that could not be detected in the absence of a particular environmental stimulus. Thus, differential GI network analysis appears to highlight subsets of genes with condition-specific interactions, uncovering dynamic connections between pathways and protein complexes.
However, the influence of environmental factors on overall network structure and topology and how the global network changes in response to different environments remains unclear. A theoretical analysis based on metabolic models and flux balance analysis predicted that only a fraction of all GIs may be revealed when examining a single environmental condition (Harrison et al., 2007). Indeed, while the average nonessential gene shares a negative GI with ~2% of other nonessential ORFs in the yeast genome, genes important for metabolic processes are statistically underrepresented in the GI network (Costanzo et al., 2010). However, careful scrutiny of GIs involving genes functioning in specific biological processes, such as DNA replication and repair, showed relatively few new DNA damage-specific negative interactions compared to the number of negative GIs identified for the same set of genes under non-DNA damaging conditions (Bandyopadhyay et al., 2010; Guenole et al., 2013). Thus, while genes in specific bioprocesses may undergo local rewiring by gaining or losing functionally relevant GIs in a condition-specific manner, it is possible that the overall structure and topology of the global genetic network remains relatively robust to conditional and environmental influences, a concept the remains to be systematically explored.
Chemical genetic interactions
A chemical-genetic interaction occurs when a specific mutant is either hypersensitive or resistant to a compound when compared to a wild-type control. If a compound precisely inhibits a target protein that is required for yeast growth, loss-of-function mutations in the target gene should model the cell’s physiological response to the compound (Marton et al., 1998). A genome-wide set of yeast mutants, including deletion alleles of nonessential genes and partial loss-of-function alleles of essential genes, can be scored for hypersensitivity or resistance to a specific compound to generate a chemical-genetic profile (Hoepfner et al., 2014; Lee et al., 2014; Piotrowski et al., 2017). By similarity analysis, a global network of GI profiles provides a guide for interpreting chemical-genetic interaction profiles and thereby linking compounds to their target pathway (Costanzo et al., 2010; Piotrowski et al., 2017). For example, the GI profile associated with a partial loss-of-function mutation in the essential gene ERG11, which encodes the target of fluconazole, closely resembles the chemical-genetic interaction profile of fluconazole.
With barcoded yeast strains, thousands of mutants can be analyzed for chemical-genetic interactions in parallel and a small diagnostic set of yeast mutants enables high throughput profiling of large compound libraries providing an unbiased method for functional characterization of entire libraries (Piotrowski et al., 2017). Target prediction is resolved to the level of the bioprocess, such that compounds can be linked to cellular functions such as ‘Cell Polarity and Morphogenesis’ or ‘Mitosis and Chromosome Segregation’ (Piotrowski et al., 2017). Compounds that are associated with specific bioprocesses can be examined in more detail with genome-wide mutant pools, which may lead to a more refined pathway-level of functional resolution.
Haploinsufficiency chemical-genetic profiling has the potential to link a compound to its target gene directly (Giaever et al., 1999). In yeast, diploid cells can generally tolerate the deletion of one copy of most essential genes, and the resultant heterozygous deletion mutants usually fail to show a fitness defect. Heterozygous diploids deleted for one copy of a specific gene, should generally have less of the corresponding gene product, and therefore be sensitive to compounds that inhibit the target gene product (Giaever et al., 1999). For example, an erg11/ERG11 heterozygous diploid is more sensitive to fluconazole than any other diploid heterozygous for deletion of an essential gene. A complete set of barcoded, heterozygotes spanning all ~1000 yeast essential genes enables comprehensive and high-throughput screens for compounds targeting this highly conserved gene set (Hoepfner et al., 2014; Lee et al., 2014).
[D]. Genetic suppression and modifier effects
As noted above, genetic suppression is an extreme type of positive GI (Fig. 4A), such that the fitness defect of a query mutant allele is partially or fully compensated by a suppressor mutation. Suppressor mutations can either be intragenic, occurring in the same gene as the original query mutation, or extragenic, involving mutations in two different genes. In model organisms, suppression interactions can be identified using standard methods for identifying GIs between two pre-existing mutant alleles, by isolating spontaneous or chemically-induced mutations that rescue the fitness defect of a mutant of interest, or by systematically overexpressing all genes in the genome individually to identify genes that can rescue a mutant phenotype (dosage suppression).
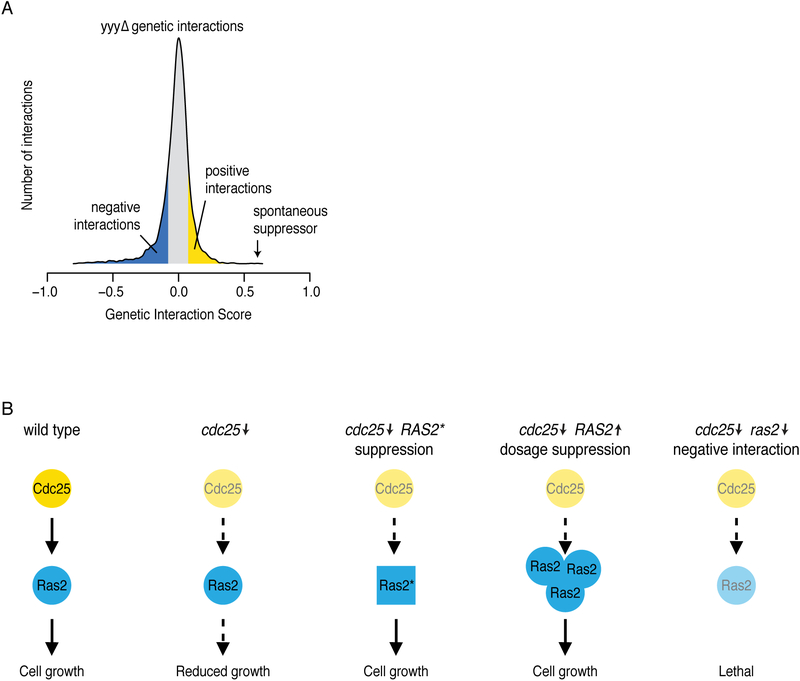
Figure 4. Genetic suppression interactions.
(A) Example of a distribution of negative (blue) and positive (yellow) genetic interactions of mutant yyyΔ determined by a genome-wide screen. The genetic interaction score is defined as the difference between the observed and the expected double mutant fitness (see Figure 1). Spontaneous suppressor mutations often represent the most extreme, strong positive genetic interactions, as indicated. (B) An example of a gene pair (CDC25 and RAS2) illustrating suppression, dosage suppression, and negative genetic interactions. Lightly shaded proteins are encoded by partial loss-of-function alleles with reduced signaling activity, Ras2* is encoded by a gain-of-function allele with increased signaling activity. Adapted from (van Leeuwen et al., 2016).
Extragenic suppression interactions are extremely rich in functional information compared to other interaction types, and mostly occur between genes that have a close functional connection, such as those encoding protein or pathway members. These interactions often involve specific genetic alterations, such as allele-specific interactions, and can be used to assign function to uncharacterized genes, to order pathway components and to understand phenotypic variability in natural populations [see (Matsui et al., 2017) for review]. Mechanistically, suppression GIs can involve gain-of-function phenotypes that compensate for a missing activity in absence of the query gene, or loss-of-function phenotypes that may antagonize query gene function or eliminate toxicity associated with absence of the query gene. For instance, in yeast, the growth defect associated with loss-of-function mutations in CDC25, which encodes the guanine nucleotide exchange factor that activates Ras2, can be suppressed by either gain-of-function mutations in RAS2, or loss-of-function mutations in IRA1, which encodes the GTPase activating protein that negatively regulates Ras2 (Fig. 4B)(van Leeuwen et al., 2016).
In addition to functionally-related mechanisms, suppression can also occur via general control systems that are shared among numerous genes with diverse functions (Magtanong et al., 2011; Patra et al., 2017). General suppressors can affect the translation of the query mutation (informational suppressors), the expression of the query gene, the stability of its gene-product, or the general animal physiology in multicellular organisms. For example, loss-of-function mutations in members of the mRNA or protein degradation pathways can rescue the fitness defect of partial loss-of-function mutants in yeast by increasing the levels of query mutant mRNA or protein (van Leeuwen et al., 2016). Systematic studies of both genomic and dosage suppression have highlighted the importance of general mechanisms of suppression, which explain nearly half of all suppressors identified for specific point-mutant alleles in yeast (Magtanong et al., 2011; Patra et al., 2017; van Leeuwen et al., 2016).
Most studies of suppression have been performed in model organisms, where the environment and other variables are highly controlled. It is clear, however, that genetic modifiers including those that show suppression GIs, complicate the connection between genotype and phenotype in natural outbred populations, including humans (Riordan and Nadeau, 2017). Genetic modifiers may influence: [1] the penetrance of a trait, or the proportion of individuals carrying a particular causative mutation that are phenotypically affected and; [2] trait expressivity, or the quantitative variation in phenotypic severity among individuals where a particular trait is penetrant. A notable example of variable expressivity is the recent discovery of resilient individuals that are healthy despite carrying mutations that have been associated with severe, early-onset, Mendelian diseases (Chen et al., 2016). A possible explanation is that these individuals carry additional mutations elsewhere in the genome that can function as suppressors and can overcome the detrimental effects of the disease mutation. For instance, a dominant suppressor locus, DFNM1, has been identified that protects individuals that are homozygous for the recessive non-syndromic deafness locus DFNB26 against hearing loss (Riazuddin et al., 2000). Similar suppressive interactions between mutations have been described in cancer, whereby suppressor mutations increase cell proliferation of otherwise slowly proliferating cancer cell mutants are associated with a poorer prognosis. For example, loss of 53BP1 in tumors lacking a functional copy of the tumor suppressor gene BRCA1 is usually associated with aggressive tumors, low survival rates, and increased resistance to chemotherapy [see (Aly and Ganesan, 2011) for review]. Mechanistically, this interaction represents the re-activation of the homologous recombination (HR) machinery, which is compromised in the absence of BRCA1, by inactivating the negative HR-regulator, 53BP1 (Aly and Ganesan, 2011).
In addition to functional suppressors, general suppression mechanisms may occur among human genomic variants and affect the expressivity or penetrance of disease. For instance, as described above, a frequently observed mechanism of general suppression in yeast involves loss-of-function mutations in mRNA decay pathway members that can suppress partial loss-of-function alleles of functionally diverse genes (van Leeuwen et al., 2016). Many diseases arise as a consequence of a premature termination codon, and thus partial inactivation of nonsense mediated mRNA decay (NMD) could be beneficial in these cases by preventing degradation of the mutated mRNA and allowing translation into a truncated peptide that could retain residual activity (Miller and Pearce, 2014). Indeed, in certain genetic disorders, such as Duchenne muscular dystrophy, treatment options are focused on increasing translation of the disease-associated mutant mRNA (Finkel, 2010).
These examples show the importance of GIs in determining human disease severity, and suggest that an understanding of genetic suppression may shed light on the mechanisms underlying the disease, which could highlight new therapeutic strategies.
[E]. Complex genetic interactions
Complex GIs involve three or more genes, and likely play a major role in the genotype to phenotype relationship. For example, while there are 18 million possible gene pairs in the yeast genome, there are 36 billion gene triplets. Although the scope of the problem is immense, there have been efforts to systematically quantify complex GIs in yeast, applying modifications of methods used to survey double mutant GIs (Haber et al., 2013; Kuzmin et al., 2018; Weinreich et al., 2013). Specifically, an extension of the digenic multiplicative model (Baryshnikova et al., 2010) has been utilized to measure yeast colony size for triple mutants, such that the expected fitness of a triple mutant accounts for all three possible digenic interactions and single mutant effects, with any remaining deviation in fitness ascribed to the simultaneous perturbation of all three genes (Kuzmin et al., 2018). To survey the extent of triple mutant ‘interaction space’, a selected set of ~200,000 triple yeast mutants was examined for fitness defects along with their embedded double mutants to identify 3,000 trigenic interactions (1.6% of tested triplets) and 9,000 digenic interactions (2.3% of tested pairs). This survey included combinations of both deletion alleles of non-essential genes and conditional TS alleles of essential genes (Kuzmin et al., 2018). Analysis of the triple mutant GI profiles revealed that, like digenic interactions, trigenic interactions tend to involve functionally related genes and the potential for a digenic query strain to show trigenic interactions correlates with a number of quantitative features of the digenic network (Costanzo et al., 2016). In particular, three digenic network features, the negative GI score between two query genes, the interaction degrees of the query genes, and the similarity of the query gene digenic interaction profiles, correlate positively with trigenic interaction degree (Costanzo et al., 2016; Kuzmin et al., 2018). Moreover, ~2/3 of the trigenic interactions overlap with a digenic interaction on the global GI network and negative trigenic interactions are generally weaker than digenic interactions, emphasizing the potential for reference double mutant networks to reveal an underlying scaffold for complex GIs. However, compared to digenic profiles, trigenic profiles show a significantly higher tendency to include genes involved in more diverse biological processes, indicating that digenic query strains are perturbed for a wider range of cellular functions than single mutant query strains (Kuzmin et al., 2018).
While trigenic interactions occur less frequently than digenic interactions, the relative number of possible triple mutant combinations is 2000-fold larger (see above), and modeling revealed that the expected number of trigenic interactions in the yeast genome is 100-fold more than the number of digenic interactions (Kuzmin et al., 2018). The global yeast digenic network encompasses ~500,000 negative GIs, while the trigenic network is predicted to encompass ~100 million negative GIs. The expanse of the global trigenic interaction network thus highlights the potential for complex GIs to affect the biology of inheritance. For example, the relative frequency and strength of digenic and trigenic interactions suggest that deleterious trigenic interactions, also termed Dobzhansky-Muller incompatibilities, play a major role in the evolution of hybrid inviability and speciation (Foley et al., 2013). Indeed, a trigenic interaction among three autosomal quantitative trail loci (QTLs) causes hybrid male sterility between two fly species, D. persimilis and D. pseudoobscura bogotana (Chang and Noor, 2010).
An important goal of genetic network analysis is to discover GIs driven by natural variation, which growing evidence suggests contributes to the genotype-to-phenotype relationship for individuals. For example, an early study examined natural variation influencing sporulation efficiency in offspring of a cross between an oak yeast strain (YPS606) and a vineyard yeast strain (BC187) (Gerke et al., 2006). In total, 5 QTLs, involving both coding and non-coding regions of transcription factor genes, were linked to meiosis efficiency, and these loci participated in both digenic and more complex GIs involving three and four loci. In another study of yeast colony morphology, a cross between two genetically diverse yeasts identified a complex GI involving 5 genes (Taylor and Ehrenreich, 2014). More general studies have surveyed quantitative trait variation in ~4,000 recombinant offspring from a cross between the laboratory yeast strain (S288c) and a vineyard strain (RM11) (Forsberg et al., 2017). Genotyping of each offspring for ~28,000 SNPs and phenotyping their association for 20 endpoint growth traits, showed that most extreme estimates from additive models of QTL effects were often inaccurate and biased. However, incorporating GI terms for pairwise and higher-order GIs significantly improved the accuracy of predicting phenotypes from genotypes. New theoretical approaches, which account for the combined effects of deleterious and beneficial mutations, are being developed to model the influence of such higher-order genetic interactions on the dynamics of evolution (Agarwala and Fisher, 2018).
In addition to QTL studies, systematic gene deletion analysis can be used to explore GIs involving natural variation and their role in the genotype-to-phenotype relationship. For example, one study involved construction of a genome-wide deletion collection in the yeast strain Σ1278b (Ryan et al., 2012), which is capable of pseudohyphal growth and differs from the reference yeast (S288C) genome by thousands of SNP loci, but can efficiently mate and generate viable hybrid progeny. This approach revealed a specific subset of genes that were essential in one genetic background but not the other, a phenotype termed conditional essentiality (Dowell et al., 2010). Detailed analysis of the genetics underlying the conditional essentiality revealed that, while it can be associated with non-chromosomal elements (Edwards et al., 2014) or digenic synthetic lethal interactions, it often involves multiple modifiers associated with a complex GI (Dowell et al., 2010). Another study genotyped ~1400 progeny from a yeast cross of two genetically diverse strains, scoring the phenotypes of deletions in 7 different chromatin remodeling genes across 10 environmental conditions (Mullis et al., 2018), revealing that the gene deletions showed between ~70 and ~540 GIs, with ~90% involving complex GIs.
While di- and trigenic interactions can be mapped genome-wide using available tools and methods, the analysis of higher order GIs becomes increasingly difficult, and is perhaps best examined using highly specific model systems. In particular, assessment of complex GIs involving mutations in a single gene have been remarkably informative. In one study, the functional consequences of mutations and combinations of mutations in the green fluorescent protein from Aequorea victoria (avGFP) expressed in bacteria were systematically assessed (Sarkisyan et al., 2016). Both fluorescence and protein stability were scored for 50,000 genotypes of avGFP, testing up to 15 missense mutations per sequence, revealing both simple and complex GIs. Mutants with over 7 mutations showed a decrease in the prevalence of GIs because the interactions started to overlap single gene and additive effects, indicating a saturation point for higher-order GIs. Another recent study focused on variation in a single tRNA gene in yeast to test the influence of GIs on gene function (Domingo et al., 2018). Here, natural variation across 10 residues of a tRNA gene in post-whole-genome-duplication yeast species was used as the foundation for building a library of ~5,000 tRNA genotypes involving 14 different nucleotide substitutions in the divergent residues, encompassing all possible genotypes that differ from the Saccharomyces cerevisiae tRNA sequence. This exhaustive mutagenesis captured up to 8th order interactions and revealed that the fitness phenotype associated with a mutation is modified by the presence of other mutations in the same tRNA molecule. In this specific case, each mutation was capable of causing both detrimental and beneficial effects in a substantial number of different genetic backgrounds. Thus, to accurately predict the effect of a mutation on fitness, it is imperative to measure mutation effects across different genetic backgrounds and account for higher-order GIs. In this context, a novel method to predict effects resulting from mutations across diverse classes of RNA and protein has been developed using a deep latent-variable model that incorporates higher-order constraints, driven by complex GIs (Riesselman et al., 2018).
These and other studies clearly illustrate the profound effect that complex GIs can have on phenotypes and underscore the need to understand complex GIs in our efforts to predict the consequences of genome variation. In particular, complex GIs should also be taken into account for efficient design of functional synthetic genomes. Indeed, the minimal set of genes required to sustain a living yeast cell is much larger than the essential gene set due to both digenic and trigenic interactions (Kuzmin et al., 2018). In fact, GIs were reported to constrain the design of the minimal genome of Mycoplasma mycoides bacteria (Hutchison et al., 2016). The construction of a viable genome was possible only after resolving synthetic lethal GIs involving non-essential genes (Hutchison et al., 2016). Thus, the essential gene set which generally comprises 20% of the eukaryotic genome (Blomen et al., 2015; Hart et al., 2015; Wang et al., 2015; Winzeler et al., 1999) is necessary but not sufficient to construct a viable organism with a minimal genome due to the phenotypic consequences of GIs on fitness.
[F]. Human genetic interaction networks and cancer
Breakthroughs in gene editing technologies over the past decade have transformed mammalian cell genetics enabling genome-scale, reverse genetic screens in diverse human cell line model systems. Similar to the impact of the arrayed collection of yeast deletion mutants (Winzeler et al., 1999), development of high-complexity genome-scale CRISPR (clustered regularly interspaced short palindromic repeats) libraries promise unprecedented functional characterization of the human genome. For example, genome-wide pooled CRISPR and transposon mutagenesis screens have defined a core set of essential genes that are required for human cell proliferation and that share functional, evolutionary and physiological properties with essential genes in other model systems (Blomen et al., 2015; Hart et al., 2015; Wang et al., 2015). These studies have laid the foundation for a new wave of functional genomics for characterizing the role of essential genes, how gene essentiality depends on genetic and tissue contexts, and how essential genes evolve (Bartha et al., 2018; Rancati et al., 2018).
Continued genome-scale, loss-of-function genetic screens in diverse cancer cell lines have begun to resolve the human essential gene set in greater detail (https://depmap.org/portal/)(Rauscher et al., 2018; Tsherniak et al., 2017). In addition to core essential genes, comparison of essential gene profiles identified subsets of genes that are specifically required for viability of particular types of cancer cells (Hart et al., 2015; Tsherniak et al., 2017; Wang et al., 2015; Wang et al., 2017a). These differential essential genes are directly relevant to our understanding of GIs in humans. Cancer is a heterogeneous disease encompassing hundreds of distinct subtypes and different genetic backgrounds. Thus, similar to conditional essential genes in model organisms (Dowell et al., 2010), the requirement of a given gene for viability of some cancer cell lines but not others may be caused by underlying modifiers and their GIs with the differential essential gene or genetic variant(s) inherent to the particular cancer subtype (Fig. 5A). Indeed, as predicted from genetic network analysis in yeast (Costanzo et al., 2016), cancer cell line-specific essentiality profiles can uncover functional relationships because genes belonging to the same pathway or protein complex are often essential in the same subset of cancer cell lines (Pan et al., 2018; Tsherniak et al., 2017; Wang et al., 2017a). Moreover, differential essential genes represent potential therapeutic targets which, when genetically or chemically inhibited, can kill cells that harbor a second cancer-specific alteration due to a synthetic lethal interaction, but spares otherwise identical cells lacking the alteration (Ashworth and Lord, 2018), an idea originally proposed by Hartwell and colleagues (Hartwell et al., 1997). For example, breast and ovarian cancer cells with mutations in BRCA1 or BRCA2 are extremely susceptible to inhibition of poly(ADP)ribose polymerase 1 (PARP1) (Ashworth and Lord, 2018).
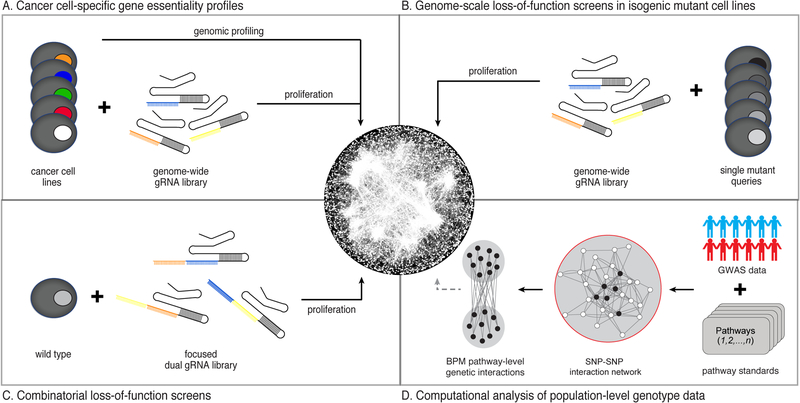
Figure 5. Strategies for mapping genetic interactions in human cells.
(A) A genome-scale gene editing approach (e.g using CRISPR-Cas9 and genome-wide gRNA library) to identify essential genes in a cancer cell-specific manner. Different cancer cell lines are represented by coloured nuclei. Additional genomic analysis is required to identify secondary mutations that interact with a particular gene required for viability across subsets of cancer cell lines. (B) Large-scale CRISPR-Cas9 screens to systematically introduce a second, defined mutation, presented by nuclei with different shades of grey, into a set of isogenic cell lines, each carrying a stable mutation in a specific query gene of interest. Genes that result in a fitness defect when targeted in a particular query mutant cell line identify potential genetic interactions. (C) A combinatorial approach enabling perturbation of two different genes, simultaneously within a specific cell line, to identify pairs of genes that, when mutated in the same cell, result in an unexpected growth phenotype. (D) Computational analysis of large-scale human genotype data that leverages pathway/functional module information as prior knowledge to aggregate genetic variants to discover pairs of pathways/functional modules that result in increased or decreased disease risk when both mutated in defined group of individuals, such as disease cohort (blue people), when compared to a control group, such as unaffected individuals (red people). Filled circles represent genes annotated to a particular pathway (modified from (Wang et al., 2017b)).
Although a comprehensive understanding of the molecular vulnerabilities of different cancer cell types will undoubtedly provide a powerful roadmap to guide GI studies and therapeutic approaches, identifying relevant endogenous second site mutation(s) that underlie differential essentiality remains a major challenge. Complementary approaches involve systematic testing for GIs between defined pairs of genes. One approach uses gene editing methods to systematically introduce a second mutation into an engineered cell line carrying a stable ‘query’ mutation of interest and subsequently identifies GIs based on relative fitness of the resultant double mutant cells (Fig. 5B)(Blomen et al., 2015; Vizeacoumar et al., 2013). For example, RNA interference (RNAi) screens using pooled short-hairpin RNA (shRNA) libraries led to the construction of a genome-scale genetic network describing GIs associated with mutation of 5 genes in an otherwise isogenic HCT116 cancer cell line (Vizeacoumar et al., 2013). The resultant network was complex and composed of densely connected functional modules with individual genes participating in multiple interactions. Similar to the networks mapped for model organisms, most GIs in the cancer genetic network identified connections between functionally related genes, confirming that GIs identify functional relationships. The cancer genetic network also uncovered examples of conserved GIs, functions for uncharacterized genes, and targetable vulnerabilities (Vizeacoumar et al., 2013). A more recent study used transposon-mutagenesis to identify GIs for six different haploid query mutant cell lines and map a preliminary genetic network connecting human secretory and vesicle traffic pathways (Blomen et al., 2015).
A second approach for combinatorial GI screens involves simultaneous perturbation of two or more genes in the same cell (Fig. 5C). Several RNAi and CRISPR technologies based on multiplexing shRNAs and guide RNAs (gRNAs), respectively, have been developed and applied to identify GIs among select subsets of genes including known drug targets, genes implicated in cancer and chromatin regulators (Bassik et al., 2013; Boettcher et al., 2018; Du et al., 2017; Han et al., 2017; Najm et al., 2018; Shen et al., 2017; Wong et al., 2016). While genome-scale application of these combinatorial approaches remains technically challenging, a recent study used CRISPR interference (CRISPRi) to identify GIs among more than 220,000 pairs of genes suggesting that large-scale GI analysis in human cells should be feasible (Horlbeck et al., 2018). Genes belonging to the same pathway or complex are often connected in the resultant network, demonstrating that human genetic networks, like yeast networks, will be rich in functional information, and provide a powerful approach for annotating human gene function (Horlbeck et al., 2018).
[G]. Translating insights from model system genetic networks to human populations
Understanding the complex relationship between genotype and phenotype to ultimately predict trait heritability remains a primary challenge of modern genetics. Application of next generation sequencing technologies to catalog millions of human genetic variants combined with the implementation of electronic health records and population-based registries (e.g. UK BioBank etc.) are generating unprecedented genomic and phenomic resources to explore the genotype-phenotype problem in the context of human disease (reviewed in (Ritchie, 2018)). Genome-wide association studies (GWAS) have leveraged these data to link thousands of genetic loci to numerous different traits. As of 2018, the NHGRI (National Human Genome Research Institute)-EBI (European Bioinformatics Institute) Catalog reported >50,000 associated loci for >3000 unique traits (https://www.ebi.ac.uk/gwas/). Despite statistical evidence linking a remarkable number of candidate variants to a given disease, locus association alone is not predictive of disease risk and there remains a substantial disparity between the disease risk explained by the genetic loci discovered by GWAS and the estimated total heritable disease risk based on familial aggregation (Eichler et al., 2010; Zuk et al., 2012; Zuk et al., 2014). Several reasons have been proposed to explain this so-called “missing heritability”, including the existence of a large number of modifier loci, each having a relatively small effect or rare variants that cannot be easily detected using traditional approaches (Eichler et al., 2010; Stahl et al., 2012; Zuk et al., 2014).
As discussed earlier, another possibility, supported by experiments in yeast, is that GIs between different combinations of common and/or rare genetic variants may be responsible for a component of trait heritability (Forsberg et al., 2017; Zuk et al., 2012). In fact, GIs are known to affect susceptibility to and/or onset of several different complex diseases (Eichler et al., 2010; Prabhu and Pe’er, 2012; Zuk et al., 2012). Unfortunately, most methods lack the statistical power to detect GIs in large-scale genotype datasets due to the vast number of possible gene-gene combinations within the human genome. For example, every individual in a typical GWAS is genotyped at ~500,000 different genetic loci. Thus, an unbiased and systematic search for GIs involves analysis of more than 1011 pairwise gene combinations. Such an analysis is not feasible because there is not enough statistical power to assess this many hypothesis tests. Indeed, a theoretical study estimated through simulations that as many as 500,000 subjects would be needed to detect a significant GI between a single pair of genes under reasonable assumptions (Zuk et al., 2012).
While it is statistically difficult to detect significant GIs between individual human gene-gene pairs, the systematic genetic network analyses in yeast and other model systems provide important relevant insights. Specifically, as discussed earlier, the yeast GI network has a highly-organized structure in which genes are grouped together to form discrete network modules, and several studies have leveraged pathway or network enrichment analyses to identify single loci associated with specific human diseases (Califano et al., 2012; Wang et al., 2010; Wu et al., 2011; Zhang et al., 2014; Zuk et al., 2014). Importantly, the prevalence and coherence of negative interactions occurring within the same functional module in a WPM structure, or between pairs of modules in a BPM structure, as observed in the yeast genetic network (Bellay et al., 2011; Costanzo et al., 2016), suggests that these features can be exploited to discover GIs between pairs of functional modules instead of individual gene-gene or SNP-SNP pairs. In particular, the tendency of functional modules to form WPM and BPM genetic network structures provides prior knowledge that can reduce the statistical burden required to detect GIs in human populations by scanning human genotyping datasets for an enrichment of pairs of variants that occur within or between known pathways and complexes (Fig. 5D). A recent study used curated biological pathway standards as prior information to aggregate genetic variants and identify GIs between combinations of human pathways in breast cancer cohorts that are associated with either increased or decreased risk of disease (Wang et al., 2017b).
Ultimately, as has been done with yeast, sustained efforts to systematically map GIs in model human cell lines will generate a global genetic network for different human cells, providing a powerful data-driven resource that defines a functional wiring diagram for humans. Exploiting GIs to define functional modules and mapping their relationships should provide key knowledge for enabling systematic discovery of pathway-level GIs from human genotyping data, leading to a new level of understanding of human biology, enhancing our knowledge of the genotype to phenotype relationship (Costanzo et al., 2016; Ma et al., 2018).
Acknowledgements
We thank Jing Hou and Benjamin VanderSluis for comments. Work on genetic networks in the Andrews, Boone and Moffat laboratories is supported by grants from the Canadian Institutes of Health Research, a grant from the National Institutes of Health [R01HG005853], and the Ontario Research Fund.
References
- Agarwala A, and Fisher DS (2018). Adaptive walks on high-dimensional fitness landscapes and seascapes with distance-dependent statistics. bioRxiv. [DOI] [PubMed] [Google Scholar]
- Aly A, and Ganesan S (2011). BRCA1, PARP, and 53BP1: conditional synthetic lethality and synthetic viability. J Mol Cell Biol 3, 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth A, and Lord CJ (2018). Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nature Reviews Clinical Oncology 15, 564–576. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Kelley R, Krogan NJ, and Ideker T (2008). Functional maps of protein complexes from quantitative genetic interaction data. PLoS Comput Biol 4, e1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, et al. (2010). Rewiring of genetic networks in response to DNA damage. Science 330, 1385–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartha I, di Iulio J, Venter JC, and Telenti A (2018). Human gene essentiality. Nat Rev Genet 19, 51–62. [DOI] [PubMed] [Google Scholar]
- Baryshnikova A (2016). Systematic Functional Annotation and Visualization of Biological Networks. Cell Syst. [DOI] [PubMed] [Google Scholar]
- Baryshnikova A, Costanzo M, Kim Y, Ding H, Koh J, Toufighi K, Youn JY, Ou J, San Luis BJ, Bandyopadhyay S, et al. (2010). Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods 7, 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, et al. (2013). A systematic Mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152, 909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellay J, Atluri G, Sing TL, Toufighi K, Costanzo M, Ribeiro PS, Pandey G, Baller J, VanderSluis B, Michaut M, et al. (2011). Putting genetic interactions in context through a global modular decomposition. Genome Research 21, 1375–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, and Pringle JR (1991). Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol 11, 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, and Park HO (2012). Cell polarization and cytokinesis in budding yeast. Genetics 191, 347–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A, et al. (2015). Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096. [DOI] [PubMed] [Google Scholar]
- Boettcher M, Tian R, Blau JA, Markegard E, Wagner RT, Wu D, Mo X, Biton A, Zaitlen N, Fu H, et al. (2018). Dual gene activation and knockout screen reveals directional dependencies in genetic networks. Nat Biotechnol 36, 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, and Weissman JS (2008). A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods 5, 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burston HE, Maldonado-Baez L, Davey M, Montpetit B, Schluter C, Wendland B, and Conibear E (2009). Regulators of yeast endocytosis identified by systematic quantitative analysis. J Cell Biol 185, 1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butland G, Babu M, Diaz-Mejia JJ, Bohdana F, Phanse S, Gold B, Yang W, Li J, Gagarinova AG, Pogoutse O, et al. (2008). eSGA: E. coli synthetic genetic array analysis. Nat Methods 5, 789–795. [DOI] [PubMed] [Google Scholar]
- Byrne AB, Weirauch MT, Wong V, Koeva M, Dixon SJ, Stuart JM, and Roy PJ (2007). A global analysis of genetic interactions in Caenorhabditis elegans. J Biol 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano A, Butte AJ, Friend S, Ideker T, and Schadt E (2012). Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat Genet 44, 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AS, and Noor MA (2010). Epistasis modifies the dominance of loci causing hybrid male sterility in the Drosophila pseudoobscura species group. Evolution 64, 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Shi L, Hakenberg J, Naughton B, Sklar P, Zhang J, Zhou H, Tian L, Prakash O, Lemire M, et al. (2016). Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol 34, 531–538. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. (2007). Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–810. [DOI] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, and Weissman JS (2006). A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol 7, R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. (2010). The genetic landscape of a cell. Science 327, 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, et al. (2016). A global genetic interaction network maps a wiring diagram of cellular function. Science 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decourty L, Saveanu C, Zemam K, Hantraye F, Frachon E, Rousselle JC, Fromont-Racine M, and Jacquier A (2008). Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc Natl Acad Sci U S A 105, 5821–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuna A, Vetsigian K, Shoresh N, Hegreness M, Colon-Gonzalez M, Chao S, and Kishony R (2008). Exposing the fitness contribution of duplicated genes. Nat Genet 40, 676–681. [DOI] [PubMed] [Google Scholar]
- Diaz-Mejia JJ, Celaj A, Mellor JC, Cote A, Balint A, Ho B, Bansal P, Shaeri F, Gebbia M, Weile J, et al. (2018). Mapping DNA damage-dependent genetic interactions in yeast via party mating and barcode fusion genetics. Mol Syst Biol 14, e7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Fedyshyn Y, Koh JL, Prasad TS, Chahwan C, Chua G, Toufighi K, Baryshnikova A, Hayles J, Hoe KL, et al. (2008). Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proc Natl Acad Sci U S A 105, 16653–16658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T (1946). Genetics of natural populations; recombination and variability in populations of Drosophila pseudoobscura. Genetics 31, 269–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo J, Diss G, and Lehner B (2018). Pairwise and higher-order genetic interactions during the evolution of a tRNA. Nature 558, 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell RD, Ryan O, Jansen A, Cheung D, Agarwala S, Danford T, Bernstein DA, Rolfe PA, Heisler LE, Chin B, et al. (2010). Genotype to phenotype: a complex problem. Science 328, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees BL, Thorsson V, Carter GW, Rives AW, Raymond MZ, Avila-Campillo I, Shannon P, and Galitski T (2005). Derivation of genetic interaction networks from quantitative phenotype data. Genome Biol 6, R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D, Roguev A, Gordon DE, Chen M, Chen SH, Shales M, Shen JP, Ideker T, Mali P, Qi LS, et al. (2017). Genetic interaction mapping in mammalian cells using CRISPR interference. Nat Methods 14, 577–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkowski J, Kramer M, Surma MA, Balakrishnan R, Cherry JM, Krogan NJ, and Ideker T (2013). A gene ontology inferred from molecular networks. Nat Biotechnol 31, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MD, Symbor-Nagrabska A, Dollard L, Gifford DK, and Fink GR (2014). Interactions between chromosomal and nonchromosomal elements reveal missing heritability. Proc Natl Acad Sci U S A 111, 7719–7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, and Nadeau JH (2010). Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 11, 446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel RS (2010). Read-through strategies for suppression of nonsense mutations in Duchenne/ Becker muscular dystrophy: aminoglycosides and ataluren (PTC124). Journal of child neurology 25, 1158–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer B, Sandmann T, Horn T, Billmann M, Chaudhary V, Huber W, and Boutros M (2015). A map of directional genetic interactions in a metazoan cell. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA (1918). The correlation between relatives on the supposition of Mendelian inheritance. Proc R Soc Endinburgh 52, 399–433. [Google Scholar]
- Foley BR, Rose CG, Rundle DE, Leong W, and Edmands S (2013). Postzygotic isolation involves strong mitochondrial and sex-specific effects in Tigriopus californicus, a species lacking heteromorphic sex chromosomes. Heredity (Edinb) 111, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg SK, Bloom JS, Sadhu MJ, Kruglyak L, and Carlborg O (2017). Accounting for genetic interactions improves modeling of individual quantitative trait phenotypes in yeast. Nat Genet 49, 497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke JP, Chen CT, and Cohen BA (2006). Natural isolates of Saccharomyces cerevisiae display complex genetic variation in sporulation efficiency. Genetics 174, 985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, and Davis RW (1999). Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet 21, 278–283. [DOI] [PubMed] [Google Scholar]
- Guenole A, Srivas R, Vreeken K, Wang ZZ, Wang S, Krogan NJ, Ideker T, and van Attikum H (2013). Dissection of DNA damage responses using multiconditional genetic interaction maps. Molecular cell 49, 346–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE, Braberg H, Wu Q, Alexander R, Haase J, Ryan C, Lipkin-Moore Z, Franks-Skiba KE, Johnson T, Shales M, et al. (2013). Systematic triple-mutant analysis uncovers functional connectivity between pathways involved in chromosome regulation. Cell Rep 3, 2168–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Jeng EE, Hess GT, Morgens DW, Li A, and Bassik MC (2017). Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol 35, 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R, Papp B, Pal C, Oliver SG, and Delneri D (2007). Plasticity of genetic interactions in metabolic networks of yeast. Proc Natl Acad Sci U S A 104, 2307–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. (2015). High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 163, 1515–1526. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Szankasi P, Roberts CJ, Murray AW, and Friend SH (1997). Integrating genetic approaches into the discovery of anticancer drugs. Science 278, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Hoepfner D, Helliwell SB, Sadlish H, Schuierer S, Filipuzzi I, Brachat S, Bhullar B, Plikat U, Abraham Y, Altorfer M, et al. (2014). High-resolution chemical dissection of a model eukaryote reveals targets, pathways and gene functions. Microbiol Res 169, 107–120. [DOI] [PubMed] [Google Scholar]
- Horlbeck MA, Xu A, Wang M, Bennett NK, Park CY, Bogdanoff D, Adamson B, Chow ED, Kampmann M, Peterson TR, et al. (2018). Mapping the Genetic Landscape of Human Cells. Cell 174, 953–967 e922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CA 3rd, Chuang RY, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, et al. (2016). Design and synthesis of a minimal bacterial genome. Science 351, aad6253. [DOI] [PubMed] [Google Scholar]
- Jaffe M, Sherlock G, and Levy SF (2017). iSeq: A New Double-Barcode Method for Detecting Dynamic Genetic Interactions in Yeast. G3 (Bethesda) 7, 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. (2009). Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323, 1693–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CA, and Schekman R (1990). Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61, 723–733. [DOI] [PubMed] [Google Scholar]
- Kelley R, and Ideker T (2005). Systematic interpretation of genetic interactions using protein networks. Nat Biotechnol 23, 561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MH, Farre JC, Mitra K, Yu MK, Ono K, Demchak B, Licon K, Flagg M, Balakrishnan R, Cherry JM, et al. (2017). Active Interaction Mapping Reveals the Hierarchical Organization of Autophagy. Mol Cell 65, 761–774 e765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin E, VanderSluis B, Wang W, Tan G, Deshpande R, Chen Y, Usaj M, Balint A, Mattiazzi Usaj M, van Leeuwen J, et al. (2018). Systematic analysis of complex genetic interactions. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AY, St Onge RP, Proctor MJ, Wallace IM, Nile AH, Spagnuolo PA, Jitkova Y, Gronda M, Wu Y, Kim MK, et al. (2014). Mapping the cellular response to small molecules using chemogenomic fitness signatures. Science 344, 208–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Crombie C, Tischler J, Fortunato A, and Fraser AG (2006). Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet 38, 896–903. [DOI] [PubMed] [Google Scholar]
- Ma J, Yu MK, Fong S, Ono K, Sage E, Demchak B, Sharan R, and Ideker T (2018). Using deep learning to model the hierarchical structure and function of a cell. Nat Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magtanong L, Ho CH, Barker SL, Jiao W, Baryshnikova A, Bahr S, Smith AM, Heisler LE, Choy JS, Kuzmin E, et al. (2011). Dosage suppression genetic interaction networks enhance functional wiring diagrams of the cell. Nature Biotechnology 29, 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani R, St Onge RP, Hartman J.L.t., Giaever G, and Roth FP (2008). Defining genetic interaction. Proc Natl Acad Sci U S A 105, 3461–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H, Shales M, Fernandez-Pinar P, Wei P, Molina M, Fiedler D, Shokat KM, Beltrao P, Lim W, and Krogan NJ (2015). Differential genetic interactions of yeast stress response MAPK pathways. Mol Syst Biol 11, 800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton MJ, DeRisi JL, Bennett HA, Iyer VR, Meyer MR, Roberts CJ, Stoughton R, Burchard J, Slade D, Dai H, et al. (1998). Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med 4, 1293–1301. [DOI] [PubMed] [Google Scholar]
- Matsui T, Lee JT, and Ehrenreich IM (2017). Genetic suppression: Extending our knowledge from lab experiments to natural populations. Bioessays 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JN, and Pearce DA (2014). Nonsense-mediated decay in genetic disease: friend or foe? Mutat Res Rev Mutat Res 762, 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullis MN, Matsui T, Schell R, Foree R, and Ehrenreich IM (2018). The complex underpinnings of genetic background effects. Nat Commun 9, 3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm FJ, Strand C, Donovan KF, Hegde M, Sanson KR, Vaimberg EW, Sullender ME, Hartenian E, Kalani Z, Fusi N, et al. (2018). Orthologous CRISPR-Cas9 enzymes for combinatorial genetic screens. Nat Biotechnol 36, 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Osmond BC, and Botstein D (1989). Suppressors of yeast actin mutants. Genetics 121, 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Meyers RM, Michel BC, Mashtalir N, Sizemore AE, Wells JN, Cassel SH, Vazquez F, Weir BA, Hahn WC, et al. (2018). Interrogation of Mammalian Protein Complex Structure, Function, and Membership Using Genome-Scale Fitness Screens. Cell Syst 6, 555–568 e557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, and Boeke JD (2006). A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124, 1069–1081. [DOI] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, and Boeke JD (2004). A robust toolkit for functional profiling of the yeast genome. Mol Cell 16, 487–496. [DOI] [PubMed] [Google Scholar]
- Patra B, Kon Y, Yadav G, Sevold AW, Frumkin JP, Vallabhajosyula RR, Hintze A, Ostman B, Schossau J, Bhan A, et al. (2017). A genome wide dosage suppressor network reveals genomic robustness. Nucleic Acids Res 45, 255–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC, Otto SP, and Whitlock MC (2000). Beyond the average. The evolutionary importance of gene interactions and variability of epistatic effects In Epistasis and the evolutionary process, Wolf JB, Brodie EDI, and Wade MJ, eds. (Oxford University Press; ). [Google Scholar]
- Piotrowski JS, Li SC, Deshpande R, Simpkins SW, Nelson J, Yashiroda Y, Barber JM, Safizadeh H, Wilson E, Okada H, et al. (2017). Functional annotation of chemical libraries across diverse biological processes. Nat Chem Biol 13, 982–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S, and Pe’er I (2012). Ultrafast genome-wide scan for SNP-SNP interactions in common complex disease. Genome Res 22, 2230–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancati G, Moffat J, Typas A, and Pavelka N (2018). Emerging and evolving concepts in gene essentiality. Nat Rev Genet 19, 34–49. [DOI] [PubMed] [Google Scholar]
- Rauscher B, Heigwer F, Henkel L, Hielscher T, Voloshanenko O, and Boutros M (2018). Toward an integrated map of genetic interactions in cancer cells. Mol Syst Biol 14, e7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S, Castelein CM, Ahmed ZM, Lalwani AK, Mastroianni MA, Naz S, Smith TN, Liburd NA, Friedman TB, Griffith AJ, et al. (2000). Dominant modifier DFNM1 suppresses recessive deafness DFNB26. Nat Genet 26, 431–434. [DOI] [PubMed] [Google Scholar]
- Riesselman AJ, Ingraham JB, and Marks DS (2018). Deep generative models of genetic variation capture the effects of mutations. Nat Methods 15, 816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan JD, and Nadeau JH (2017). From Peas to Disease: Modifier Genes, Network Resilience, and the Genetics of Health. Am J Hum Genet 101, 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD (2018). Large-Scale Analysis of Genetic and Clinical Patient Data. 1, 263–274. [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, et al. (2008). Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 322, 405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan O, Shapiro RS, Kurat CF, Mayhew D, Baryshnikova A, Chin B, Lin ZY, Cox MJ, Vizeacoumar F, Cheung D, et al. (2012). Global gene deletion analysis exploring yeast filamentous growth. Science 337, 1353–1356. [DOI] [PubMed] [Google Scholar]
- Sarkisyan KS, Bolotin DA, Meer MV, Usmanova DR, Mishin AS, Sharonov GV, Ivankov DN, Bozhanova NG, Baranov MS, Soylemez O, et al. (2016). Local fitness landscape of the green fluorescent protein. Nature 533, 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. (2005). Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123, 507–519. [DOI] [PubMed] [Google Scholar]
- Segre D, Deluna A, Church GM, and Kishony R (2005). Modular epistasis in yeast metabolism. Nat Genet 37, 77–83. [DOI] [PubMed] [Google Scholar]
- Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN, et al. (2017). Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat Methods 14, 573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, and Ideker T (2011). Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis RW, Nislow C, Roth FP, and Giaever G (2007). Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet 39, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Wegmann D, Trynka G, Gutierrez-Achury J, Do R, Voight BF, Kraft P, Chen R, Kallberg HJ, Kurreeman FA, et al. (2012). Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Genet 44, 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, and Botstein D (1988). Unlinked noncomplementation: isolation of new conditional-lethal mutations in each of the tubulin genes of Saccharomyces cerevisiae. Genetics 119, 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MB, and Ehrenreich IM (2014). Genetic interactions involving five or more genes contribute to a complex trait in yeast. PLoS Genet 10, e1004324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. (2004). Global mapping of the yeast genetic interaction network. Science 303, 808–813. [DOI] [PubMed] [Google Scholar]
- Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, et al. (2017). Defining a Cancer Dependency Map. Cell 170, 564–576 e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Nichols RJ, Siegele DA, Shales M, Collins SR, Lim B, Braberg H, Yamamoto N, Takeuchi R, Wanner BL, et al. (2008). High-throughput, quantitative analyses of genetic interactions in E. coli. Nat Methods 5, 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen J, Pons C, Mellor JC, Yamaguchi TN, Friesen H, Koschwanez J, Usaj MM, Pechlaner M, Takar M, Usaj M, et al. (2016). Exploring genetic suppression interactions on a global scale. Science 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wageningen S, Kemmeren P, Lijnzaad P, Margaritis T, Benschop JJ, de Castro IJ, van Leenen D, Groot Koerkamp MJ, Ko CW, Miles AJ, et al. (2010). Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell 143, 991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizeacoumar FJ, Arnold R, Vizeacoumar FS, Chandrashekhar M, Buzina A, Young JT, Kwan JH, Sayad A, Mero P, Lawo S, et al. (2013). A negative genetic interaction map in isogenic cancer cell lines reveals cancer cell vulnerabilities. Molecular systems biology 9, 696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, and Hakonarson H (2010). Analysing biological pathways in genome-wide association studies. Nat Rev Genet 11, 843–854. [DOI] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, and Sabatini DM (2015). Identification and characterization of essential genes in the human genome. Science 350, 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, Chen WW, Lander ES, and Sabatini DM (2017a). Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xu ZZ, Costanzo M, Boone C, Lange CA, and Myers CL (2017b). Pathway-based discovery of genetic interactions in breast cancer. PLoS Genet 13, e1006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich DM, Lan Y, Wylie CS, and Heckendorn RB (2013). Should evolutionary geneticists worry about higher-order epistasis? Curr Opin Genet Dev 23, 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Wong ASL, Choi GCG, Cui CH, Pregernig G, Milani P, Adam M, Perli SD, Kazer SW, Gaillard A, Hermann M, et al. (2016). Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proceedings of the National Academy of Sciences 113, 2544–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, and Lin X (2011). Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 89, 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Boerwinkle E, and Xiong M (2014). Epistasis analysis for quantitative traits by functional regression model. Genome Res 24, 989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, and Lander ES (2012). The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A 109, 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, Daly MJ, Neale BM, Sunyaev SR, and Lander ES (2014). Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A 111, E455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]