Abstract
Transformed root cultures (TRC) are used to mass produce arbuscular mycorrhizal (AM) fungal propagules in vitro. These propagules are then used in research, agriculture, and ecological restoration. There are many examples from other microbial systems that long-term in vitro propagation leads to domesticated strains that differ genetically and functionally. Here, we discuss potential consequences of in TRC propagation on AM fungal traits, and how this may affect their functionality. We examine weather domestication of AM fungi has already happened and finally, we explore whether it is possible to overcome TRC-induced domestication.
Keywords: arbuscular mycorrhizal fungi, fungal domestication, fungal evolution, in vitro propagation, transformed root cultures
Introduction
Domestication of plants and animals has been a hallmark of the Anthropocene (Zeder, 2006), resulting in altered morphology, decreased genetic diversity, altered behavior, and altered function in the domesticant. Such far reaching changes are necessary to maintain the domesticated state but can present a risk to food security (Chen et al., 2015; Whitehead et al., 2017; Egan et al., 2018), and in some cases, to the status of the domesticated species (Pearce, 2003; Ploetz, 2003). For example, domestication of wild bananas led to a sterile and genetically homogeneous cultivar that now faces extinction (Pearce, 2003; Ploetz, 2003). Similarly, decreases in genetic diversity as a result of domestication has been well documented in many crops, including common bean (Bitocchi et al., 2013), rice (Ram et al., 2007), wheat (Reif et al., 2005), soybean (Hyten et al., 2006), and pear (Nishio et al., 2016). This decrease in genetic diversity is often associated with losses of functional traits such as herbivore resistance (Chaudhary and Bhupendra, 2013), reduced immune function (e.g., Honey bees, López-Uribe et al., 2017) or even behavioral changes such as loss of immigration ability in monarch butterflies (Tenger-Trolander et al., 2019).
The effect of domestication on ecological competence is not novel in mycology (Jinks, 1952; Roper et al., 2011); plant pathogens lose pathogenicity when kept in culture for extended periods (Naiki and Cook, 1983), and domestication of Saccharomyces cerevisiae created yeasts without the ability to reproduce sexually or survive outside of laboratory conditions (Gallone et al., 2016). Aspergillus oryzae diverged from a pathogen to become a commercially important fermenter and subsequently lost genes related to pathogenicity (Machida et al., 2005). Such losses may result from bottleneck effects and environmental selection, especially if the system used in cultivation does not represent the conditions under which they originally evolved (see review by Douglas and Klaenhammer, 2010).
Arbuscular mycorrhizal (AM) fungi are obligate biotrophs that participate in an ancient symbiosis with plants (Smith and Read, 2008; Brundrett and Brundrett, 2009). Through this symbiosis, AM fungi provide plants increased access to soil resources in return for carbon in the form of sugar and lipids (Luginbuehl et al., 2017). Besides the nutritional benefit to the plants, AM fungi can also increase plant tolerance to environmental stress [e.g., water (Ruiz-Lozano and Aroca, 2010), salinity (Porcel et al., 2012), and heavy metals (Díaz et al., 1996)]. AM fungi are known to stimulate plant photosynthetic activity (Boldt et al., 2011) and enhance plants’ disease resistance (Pozo and Azcón-Aguilar, 2007; Jung et al., 2012). Because of these benefits, considerable effort has focused on finding ways to propagate and study these fungi for potential applications including agriculture, landscaping, and landscape restoration (Sawers et al., 2008; Berruti et al., 2016). One of the most successful methods of propagating clean material employs the use of transformed root cultures (TRC) (see sidebar#1 for information on TRC) (Mosse and Hepper, 1975; Bécard and Fortin, 1988; Stockinger et al., 2009; Rosikiewicz et al., 2017). While this method is efficient for producing uncontaminated propagules, it represents a highly artificial environment and could potentially lead to domesticated AM fungal strains (Figure 1).
Sidebar#1. Transformed root cultures.
TRC involves infection of a “hairy root” culture with an AM fungal spore. A hairy root culture is the product of gene transfer from the root-inducing (Ri) plasmid of the parasite Agrobacterium rhizogenes into the genome of a host plant (Willmitzer et al., 1982). The Ri plasmid contains genes that increase rates of both cell division through increased cytokinins production and cell elongation through auxin production. The transformed root also produces opines used as a food source by the colonizing bacteria. After eliminating the bacterium with antibiotics, the resulting plant is a proliferation of “hairy roots” that grow faster and produce higher quantities of secondary metabolites than normal roots (Capone et al., 1989). The roots are kept in dark and there is no photosynthetic tissue or shoot present. This method, in addition to the industrial benefits, including reduced contamination, minimal space and maintenance requirements, and standardization, has enabled long-term laboratory cultures, effectively domesticating certain isolates.
Figure 1.
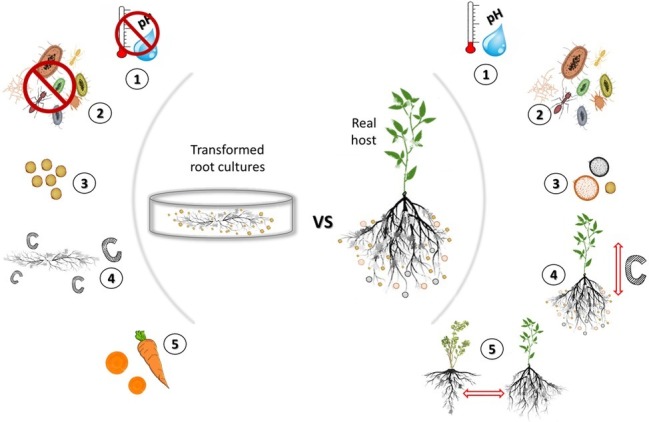
Comparison of the artificial growing environment of transformed root cultures (TRC) to a natural environment. While TRC can produce uncontaminated and abundant AM fungal propagules, they consists a highly artificial and eutrophic environment that lacks environmental stimuli. (1) Lack of abiotic stimuli: Petri dishes are kept sealed to retain moisture and are propagated in the dark at a stable temperature that benefits the AM fungal productivity (propagule production). In a natural environment, there is continuous fluctuation of temperature, and moisture as well as high spatial variation in pH but also in multiple other abiotic factors. Such abiotic factors can affect the AM fungal growth as well as the mycorrhizal response (MR) (Wang et al., 1993; Heinemeyer and Fitter, 2004) and have seen to alter the gene expression of fungi (Meyer et al., 2017). (2, 3) Lack of biotic stimuli: the main benefit of TRC is the ability to grow AM fungi under sterile conditions that allows for uncontaminated propagation of the desired species/isolate. Each TRC contains only a single isolate of AM fungi. AM fungi have antagonistic and synergetic relations with other soil microbes (Mar Vázquez et al., 2000) as well as with other AM fungi (Wilson, 1984; Engelmoer et al., 2014). The TRC environment lacks biotic interactions that when present can lead to the production of multiple chemical compounds (Naiki and Cook, 1983; Gallone et al., 2016) that confer stress resistance or can enhance the metabolic activity of the engaged microbes (Ola et al., 2013). (4) Unrestricted carbon flow: TRC lack a shoot and therefore carbon flow to the AM fungus is consistent and continuous (Fortin et al., 2002) in contrast to a real plant where carbon flow fluctuates daily and seasonally between shoot and root (Lippu, 1998). (5) Lack of host diversity: in nature, AM fungi can interact simultaneously with multiple host. Host identity can alter significantly the gene expression of AM fungi (Mateus et al., 2019) and alter the progression of the symbiosis (Angelard et al., 2010). The dramatic reduction of host diversity and continuous re-cultivation using a sole host [most commonly carrot (Daucus carota)] may affect the function and genetic diversity of the cultured AM fungi.
Currently, commercial AM fungal inocula are used both in horticulture and field applications (Berruti et al., 2016). Many of the fungal propagules used in commercial products originate from TRC. The effect of TRC on the evolution of “domesticated” AM fungi is not clear (Plenchette et al., 1996). Here, we argue that commercial production of AM fungi via TRC represents strong selection pressure on fungi and represents a form of domestication, through changes to nutrient limitations, microbial consortia, and reduced host variation. Such selection pressure may lead to reduced genetic diversity and mutualistic quality.
Luxurious Nutrient Conditions
Although AM fungi form functional mycorrhizas in TRC, the unique nutritional strategy of hairy roots may affect the quality of the symbiosis. In association with a normal plant, AM fungi grow in tandem with roots which fluctuate daily and seasonally (Lippu, 1998). In the case of transformed roots, the flow of carbon is consistent and continuous (Fortin et al., 2002), thereby promoting unrestricted fungal growth. In contrast, natural plants, which allocate most carbon to above ground shoots, impose carbon limits to root fungal symbionts. Root fungal symbionts of hairy roots are thus not carbon-limited. Unsurprisingly, such growing conditions promote spore production which demands significant carbon reserves (up to 60,000 per plate for R. irregulare DAOM197198) (Rosikiewicz et al., 2017).
In addition to luxury carbon, hairy roots experience no nutrient limitation in TRC (Bécard and Fortin, 1988). Such luxury nutrient conditions do not promote fungal provisioning of nutrients to hosts. There is considerable evidence that luxury soil nutrient status reduces both AM fungal abundance intraradically and mycorrhizal response (MR) in hosts (Menge et al., 1978; Thomson et al., 1986; Breuillin et al., 2010; Bonneau et al., 2013), which may lead to less beneficial associations. For example, increased N levels (via nitrogen fertilization) can select for rhizobia (Weese et al., 2015), and AM fungi (Johnson, 1993) that provide reduced benefit to the host plants. It is therefore possible that a highly eutrophic environment, such as TRC, may promote selection for less mutualistic AM fungi.
Talk Between Microbial Neighbors
The monoxenic environment of TRC lacks much of the hyphosphere/rhizosphere microbial consortia that play an important role in the AM symbiosis. Co-existing microbes engaged in antagonistic or synergetic interactions produce bioactive compounds which can be used in defense, to confer stress tolerance or boost metabolic activities for the producers (Ola et al., 2013). Such compounds are not produced when bacteria (Koskiniemi et al., 2012) and fungi (Naiki and Cook, 1983; Gallone et al., 2016) are maintained under axenic conditions due to lack of appropriate environmental stimuli from neighboring microbes (Marmann et al., 2014). The pathways for these signaling compounds can be lost permanently via selective gene deletion over generations of continuous propagation in vitro.
Similar to other microbes, AM fungi have antagonistic and synergetic relations with other AM fungi (Wilson, 1984; Engelmoer et al., 2014) and other soil microbes (Mar Vázquez et al., 2000). For example, it was recently demonstrated that AM fungi have the ability to indirectly increase the nitrogen (N) uptake by plants via association with soil microbes (Hestrin et al., 2019). Therefore, growing in an environment that inhibits these interactions could reduce the effectiveness of such strains in natural conditions.
In addition to the selection pressure resulting from lack of microbial cross talk, reduction or even elimination of fungal endobacteria and bacteria that reside on the hyphal or spore surface in TRC, can affect fungal function (Dearth et al., 2018) and mutualism performance (Vannini et al., 2016). Establishing AM fungi in TRC requires surface sterilization and antibiotics in order to eliminate surface bacteria (Bécard and Fortin, 1988). However, AM fungi naturally comprise a community of bacteria that reside in, and on, hyphae and spores. Abundant rhizobia and pseudomonads have been found attached on spore and hyphal surface (Bianciotto et al., 1996b; Roesti et al., 2005; Agnolucci et al., 2015), but also bacterium-like organisms (BLOs) (Bianciotto et al., 1996a; Naumann et al., 2010) and Mollicutes/mycoplasma-related endobacteria (MRE) (Desiro et al., 2014; Torres-Cortes et al., 2015; Naito et al., 2017) were detected within the cytoplasm. Some of these bacteria possess chitinolytic abilities (Roesti et al., 2005; Agnolucci et al., 2015) and their abilities to degrade spore walls can play a crucial role in spore germination (Mayo et al., 1986). Of course, the presence of such bacteria can also benefit the colonized plants via a cascade of gene activation and chemical signals (Artursson et al., 2006). Long-term in vitro culturing could negatively affect the interaction between AM fungi and their own beneficial mutualists (Lumini et al., 2007).
Plant Identity
There is increasing evidence that plant genotype can significantly affect the symbiosis (Chialva et al., 2018; Mateus et al., 2019). In the case of TRC propagation, fungi are exposed to dramatically reduced host diversity [most commonly carrot (Daucus carota) or tomato (Solanum lycopersicum)]. While gene activation in the early stages of colonization are preserved among hosts (Delaux et al., 2014), the progression of the symbiosis can be significantly altered depending on host identity both regarding the plant (Angelard et al., 2010) and fungal response (Cavagnaro et al., 2001; Koch et al., 2017). Mateus et al. (2019) observed large differences in the expression between fungal isolates growing on multiple cassava cultivars, but the differences were influenced largely by the genotype of the cultivar host. The reduction in host genetic diversity to a single genotype in TRC may lead to genetic drift and unused gene deletion for the AM fungus (Muller’s ratchet in host restricted lineages, see Moran et al., 2008). Recently, Sugiura et al. (2019) identified myristate, a fatty acid, as a usable carbon source from Rhizoglomus irregulare during the asymbiotic growth that can promote hyphal growth to the production of daughter spores in a host-free culture. While such information advances our knowledge in AM fungal metabolism, such a mechanism could also lead to host-free AM fungal propagation systems, with unknown effects on the efficacy of the symbiosis. Culturing symbionts in host-free environments has been shown to reduce symbiotic quality (Marx and Daniel, 1976; Speakman, 1982).
Is there Evidence of Domestication on Arbuscular Mycorrhizal Fungi?
Given all the opportunities for deleterious selection on AM fungi growing in TRC, is there any evidence that domestication has happened? Evidence for domestication would require reduced genetic variation as well as morphological and functional changes.
Reduced Genetic Variation
There is evidence that controlled conditions such as TRC can lead to loss of genetic diversity among some AM fungal isolates For example, Wyss and Bonfante (1993) showed genotypic changes among isolates of a single species (Funneliformis mosseae BEG12) when maintained under long-term lab conditions. In addition, there is evidence of sequence loss in spores of an isolate of a Glomus coronatum when maintained in cultures compared to field originated spores (Clapp et al., 2001) and reduced allelic variation in spores of Claroideoglomus etunicatum compared to the parent isolate following single spore inoculations (Boon et al., 2013).
Morphological and Functional Alterations
Regardless the mechanism leading to genotype changes, there is evidence that in vitro cultivation affects AM fungal functional traits. In vitro cultivation has led to increased germination rates (Kokkoris et al., 2019b) and reduced in propagule size (Pawlowska et al., 1999; Calvet et al., 2013). Plenchette et al. (1996) found that in vitro produced spores of Glomus versiforme were significantly less infective, even only after three successive generation in vitro. Calvet et al. (2013) observed that in vitro colonization of AM fungi reduced host nutritional benefit. Similarly, Kokkoris and Hart (2019) showed that in vitro propagation resulted in a trade-off between spore production and phosphorus (P) benefit. Copious spore production over nutritional benefit is a trade-off that seems to be preserved even when this isolate is grown in pots with a variety of different hosts (Kokkoris et al., 2019a).
Loss of Endobacterial Symbionts
In vitro cultivation may affect the endocellular bacteria associated with fungal spores. Candidatus Glomeribacter gigasporarum is a bacterium that resides in spores of Gigaspora margarita. In vitro, this bacterium experiences populational dilution and eventually disappears leading to “pure” spores over successive generation in vitro (Lumini et al., 2007). Although the bacterium is not required for G. margarita to complete its life cycle, its absence alters spore’s morphology and negatively affects germination and growth (Lumini et al., 2007) and can significantly alter the fungal activity (including protein expression, and quality and quantity of lipidic profile) (Salvioli et al., 2010, 2016).
Incompatibility Between Isolates?
One potential consequence of TRC cultivation may affect hyphal fusion among compatible fungi. For example, in vivo cultivation for 20 years led to vegetative incompatibility for F. mosseae (Sbrana et al., 2018). If long-term culturing in TRC inhibits the ability of anastomosis, then domesticated isolates might be unable to interact with other isolates in nature. Incompatibility could even lead to a permanent homokaryotic stage, preventing genetic information exchange between compatible isolates and thus adaptation to novel conditions (see sidebar#2 for the importance of anastomosis on AM fungi). It could also pose a survivorship disadvantage for such an isolate if used for field inoculations due to the isolation from the natural hyphal network (Sbrana et al., 2011). Loss of anastomosis might be the reason why in vitro produced strains often fail to establish and persist in natural environments post inoculation (Corkidi et al., 2004; Farmer et al., 2007; Tarbell and Koske, 2007).
Sidebar#2. The role of anastomosis in arbuscular mycorrhizal fungi.
Heterokaryosis, first coined by Burgeff (1914), is common in the fungal kingdom. It occurs when compatible hyphae fuse (anastomosis), but do not undergo karyogamy. Heterokaryosis is observed in earlier-diverging lineages of fungi such as Phycomyces (Mucoromycota), where most sexual spores contain three to four nuclei, which can be genetically similar or different (Mehta and Cerdá-Olmedo, 2001). The classical understanding of heterokaryosis emphasizes the dikaryon as a precursor to karyogamy, or the eventual fusing of two nuclei to form a monokaryotic, diploid cell, which then undergoes meiosis leading to the production of haploid, sexual spores. However, there are many examples of fungi for which the heterokaryon plays a role beyond sexual recombination [Jinks, 1952 (Penicillium); James et al., 2008 (Heterobasidion); Roper et al., 2011; Samils et al., 2014 (Neurospora)] Diverse nucleotypes in spores may prevent the loss of genetic variation and functional diversity in the case of population reduction (i.e., prevent genetic drift through bottleneck effects) or ensure maintenance of genetic information when new populations are established from few propagules (prevent genetic drift through founder effects).
Similar to other heterothallic fungal groups (Asco- and Basidiomycotan), nucleotype diversity may be maintained through anastomosis in wild populations of closely related AMF (Giovannetti et al., 1999, 2001, 2003; Croll et al., 2008). For AM fungi, anastomosis has been observed among hyphae of the same isolates during the asymbiotic and symbiotic stage even when growing in different systems but also been observed between closely related isolates (Giovannetti et al., 2001; Croll et al., 2009; Purin and Morton, 2011; De la Providencia et al., 2013; de Novais et al., 2013; Barreto De Novais et al., 2017). During anastomosis, plasmogamy can occur, and mitochondria (De la Providencia et al., 2013) and nuclei (Croll et al., 2008) can be shared between partners. Although anastomosis is recognized primarily as a healing mechanism post disturbance (de la Providencia et al., 2005) especially for Gigaspora sp., also plays an important ecological role, since newly germinating spores can connect to the pre-established network prior interacting with a host, gaining an important survivorship benefit (Sbrana et al., 2011).
While debate about their status as homo- versus heterokaryons has circulated for nearly two decades (Kuhn et al., 2001; Hijri and Sanders, 2004; Pawlowska and Taylor, 2004; Croll et al., 2008; Tisserant et al., 2013; Lin et al., 2014; Boon et al., 2015), recent evidence shows that AM fungi can be haploid and homokaryotic (meaning that they contain thousands of genetically similar nuclei with haploid number of chromosomes) or haploid and dikaryotic (meaning that they contain two genetically distinct type of nuclei with in equal proportions, with a haploid number of chromosomes) (Ropars et al., 2016; Chen et al., 2018a). Corradi and Brachmann (2017) proposed that compatible AM fungi have the ability to mix their nuclei continually in the field, through anastomosis, effectively creating genetically novel isolates via karyogamy and meiosis. Although it is still not yet clear under which conditions the dikaryons proceed to karyogamy and meiosis aka to sexual reproduction (Chen et al., 2018b), hyphal fusion between compatible isolates seem to represent a vital step for plasmogamy and exchange of genetic information.
Can We Overcome Transformed Root Cultures-Induced Domestication?
If TRC propagation of AM fungi produces inferior mutualists, it is reasonable to wonder whether specific practices in TRC production could prevent such unwanted changes. Such practices, like co-cultivation, medium modifications and re-association with natural hosts, already exist and applied in other microbial systems.
Co-Cultivation with Microbes
Axenic, and in case of AM fungi, monoxenic growing conditions can reduce the chemical diversity of the produced compounds due to lack of environmental stimuli. Co-cultivation of microbes can activate silent gene clusters of the microbial partners (Brakhage et al., 2008), protecting fungi against genetic drift and gene deletion. For example co-culture of bacteria and fungi (Fusarium tricinctum Bacillus subtilis) showed a 78-fold increase in fungal metabolite production compared to the pure culture of the fungus (Ola et al., 2013). Similarly, a fungal co-culture (Coprinopsis cinerea and Gongronella sp.) produced 900 times increased oxidation activity compared to pure cultures (Pan et al., 2014). Growing AM fungi with two Paenibacillus validus bacterial isolates increased fungal growth even in absence of a host (Hildebrandt et al., 2002). Co-cultivation of AM fungi with diverse microorganisms may be a way to maintain genetic variation and function by activating AM fungal genes that would otherwise be silent, due to reduced environmental stimuli, and prone to deletion if maintained long term in TRC.
Culturing Conditions
The most commonly used TRC medium is the M medium proposed by Bécard and Fortin (1988). While no major modifications have been made on M medium, addition of simulative chemical molecules could compensate for lack of microbial associates and trigger gene activation for secondary metabolite production. For example, the addition of fatty acids (signal from P. validus, see previous section) can induce colonization ability and stimulate the spore production of AM fungi (Kameoka et al., 2019). In addition, chemical effectors responsible for promoting hyphal branching, mycorrhization, and the efficiency of the symbiosis have been identified (e.g., strigolactones) (Akiyama et al., 2005; Besserer et al., 2006), which may lead to increased gene activation and help maintaining the genetic and functional variation in TRC.
Additional changes in the medium or in the growing conditions may stimulate recombination in AM fungi and encourage the production of novel genotypes. For example, in Coprinus congregatus, a Basidiomycete, the quantity of arginine in the medium controls the expression of the mating type genes and ultimately the growth of the fungus as a homo or dikaryon (Ross et al., 1991). We need to identify environmental controls of AM fungal mating behavior to optimize growing conditions that will choose for efficient symbionts and not just copious spore producers.
Re-Association with Compatible Hosts
Ectomycorrhizal (ECM) fungi can lose their symbiotic ability and eventually fail to colonize plant roots if maintained in vitro long term (Marx and Daniel, 1976). Growing strains via host passage (association with a compatible host) every 4 years alleviates this bottleneck (Marx, 1981). The re-isolated strain from the colonized roots shows increased colonization ability but also increased symbiotic quality compared to solely in vitro retained strains (Thomson et al., 1993). Similarly for pathogenic fungi, pathogenicity can be lost with long-term in vitro cultivation and “passaging” the strains trough a compatible host and re-isolating and can revitalize their infective abilities (Speakman, 1982). Furthermore, the “asexual” yeast Candida albicans was stimulated to mate when injected into a mammalian host (Hull et al., 1999) showing the significant role an appropriate host can have even for sexual reproduction. While the presence of a root system is a prerequisite for AM fungal cultures, the important differences between TRC and real plant system may alter the AM fungal function. Passage through real host or even community of hosts could retain the functionality of the domesticated strains.
Conclusions
There is clear evidence that continuous in vitro propagation alters AM fungal morphology, genetics, and functioning, meaning that domestication of such strains is in progress or has occurred. While mass production of AM fungal propagules is needed for a sustainable inoculant industry, in vitro propagation may bring unwanted changes to the cultured isolates. If domestication reduces the isolate’s ability to anastomose, these fungi would have a fitness disadvantage in the field. Alternatively, if the unnatural environment of TRC creates strains that are less beneficial in natural conditions, but these isolates are still able to anastomose with native fungi, such isolates may impact negatively on the gene pool of natural populations. It is important to further examine the effects of domestication on AM fungi and predict how changes could greatly affect the environment following inoculation with such strains.
Data Availability Statement
No datasets were generated or analyzed for this study.
Author Contributions
MH and VK conceptualized the work and shared the writing and revision of the MS. MH and VK approved the publication of the MS in its current form. MH and VK agreed to be accountable for all aspects of the work including accuracy or integrity of any part of the work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- AM
Arbuscular mycorrhizas/arbuscular mycorrhizal
- BLOs
Bacterial like organisms
- ECM
Ectomycorrhizas/ectomycorrhizal
- MRE
Mollicutes/mycoplasma-related endobacteria
- MR
Mycorrhizal response
- N
Nitrogen
- P
Phosphorus
- Ri
Root inducing
- TRC
Transformed root cultures.
Footnotes
Funding. MH was funded by the NSERC Discovery Grant.
References
- Agnolucci M., Battini F., Cristani C., Giovannetti M. (2015). Diverse bacterial communities are recruited on spores of different arbuscular mycorrhizal fungal isolates. Biol. Fertil. Soils 51, 379–389. 10.1007/s00374-014-0989-5 [DOI] [Google Scholar]
- Akiyama K., Matsuzaki K., Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827. 10.1038/nature03608, PMID: [DOI] [PubMed] [Google Scholar]
- Angelard C., Colard A., Niculita-Hirzel H., Croll D., Sanders I. R. (2010). Segregation in a mycorrhizal fungus alters Rice growth and symbiosis-specific gene transcription. Curr. Biol. 20, 1216–1221. 10.1016/j.cub.2010.05.031, PMID: [DOI] [PubMed] [Google Scholar]
- Artursson V., Finlay R. D., Jansson J. K. (2006). Interactions between arbuscular mycorrhizal fungi and bacteria and their potential for stimulating plant growth. Environ. Microbiol. 8, 1–10. 10.1111/j.1462-2920.2005.00942.x, PMID: [DOI] [PubMed] [Google Scholar]
- Barreto De Novais C., Pepe A., Siqueira J. O., Giovannetti M., Sbrana C., Brazil M. (2017). Scientia agricola compatibility and incompatibility in hyphal anastomosis of arbuscular mycorrhizal. Sci. Agric. 74, 411–416. 10.1590/1678-992x-2016-0243 [DOI] [Google Scholar]
- Bécard G., Fortin J. A. (1988). Early events of vesicular–arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol. 108, 211–218. 10.1111/j.1469-8137.1988.tb03698.x [DOI] [PubMed] [Google Scholar]
- Berruti A., Lumini E., Balestrini R., Bianciotto V. (2016). Arbuscular mycorrhizal fungi as natural biofertilizers: let’s benefit from past successes. Front. Microbiol. 6:1559. 10.3389/fmicb.2015.01559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A., Puech-Pagè V., Kiefer P., Gomez-Roldan V., Jauneau A., Bastien Roy S., et al. (2006). Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4:e226. 10.1371/journal.pbio.0040226, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianciotto V., Bandi C., Minerdi D., Sironi M., Tichy H. V., Bonfante P. (1996a). An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. Appl. Environ. Microbiol. 62, 3005–3010. Available at: http://www.ncbi.nlm.nih.gov/pubmed/8702293 [Accessed July 11, 2019] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianciotto V., Minerdi D., Perotto S., Bonfante P. (1996b). Cellular interactions between arbuscular mycorrhizal fungi and rhizosphere bacteria. Protoplasma 193, 123–131. 10.1007/BF01276640 [DOI] [Google Scholar]
- Bitocchi E., Bellucci E., Giardini A., Rau D., Rodriguez M., Biagetti E., et al. (2013). Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 197, 300–313. 10.1111/j.1469-8137.2012.04377.x, PMID: [DOI] [PubMed] [Google Scholar]
- Boldt K., Pörs Y., Haupt B., Bitterlich M., Kühn C., Grimm B., et al. (2011). Photochemical processes, carbon assimilation and RNA accumulation of sucrose transporter genes in tomato arbuscular mycorrhiza. J. Plant Physiol. 168, 1256–1263. 10.1016/j.jplph.2011.01.026, PMID: [DOI] [PubMed] [Google Scholar]
- Bonneau L., Huguet S., Wipf D., Pauly N., Truong H.-N. (2013). Combined phosphate and nitrogen limitation generates a nutrient stress transcriptome favorable for arbuscular mycorrhizal symbiosis in Medicago truncatula. New Phytol. 199, 188–202. 10.1111/nph.12234, PMID: [DOI] [PubMed] [Google Scholar]
- Boon E., Halary S., Bapteste E., Hijri M. (2015). Studying genome heterogeneity within the arbuscular mycorrhizal fungal cytoplasm. Genome Biol. Evol. 7, 505–521. 10.1093/gbe/evv002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon E., Zimmerman E., St-Arnaud M., Hijri M., Harris S., Hijri Zimmerman M. E., et al. (2013). Allelic differences within and among sister spores of the arbuscular mycorrhizal fungus Glomus etunicatum suggest segregation at sporulation. PLoS One 8:e83301. 10.1371/journal.pone.0083301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakhage A. A., Schuemann J., Bergmann S., Scherlach K., Schroeckh V., Hertweck C. (2008). “Activation of fungal silent gene clusters: A new avenue to drug discovery” in Natural Compounds as Drugs. eds. Petersen F., Amstutz R. (Basel: Birkhäuser Basel; ), 1–12. 10.1007/978-3-7643-8595-8_1 [DOI] [PubMed] [Google Scholar]
- Breuillin F., Schramm J., Hajirezaei M., Ahkami A., Favre P., Druege U., et al. (2010). Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 64, 1002–1017. 10.1111/j.1365-313X.2010.04385.x, PMID: [DOI] [PubMed] [Google Scholar]
- Brundrett M. C., Brundrett M. C. (2009). Mycorrhizal associations and other means of nutrition of vascular plants: understanding the global diversity of host plants by resolving conflicting information and developing reliable means of diagnosis. Plant Soil 320, 37–77. 10.1007/s11104-008-9877-9 [DOI] [Google Scholar]
- Burgeff H. (1914). Untersuchungen über Variabilität, Sexualität und Erblichkeit bei Phycomyces nitens Kunze. Flora oder Allg. Bot. Zeitung 107, 259–P4. 10.1016/S0367-1615(17)32823-9 [DOI] [Google Scholar]
- Calvet C., Camprubi A., Pérez-Hernández A., Lovato P. E. (2013). Plant growth stimulation and root colonization potential of in vivo versus in vitro arbuscular mycorrhizal inocula: American Society for Horticultural Science; Available at: http://hortsci.ashspublications.org/content/48/7/897.short [Accessed January 20, 2019]. [Google Scholar]
- Capone I., Spanò L., Cardarelli M., Bellincampi D., Petit A., Costantino P. (1989). Induction and growth properties of carrot roots with different complements of Agrobacterium rhizogenes T-DNA. Plant Mol. Biol. 13, 43–52. 10.1007/BF00027334, PMID: [DOI] [PubMed] [Google Scholar]
- Cavagnaro T. R., Gao L. L., Smith F. A., Smith S. E. (2001). Morphology of arbuscular mycorrhizas is influenced by fungal identity. New Phytol. 151, 469–475. 10.1046/j.0028-646x.2001.00191.x [DOI] [Google Scholar]
- Chaudhary B., Bhupendra (2013). Plant domestication and resistance to herbivory. Int. J. Plant Genomics 2013:572784. 10.1155/2013/572784, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. C., Mathieu S., Hoffrichter A., Sedzielewska-Toro K., Peart M., Pelin A., et al. (2018b). Single nucleus sequencing reveals evidence of inter-nucleus recombination in arbuscular mycorrhizal fungi. elife 7:e39813. 10.7554/eLife.39813.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. C. H., Morin E., Beaudet D., Noel J., Yildirir G., Ndikumana S., et al. (2018a). High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytol. 220, 1161–1171. 10.1111/nph.14989 [DOI] [PubMed] [Google Scholar]
- Chen Y. H., Gols R., Benrey B. (2015). Crop domestication and its impact on naturally selected trophic interactions. Annu. Rev. Entomol. 60, 35–58. 10.1146/annurev-ento-010814-020601, PMID: [DOI] [PubMed] [Google Scholar]
- Chialva M., Salvioli di Fossalunga A., Daghino S., Ghignone S., Bagnaresi P., Chiapello M., et al. (2018). Native soils with their microbiotas elicit a state of alert in tomato plants. New Phytol. 220, 1296–1308. 10.1111/nph.15014, PMID: [DOI] [PubMed] [Google Scholar]
- Clapp J. P., Rodriguez A., Dodd J. C. (2001). Inter- and intra-isolate rRNA large subunit variation in Glomus coronatum spores. New Phytol. 149, 539–554. 10.1046/j.1469-8137.2001.00060.x [DOI] [PubMed] [Google Scholar]
- Corkidi L., Allen E. B., Merhaut D., Allen M. F., Downer J., Bohn J., et al. (2004). Assessing the infectivity of commercial mycorrhizal inoculants in plant nursery conditions. Available at: http://www.anla.org [Accessed October 2, 2018].
- Corradi N., Brachmann A. (2017). Fungal mating in the most widespread plant symbionts? Trends Plant Sci. 22, 175–183. 10.1016/j.tplants.2016.10.010, PMID: [DOI] [PubMed] [Google Scholar]
- Croll D., Corradi N., Gamper H. A., Sanders I. R. (2008). Multilocus genotyping of arbuscular mycorrhizal fungi and marker suitability for population genetics. New Phytol. 180, 564–568. 10.1111/j.1469-8137.2008.02602.x, PMID: [DOI] [PubMed] [Google Scholar]
- Croll D., Giovannetti M., Koch A. M., Sbrana C., Ehinger M., Lammers P. J., et al. (2009). Nonself vegetative fusion and genetic exchange in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 181, 924–937. 10.1111/j.1469-8137.2008.02726.x, PMID: [DOI] [PubMed] [Google Scholar]
- de la Providencia I. E., de Souza F. A., Fernandez F., Delmas N. S., Declerck S. (2005). Arbuscular mycorrhizal fungi reveal distinct patterns of anastomosis formation and hyphal healing mechanisms between different phylogenic groups. New Phytol. 165, 261–271. 10.1111/j.1469-8137.2004.01236.x, PMID: [DOI] [PubMed] [Google Scholar]
- De la Providencia I. E., Nadimi M., Beaudet D., Morales G. R., Hijri M., Rodriguez Morales G., et al. (2013). Detection of a transient mitochondrial DNA heteroplasmy in the progeny of crossed genetically divergent isolates of arbuscular mycorrhizal fungi. New Phytol. 200, 211–221. 10.1111/nph.12372, PMID: [DOI] [PubMed] [Google Scholar]
- de Novais C. B. C. B., Sbrana C., Júnior O. J. S., Siqueira J. O. J. O., Giovannetti M., Saggin Junior O. J., et al. (2013). Vegetative compatibility and anastomosis formation within and among individual germlings of tropical isolates of arbuscular mycorrhizal fungi (Glomeromycota). Mycorrhiza 23, 325–331. 10.1007/s00572-013-0478-y, PMID: [DOI] [PubMed] [Google Scholar]
- Dearth S. P., Castro H. F., Venice F., Tague E. D., Novero M., Bonfante P., et al. (2018). Metabolome changes are induced in the arbuscular mycorrhizal fungus Gigaspora margarita by germination and by its bacterial endosymbiont. Mycorrhiza 28, 421–433. 10.1007/s00572-018-0838-8, PMID: [DOI] [PubMed] [Google Scholar]
- Delaux P.-M., Varala K., Edger P. P., Coruzzi G. M., Pires J. C., Ané J.-M. (2014). Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet. 10:e1004487. 10.1371/journal.pgen.1004487, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiro A., Salvioli A., Ngonkeu E. L., Mondo S. J., Epis S., Faccio A., et al. (2014). Detection of a novel intracellular microbiome hosted in arbuscular mycorrhizal fungi. ISME J. 8, 257–270. 10.1038/ismej.2013.151, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz G., Azcón-Aguilar C., Honrubia M. (1996). Influence of arbuscular mycorrhizae on heavy metal (Zn and Pb) uptake and growth of Lygeum spartum and Anthyllis cytisoides. Plant Soil 180, 241–249. 10.1007/BF00015307 [DOI] [Google Scholar]
- Douglas G. L., Klaenhammer T. R. (2010). Genomic evolution of domesticated microorganisms. Annu. Rev. Food Sci. Technol. 1, 397–414. 10.1146/annurev.food.102308.124134, PMID: [DOI] [PubMed] [Google Scholar]
- Egan P. A., Adler L. S., Irwin R. E., Farrell I. W., Palmer-Young E. C., Stevenson P. C. (2018). Crop domestication alters floral reward chemistry with potential consequences for pollinator health. Front. Plant Sci. 9:1357. 10.3389/fpls.2018.01357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmoer D. J. P., Behm J. E., Toby Kiers E. (2014). Intense competition between arbuscular mycorrhizal mutualists in an in vitro root microbiome negatively affects total fungal abundance. Mol. Ecol. 23, 1584–1593. 10.1111/mec.12451, PMID: [DOI] [PubMed] [Google Scholar]
- Farmer M. J., Li X., Feng G., Zhao B., Chatagnier O., Gianinazzi S., et al. (2007). Molecular monitoring of field-inoculated AMF to evaluate persistence in sweet potato crops in China. Appl. Soil Ecol. 35, 599–609. 10.1016/j.apsoil.2006.09.012 [DOI] [Google Scholar]
- Fortin J. A., Bécard G., Declerck S., Dalpé Y., St-Arnaud M., Coughlan A. P., et al. (2002). Arbuscular mycorrhiza on root-organ cultures. Can. J. Bot. 80, 1–20. 10.1139/b01-139 [DOI] [Google Scholar]
- Gallone B., Steensels J., Prahl T., Soriaga L., Saels V., Herrera-Malaver B., et al. (2016). Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 166, 1397–1410.e16. 10.1016/j.cell.2016.08.020, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti M., Azzolini D., Citernesi A. S. (1999). Anastomosis formation and nuclear and protoplasmic exchange in arbuscular mycorrhizal fungi. Appl. Environ. Microbiol. 65, 5571–5575. Available at: http://www.ncbi.nlm.nih.gov/pubmed/10584019 [Accessed April 3, 2018].. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti M., Fortuna P., Citernesi A. S., Morini S., Nuti M. P. (2001). The occurrence of anastomosis formation and nuclear exchange in intact arbuscular mycorrhizal networks. New Phytol. 151, 717–724. 10.1046/j.0028-646x.2001.00216.x [DOI] [PubMed] [Google Scholar]
- Giovannetti M., Sbrana C., Strani P., Agnolucci M., Rinaudo V., Avio L. (2003). Genetic diversity of isolates of Glomus mosseae from different geographic areas detected by vegetative compatibility testing and biochemical and molecular analysis. Appl. Environ. Microbiol. 69, 616–624. 10.1128/AEM.69.1.616-624.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer A., Fitter A. H. (2004). Impact of temperature on the arbuscular mycorrhizal (AM) symbiosis: growth responses of the host plant and its AM fungal partner. J. Exp. Bot. 55, 525–534. 10.1093/jxb/erh049, PMID: [DOI] [PubMed] [Google Scholar]
- Hestrin R., Hammer E. C., Mueller C. W., Lehmann J. (2019). Synergies between mycorrhizal fungi and soil microbial communities increase plant nitrogen acquisition. Commun. Biol. 2:233. 10.1038/s42003-019-0481-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijri M., Sanders I. R. (2004). The arbuscular mycorrhizal fungus Glomus intraradices is haploid and has a small genome size in the lower limit of eukaryotes. Fungal Genet. Biol. 41, 253–261. 10.1016/j.fgb.2003.10.011, PMID: [DOI] [PubMed] [Google Scholar]
- Hildebrandt U., Janetta K., Bothe H. (2002). Towards growth of arbuscular mycorrhizal fungi independent of a plant host. Appl. Environ. Microbiol. 68, 1919–1924. 10.1128/AEM.68.4.1919-1924.2002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull C. M., Johnson A. D., Johnson A. D. (1999). Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285, 1271–1275. 10.1126/science.285.5431.1271, PMID: [DOI] [PubMed] [Google Scholar]
- Hyten D. L., Song Q., Zhu Y., Choi I.-Y., Nelson R. L., Costa J. M., et al. (2006). Impacts of genetic bottlenecks on soybean genome diversity. Available at: www.pnas.orgcgidoi10.1073pnas.0604379103 [Accessed July 11, 2019]. [DOI] [PMC free article] [PubMed]
- James T. Y., Stenlid J., Olson Å., Johannesson H. (2008). Evolutionary significance of imbalanced nuclear ratios within heterokaryons of the basidiomycete fungus Heterobasidion parviporum. Evolution 62, 2279–2296. 10.1111/j.1558-5646.2008.00462.x, PMID: [DOI] [PubMed] [Google Scholar]
- Jinks J. L. (1952). Heterokaryosis; a system of adaption in wild fungi. Proc. R. Soc. Lond. Ser. B Biol. Sci. 140, 83–99. 10.1098/RSPB.1952.0046 [DOI] [PubMed] [Google Scholar]
- Johnson N. C. (1993). Can fertilization of soil select less mutualistic mycorrhizae? Ecol. Appl. 3, 749–757. 10.2307/1942106, PMID: [DOI] [PubMed] [Google Scholar]
- Jung S. C., Martinez-Medina A., Lopez-Raez J. A., Pozo M. J. (2012). Mycorrhiza-induced resistance and priming of plant defenses. J. Chem. Ecol. 38, 651–664. 10.1007/s10886-012-0134-6, PMID: [DOI] [PubMed] [Google Scholar]
- Kameoka H., Tsutsui I., Saito K., Kikuchi Y., Handa Y., Ezawa T., et al. (2019). Stimulation of asymbiotic sporulation in arbuscular mycorrhizal fungi by fatty acids. Nat. Microbiol. 4, 1654–1660. 10.1038/s41564-019-0485-7, PMID: [DOI] [PubMed] [Google Scholar]
- Koch A. M., Antunes P. M., Maherali H., Hart M. M., Klironomos J. N. (2017). Evolutionary asymmetry in the arbuscular mycorrhizal symbiosis: conservatism in fungal morphology does not predict host plant growth. New Phytol. 214, 1330–1337. 10.1111/nph.14465, PMID: [DOI] [PubMed] [Google Scholar]
- Kokkoris V., Hamel C., Hart M. M. (2019a). Mycorrhizal response in crop versus wild plants. PLoS One 14:e0221037. 10.1371/journal.pone.0221037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkoris V., Hart M. M. (2019). The role of in vitro cultivation on symbiotic trait and function variation in a single species of arbuscular mycorrhizal fungus. Fungal Biol. 123, 732–744. 10.1016/j.funbio.2019.06.009, PMID: [DOI] [PubMed] [Google Scholar]
- Kokkoris V., Miles T., Hart M. M. (2019b). The role of in vitro cultivation on asymbiotic trait variation in a single species of arbuscular mycorrhizal fungus. Fungal Biol. 123, 307–317. 10.1016/j.funbio.2019.01.005 [DOI] [PubMed] [Google Scholar]
- Koskiniemi S., Sun S., Berg O. G., Andersson D. I. (2012). Selection-driven gene loss in bacteria. PLoS Genet. 8:e1002787. 10.1371/journal.pgen.1002787, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn G., Hijri M., Sanders I. R. (2001). Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature 414, 745–748. 10.1038/414745a, PMID: [DOI] [PubMed] [Google Scholar]
- Lin K., Limpens E., Zhang Z., Ivanov S., Saunders D. G. O., Mu D., et al. (2014). Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet. 10:e1004078. 10.1371/journal.pgen.1004078, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippu J. (1998, 1998). Redistribution of 14C-labelled reserve carbon in Pinus sylvestris seedlings during shoot elongation. Silva Fennica 32:696. 10.14214/sf.696 [DOI] [Google Scholar]
- López-Uribe M. M., Appler R. H., Youngsteadt E., Dunn R. R., Frank S. D., Tarpy D. R. (2017). Higher immunocompetence is associated with higher genetic diversity in feral honey bee colonies (Apis mellifera). Conserv. Genet. 18, 659–666. 10.1007/s10592-017-0942-x [DOI] [Google Scholar]
- Luginbuehl L. H., Menard G. N., Kurup S., Van Erp H., Radhakrishnan G. V., Breakspear A., et al. (2017). Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356, 1175–1178. 10.1126/science.aan0081, PMID: [DOI] [PubMed] [Google Scholar]
- Lumini E., Bianciotto V., Jargeat P., Novero M., Salvioli A., Faccio A., et al. (2007). Presymbiotic growth and sporal morphology are affected in the arbuscular mycorrhizal fungus Gigaspora margarita cured of its endobacteria. Cell. Microbiol. 9, 1716–1729. 10.1111/j.1462-5822.2007.00907.x, PMID: [DOI] [PubMed] [Google Scholar]
- Machida M., Asai K., Sano M., Tanaka T., Kumagai T., Terai G., et al. (2005). Genome sequencing and analysis of Aspergillus oryzae. Nature 438, 1157–1161. 10.1038/nature04300, PMID: [DOI] [PubMed] [Google Scholar]
- Mar Vázquez M., César S., Azcón R., Barea J. M. (2000). Interactions between arbuscular mycorrhizal fungi and other microbial inoculants (Azospirillum, Pseudomonas, Trichoderma) and their effects on microbial population and enzyme activities in the rhizosphere of maize plants. Appl. Soil Ecol. 15, 261–272. 10.1016/S0929-1393(00)00075-5 [DOI] [Google Scholar]
- Marmann A., Aly A., Lin W., Wang B., Proksch P., Marmann A., et al. (2014). Co-cultivation—a powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar. Drugs 12, 1043–1065. 10.3390/md12021043, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx D. H. (1981). Variability in ectomycorrhizal development and growth among isolates of Pisolithus tinctorius as affected by source, age, and reisolation. Can. J. For. Res. 11, 168–174. 10.1139/x81-022 [DOI] [Google Scholar]
- Marx D. H., Daniel W. J. (1976). Maintaining cultures of ectomycorrhizal and plant pathogenic fungi in sterile water cold storage. Can. J. Microbiol. 22, 338–341. 10.1139/m76-051, PMID: [DOI] [PubMed] [Google Scholar]
- Mateus I. D., Masclaux F. G., Aletti C., Rojas E. C., Savary R., Dupuis C., et al. (2019). Dual RNA-seq reveals large-scale non-conserved genotype×genotype-specific genetic reprograming and molecular crosstalk in the mycorrhizal symbiosis. ISME J. 13, 1226–1238. 10.1038/s41396-018-0342-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo K., Davis R. E., Motta J. (1986). Stimulation of germination of spores of Glomus versiforme by spore-associated bacteria. Mycologia 78, 426–431. 10.1080/00275514.1986.12025265 [DOI] [Google Scholar]
- Mehta B. J., Cerdá-Olmedo E. (2001). Intersexual partial diploids of Phycomyces Available at: https://www.genetics.org/content/genetics/158/2/635.full.pdf [Accessed July 11, 2019]. [DOI] [PMC free article] [PubMed]
- Menge J. A., Steirle D., Bagyaraj D. J., Johnson E. L. V., Leonard R. T. (1978). Phosphorus concentrations in plants responsible for inhibition of mycorrhizal infection. New Phytol. 80, 575–578. 10.1111/j.1469-8137.1978.tb01589.x [DOI] [Google Scholar]
- Meyer M., Bourras S., Gervais J., Labadie K., Cruaud C., Balesdent M.-H., et al. (2017). Impact of biotic and abiotic factors on the expression of fungal effector-encoding genes in axenic growth conditions. Fungal Genet. Biol. 99, 1–12. 10.1016/j.fgb.2016.12.008, PMID: [DOI] [PubMed] [Google Scholar]
- Moran N. A., McCutcheon J. P., Nakabachi A. (2008). Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190. 10.1146/annurev.genet.41.110306.130119, PMID: [DOI] [PubMed] [Google Scholar]
- Mosse B., Hepper H. (1975). Vesicular-arbuscular mycorrhizal infections in root organ cultures. Physiol. Plant Pathol. 5, 215–223. 10.1016/0048-4059(75)90088-0 [DOI] [Google Scholar]
- Naiki T., Cook R. J. (1983). Factors in loss of pathogenicity in Gaeumannomyces graminis var. tritici. Available at: https://www.apsnet.org/publications/phytopathology/backissues/Documents/1983Articles/Phyto73n12_1652.PDF [Accessed October 14, 2017].
- Naito M., Desirò A., González J. B., Tao G., Morton J. B., Bonfante P., et al. (2017). Candidatus Moeniiplasma glomeromycotorum’, an endobacterium of arbuscular mycorrhizal fungi. Int. J. Syst. Evol. Microbiol. 67, 1177–1184. 10.1099/ijsem.0.001785, PMID: [DOI] [PubMed] [Google Scholar]
- Naumann M., Schübler A., Bonfante P. (2010). The obligate endobacteria of arbuscular mycorrhizal fungi are ancient heritable components related to the Mollicutes. ISME J. 4, 862–871. 10.1038/ismej.2010.21, PMID: [DOI] [PubMed] [Google Scholar]
- Nishio S., Takada N., Saito T., Yamamoto T., Iketani H. (2016). Estimation of loss of genetic diversity in modern Japanese cultivars by comparison of diverse genetic resources in Asian pear (Pyrus spp.). BMC Genet. 17:81. 10.1186/s12863-016-0380-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ola A. R. B., Thomy D., Lai D., Brötz-Oesterhelt H., Proksch P. (2013). Inducing secondary metabolite production by the endophytic fungus Fusarium tricinctum through coculture with Bacillus subtilis. J. Nat. Prod. 76, 2094–2099. 10.1021/np400589h, PMID: [DOI] [PubMed] [Google Scholar]
- Pan K., Zhao N., Yin Q., Zhang T., Xu X., Fang W., et al. (2014). Induction of a laccase Lcc9 from Coprinopsis cinerea by fungal coculture and its application on indigo dye decolorization. Bioresour. Technol. 162, 45–52. 10.1016/j.biortech.2014.03.116, PMID: [DOI] [PubMed] [Google Scholar]
- Pawlowska T. E., Douds D. D., Charvat I. (1999). In vitro propagation and life cycle of the arbuscular mycorrhizal fungus Glomus etunicatum. Mycol. Res. 103, 1549–1556. 10.1017/S0953756299008801 [DOI] [Google Scholar]
- Pawlowska T. E., Taylor J. W. (2004). Organization of genetic variation in individuals of arbuscular mycorrhizal fungi. Nature 427, 733–737. 10.1038/nature02290, PMID: [DOI] [PubMed] [Google Scholar]
- Pearce F. (2003). Databases selected: Multiple databases going bananas. Available at: http://courseresources.mit.usf.edu/sgs/ph6934/webpages/CC/module_5/read/going_bananas_pearce.pdf [Accessed July 11, 2019].
- Plenchette C., Declerck S., Diop T. A., Strullu D. G. (1996). Infectivity of monoaxenic subcultures of the arbuscular mycorrhizal fungus Glomus versiforme associated with Ri-T-DNA-transformed carrot root. Appl. Microbiol. Biotechnol. 46, 545–548. 10.1007/s002530050858 [DOI] [Google Scholar]
- Ploetz R. C. (2003). “Yes. We won’t have bananas.” What realistic threats do diseases pose to banana production? Pestic. Outlook 14, 62–64. 10.1039/b303002b [DOI] [Google Scholar]
- Porcel R., Aroca R., Ruiz-Lozano J. M. (2012). Salinity stress alleviation using arbuscular mycorrhizal fungi. A review. Agron. Sustain. Dev. 32, 181–200. 10.1007/s13593-011-0029-x [DOI] [Google Scholar]
- Pozo M. J., Azcón-Aguilar C. (2007). Unraveling mycorrhiza-induced resistance. Curr. Opin. Plant Biol. 10, 393–398. 10.1016/j.pbi.2007.05.004, PMID: [DOI] [PubMed] [Google Scholar]
- Purin S., Morton J. B. (2011). In situ analysis of anastomosis in representative genera of arbuscular mycorrhizal fungi. Mycorrhiza 21, 505–514. 10.1007/s00572-010-0356-9, PMID: [DOI] [PubMed] [Google Scholar]
- Ram S. G., Thiruvengadam V., Vinod K. K. (2007). Genetic diversity among cultivars, landraces and wild relatives of rice as revealed by microsatellite markers. J. Appl. Genet. 48, 337–345. 10.1007/BF03195230, PMID: [DOI] [PubMed] [Google Scholar]
- Reif J. C., Zhang P., Dreisigacker S., Warburton M. L., van Ginkel M., Hoisington D., et al. (2005). Wheat genetic diversity trends during domestication and breeding. Theor. Appl. Genet. 110, 859–864. 10.1007/s00122-004-1881-8, PMID: [DOI] [PubMed] [Google Scholar]
- Roesti D., Ineichen K., Braissant O., Redecker D., Wiemken A., Aragno M. (2005). Bacteria associated with spores of the arbuscular mycorrhizal fungi Glomus geosporum and Glomus constrictum. Appl. Environ. Microbiol. 71, 6673–6679. 10.1128/AEM.71.11.6673-6679.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropars J., Sędzielewska Toro K., Noel J., Pelin A., Charron P., Farinelli L., et al. (2016, 2016). Evidence for the sexual origin of heterokaryosis in arbuscular mycorrhizal fungi. Nat. Microbiol. 1:16033. 10.1038/nmicrobiol.2016.33 [DOI] [PubMed] [Google Scholar]
- Roper M., Ellison C., Taylor J., N G. (2011). Nuclear and genome dynamics in multinucleate ascomycete fungi. Curr. Biol. 21, R786–R793. 10.1016/j.cub.2011.06.042, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosikiewicz P., Bonvin J., Sanders I. R. (2017). Cost-efficient production of in vitro Rhizophagus irregularis. Mycorrhiza 27, 477–486. 10.1007/s00572-017-0763-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross I. K., Loftus M. G., Foster L. M. (1991). Homokaryon-dikaryon phenotypic switching in an arginine requiring mutant of Coprinus congregatus. Mycol. Res. 95, 776–781. 10.1016/S0953-7562(09)80037-3 [DOI] [Google Scholar]
- Ruiz-Lozano J. M., Aroca R. (2010). “Host response to osmotic stresses: stomatal behaviour and water use efficiency of arbuscular mycorrhizal plants” in Arbuscular mycorrhizas: Physiology and function (The Netherlands: Springer; ), 239–256. [Google Scholar]
- Salvioli A., Chiapello M., Fontaine J., Hadj-Sahraoui A. L., Grandmougin-Ferjani A., Lanfranco L., et al. (2010). Endobacteria affect the metabolic profile of their host Gigaspora margarita, an arbuscular mycorrhizal fungus. Environ. Microbiol. 12, 2083–2095. 10.1111/j.1462-2920.2010.02246.x, PMID: [DOI] [PubMed] [Google Scholar]
- Salvioli A., Ghignone S., Novero M., Navazio L., Venice F., Bagnaresi P., et al. (2016). Symbiosis with an endobacterium increases the fitness of a mycorrhizal fungus, raising its bioenergetic potential. ISME J. 10, 130–144. 10.1038/ismej.2015.91, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samils N., Oliva J., Johannesson H. (2014). Nuclear interactions in a heterokaryon: insight from the model Neurospora tetrasperma. Proc. R. Soc. B Biol. Sci. 281, 20140084–20140084. 10.1098/rspb.2014.0084, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers R. J. H., Gutjahr C., Paszkowski U. (2008). Cereal mycorrhiza: an ancient symbiosis in modern agriculture. Trends Plant Sci. 13, 93–97. 10.1016/j.tplants.2007.11.006, PMID: [DOI] [PubMed] [Google Scholar]
- Sbrana C., Fortuna P., Giovannetti M. (2011). Plugging into the network: belowground connections between germlings and extraradical mycelium of arbuscular mycorrhizal fungi. Mycologia 103, 307–316. 10.3852/10-125 [DOI] [PubMed] [Google Scholar]
- Sbrana C., Strani P., Pepe A., de Novais C. B., Giovannetti M. (2018). Divergence of Funneliformis mosseae populations over 20 years of laboratory cultivation, as revealed by vegetative incompatibility and molecular analysis. Mycorrhiza 28, 329–341. 10.1007/s00572-018-0830-3, PMID: [DOI] [PubMed] [Google Scholar]
- Smith S. E., Read D. J. (2008). Mycorrhizal symbiosis. London: Academic Press. [Google Scholar]
- Speakman J. B. (1982). A simple, reliable method of producing perithecia of Gaeumannomyces gramnis var. tritici and its application to isolates of Phialophora spp. Trans. Br. Mycol. Soc. 79, 350–353. 10.1016/S0007-1536(82)80129-0 [DOI] [Google Scholar]
- Stockinger H., Walker C., Schuessler A. (2009). “Glomus intraradices DAOM197198”, a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol. 183, 1176–1187. 10.1111/j.1469-8137.2009.02874.x, PMID: [DOI] [PubMed] [Google Scholar]
- Sugiura Y., Akiyama R., Tanaka S., Yano K., Kameoka H., Kawaguchi M., et al. (2019). Myristate as a carbon and energy source for the asymbiotic growth of the arbuscular mycorrhizal fungus Rhizophagus irregularis. bioRxiv 731489 [Preprint]. 10.1101/731489 [DOI] [PMC free article] [PubMed]
- Tarbell T. J., Koske R. E. (2007). Evaluation of commercial arbuscular mycorrhizal inocula in a sand/peat medium. Mycorrhiza 18, 51–56. 10.1007/s00572-007-0152-3, PMID: [DOI] [PubMed] [Google Scholar]
- Tenger-Trolander A., Lu W., Noyes M., Kronforst M. R. (2019). Contemporary loss of migration in monarch butterflies. Proc. Natl. Acad. Sci. USA 116, 14671–14676. 10.1073/pnas.1904690116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson B. D., Malajczuk N., Grove T. S., Hardy G. E. S. (1993). Improving the colonization capacity and effectiveness of ectomycorrhizal fungal cultures by association with a host plant and re-isolation. Mycol. Res. 97, 839–844. 10.1016/S0953-7562(09)81159-3 [DOI] [Google Scholar]
- Thomson B. D., Robson A. D., Abbott L. K. (1986). Effects of phosphorus on the formation of mycorrhizas by Gigaspora calospora and Glomus fasciculatum in relation to root carbohydrates. New Phytol. 103, 751–765. 10.1111/j.1469-8137.1986.tb00850.x [DOI] [Google Scholar]
- Tisserant E., Malbreil M., Kuo A., Kohler A., Symeonidi A., Balestrini R., et al. (2013). Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. USA 110, 20117–20122. 10.1073/pnas.1313452110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Cortes G., Ghignone S., Bonfante P., Schussler A. (2015). Mosaic genome of endobacteria in arbuscular mycorrhizal fungi: Transkingdom gene transfer in an ancient mycoplasma-fungus association (Vol 112, pg 7785, 2015). Proc. Natl. Acad. Sci. USA 112, E5376–E5376. 10.1073/pnas.1516662112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C., Carpentieri A., Salvioli A., Novero M., Marsoni M., Testa L., et al. (2016). An interdomain network: the endobacterium of a mycorrhizal fungus promotes antioxidative responses in both fungal and plant hosts. New Phytol. 211, 265–275. 10.1111/nph.13895, PMID: [DOI] [PubMed] [Google Scholar]
- Wang G. M., Stribley D. P., Tinker P. B., Walker C. (1993). Effects of pH on arbuscular mycorrhiza I. field observations on the long-term liming experiments at Rothamsted and Woburn. New Phytol. 124, 465–472. 10.1111/j.1469-8137.1993.tb03837.x [DOI] [Google Scholar]
- Weese D. J., Heath K. D., Dentinger B. T. M., Lau J. A. (2015). Long-term nitrogen addition causes the evolution of less-cooperative mutualists. Evolution 69, 631–642. 10.1111/evo.12594, PMID: [DOI] [PubMed] [Google Scholar]
- Whitehead S. R., Turcotte M. M., Poveda K. (2017). Domestication impacts on plant–herbivore interactions: a meta-analysis. Philos. Trans. R. Soc. B Biol. Sci. 372:20160034. 10.1098/rstb.2016.0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmitzer L., Sanchez-Serrano J., Buschfeld E., Schell J. (1982). DNA from Agrobacterium rhizogenes in transferred to and expressed in axenic hairy root plant tissues. MGG Mol. Gen. Genet. 186, 16–22. 10.1007/BF00422906 [DOI] [Google Scholar]
- Wilson J. M. (1984). Comparative development of infection by 3 vesicular arbuscular mycorrhizal fungi. New Phytol. 97, 413–426. 10.1111/j.1469-8137.1984.tb03607.x [DOI] [Google Scholar]
- Wyss P., Bonfante P. (1993). Amplification of genomic DNA of arbuscular-mycorrhizal (AM) fungi by PCR using short arbitrary primers. Mycol. Res. 97, 1351–1357. 10.1016/S0953-7562(09)80169-X [DOI] [Google Scholar]
- Zeder M. A. (2006). Central questions in the domestication of plants and animals. Evol. Anthropol. Issues News Rev. 15, 105–117. 10.1002/evan.20101 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analyzed for this study.


