Abstract
Mitral valve diseases affect ∼3% of the population and are the most common reasons for valvular surgery because no drug-based treatments exist. Inheritable genetic mutations have now been established as the cause of mitral valve insufficiency, and four different missense mutations in the filamin A gene (FLNA) have been found in patients suffering from nonsyndromic mitral valve dysplasia (MVD). The filamin A (FLNA) protein is expressed, in particular, in endocardial endothelia during fetal valve morphogenesis and is key in cardiac development. The FLNA-MVD-causing mutations are clustered in the N-terminal region of FLNA. How the mutations in FLNA modify its structure and function has mostly remained elusive. In this study, using NMR spectroscopy and interaction assays, we investigated FLNA-MVD-causing V711D and H743P mutations. Our results clearly indicated that both mutations almost completely destroyed the folding of the FLNA5 domain, where the mutation is located, and also affect the folding of the neighboring FLNA4 domain. The structure of the neighboring FLNA6 domain was not affected by the mutations. These mutations also completely abolish FLNA’s interactions with protein tyrosine phosphatase nonreceptor type 12, which has been suggested to contribute to the pathogenesis of FLNA-MVD. Taken together, our results provide an essential structural and molecular framework for understanding the molecular bases of FLNA-MVD, which is crucial for the development of new therapies to replace surgery.
Significance
Mitral valve diseases are very common, affecting ∼3% of the population. Currently, the only available treatment is surgery. Four different missense mutations in the filamin A gene have been found in patients suffering from nonsyndromic mitral valve dysplasia (FLNA-MVD). The molecular mechanism of FLNA-MVD has remained elusive, but it is essential for the development of novel drug-based therapies. In this study, we investigated two FLNA-MVD-causing mutations in filamin. Our results clearly indicated that these mutations have critical structural effects on filamin and also affect filamin’s interactions with other proteins. Taken together, our results provide an essential structural and molecular framework for understanding the molecular bases of FLNA-MVD, which is crucial for the development of new therapies to replace surgical options.
Introduction
Mitral valve prolapse (MVP) is a relatively common disease that affects around 3% of the world’s population (1). It is also one of the most common indications for valvular surgery, and there are currently no drug-based treatments available. Nowadays, although genetic defects have definitively been associated with both syndromic and nonsyndromic forms of MVP, how the mutations in the translated protein modify its structure and function has remained elusive. The filamin A gene (FLNA) was first associated with inherited nonsyndromic mitral valve dysplasia (MVD) (FLNA-MVD OMIM 314400) (1, 2). Today, four missense mutations (G288R, P637Q, V711D, and H743P (Fig. 1 A)) have been identified in FLNA-MVD patients, and these mutations cause thickened myxomatous Barlow-like leaflet dystrophy (2, 3, 4). In addition to the thickening of the leaflets, a patient’s mitral valve apparatus presents with specific characteristics. The chordae are shorter, and the papillary muscles are displaced close to the mitral annulus. Echocardiographic evaluation has also unveiled a unique and homogeneous phenotype. In fact, the prolapse of the leaflet in systole is unusually associated with a restrictive motion in diastole in FLNA-MVD patients.
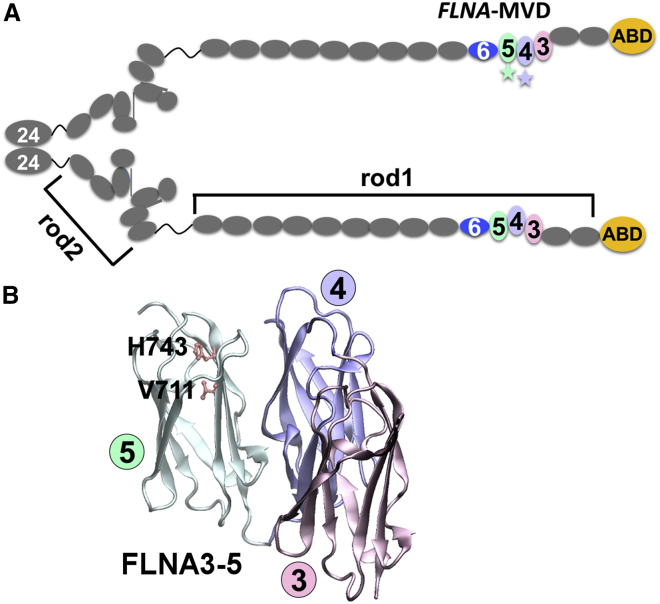
Figure 1.
An overview of the structural organization of FLNA. (A) Shown is the schematic representation of FLNA dimer having 24 Ig domains (gray) and the actin-binding domain (ABD) (yellow) in a monomer. The two monomers are dimerized via domain 24. Rod1 consists of domains 1–15, and rod2 consists of domains 16–23. The studied domains, 4–6, are colored in pale purple, pale green, and blue, respectively. The localization of FLNA-MVD P637Q mutation in domain 4 and V711D and H743P mutations in domain 5 have been highlighted with stars. The fourth missense mutation causing FLNA-MVD, G288R, is located in domain 1. (B) The crystal structure of FLNA3–5 (PDB: 4M9P) (14) is colored in pink (FLNA3), pale purple (FLNA4), and pale green (FLNA5). Residues V711 and H743, whose mutation to aspartic acid and proline residues, respectively, cause FLNA-MVD, are shown with ball-and-stick models. To see this figure in color, go online.
Filamin A (FLNA) (Fig. 1 A) is a ubiquitously expressed cytoskeletal protein (5) that is known to be one of the key proteins in cardiac development, and it is particularly expressed in endocardial endothelia during fetal valve morphogenesis. FLNA knockout leads to embryonic lethality with a pleomorphic array of cardiac malformations (6, 7, 8). FLNA is a large cytosolic protein that provides a link between the cytoskeleton and the cell surface by interacting simultaneously with both extracellular matrix-bound integrins and the actin cytoskeleton. These interactions make possible the central role of FLNA in cellular mechanotransduction (9, 10). FLNA binds to numerous proteins, including transmembrane receptors and signaling molecules (9). Therefore, FLNA has essential scaffolding functions and integrates multiple cellular behaviors during embryonic development, cellular migration, and mechanical stress responses (9, 11, 12). Structurally, FLNA is a 280 kDa homodimeric protein consisting of N-terminal actin-binding domains followed by 24 homologous immunoglobulin (Ig)-like domains, of which domains 1–15 form rod1, and domains 16–23 form rod2. Dimerization occurs via the most C-terminal Ig domain, FLNA24. The Ig domains 3–5 and 16–21 form tightly arranged compact substructures in otherwise flexible Ig domain rods (13, 14, 15). Very interestingly, FLNA-MVD-causing mutations are all clustered at the N-terminal region of rod1 (2, 3, 4) (Fig. 1).
Protein tyrosine phosphatase 12 (PTPN12) has been suggested to be one of the key FLNA-binding partners implicated in mitral valve diseases (16, 17). PTPN12 (PTP-PEST) is a ubiquitous cytosolic protein tyrosine phosphatase that consists of an N-terminal catalytic domain and a C-terminal noncatalytic domain (18). The proline-rich domain in the C-terminal noncatalytic domain has been shown to a be key element in interactions of FLNA and PTPN12 (17, 19). Our recent studies further revealed that the main binding site of PTPN12 on FLNA is the Ig domain 4, but the neighboring Ig domains 3, 5, and 6 also exhibit some binding (20). PTPN12 has been shown to be essential for cellular motility and cytoskeleton dynamics (21). PTPN12 is crucial for normal embryonic development as demonstrated by the fact that PTPN12 invalidation is embryonically lethal in mice because of important vascular defects and unsuccessful liver formation. PTPN12 is also required for integrin-mediated adhesion and migration of endothelial cells but not for their differentiation and proliferation. PTPN12 regulates Rho GTPase (18), which binds to the most C-terminal FLNA domain 24 (22). FLNA is also involved in focal adhesion signaling pathway regulation, and it has been suggested that FLNA may act as a scaffold for the spatial organization of Rho GTPase-mediated signaling pathways (23). Our earlier studies have, indeed, shown that FLNA-MVD mutations deregulate the Rho/Rac1 balance and affect cellular spreading and migration, resulting from increased Rho activity (16). Accordingly, PTPN12-FLNA interactions might contribute to the pathogenesis of FLNA-MVD.
The molecular bases behind valvular diseases are, in general, poorly understood. This has restricted the development of drug-based therapies, which are currently not available. We have recently reported the structural and functional consequences of FLNA-MVD causing a P637Q mutation located at the middle of the compact FLNA rod1 fragment 3–5 (20). This mutation was observed to change FLNA’s force resilience and abolish its interaction with protein tyrosine phosphatase PTPN12 (20). Here, by combining NMR spectroscopy with small angle x-ray scattering (SAXS) and surface plasmon resonance (SPR), we show that the missense mutations that cause FLNA-MVD, which are located on FLNA5 (V711D and H743P), destroy the folding of Ig domains 4 and 5 and abolish the ability of PTPN12 to bind to the mutated FLNA.
Materials and Methods
Recombinant proteins
FLNA domains (Swiss-Prot P21333.4, aa 574–869) were cloned into a pGTvL1-SGC vector (Structural Genomics Consortium, University of Oxford, Oxford, UK) using the ligase independent method (24), and the PTPN12 fragment (Swiss-Prot Q05209.3, aa 600–780) was cloned into a pET23b vector using the method described by Duval et al. (16). The mutations were introduced to the desired expression constructs using the QuikChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). All the expression plasmids were verified by sequencing. The production of glutathione S-transferase (GST) fusion proteins occurred in Terrific Broth (2.4% w/v yeast extract, 1.2% w/v tryptone, 0.5% w/v glycerol, 0.017 M KH2PO4, 0.072 M KH2PO4) by the addition of isopropyl β-D-1-thiogalactopyranoside to 0.4 mM at 30°C for 4–6 h using Escherichia coli BL21 Gold cells. Complete lysis of the cells was achieved using the EmulsiFlex-C3 homogenizer (Avestin, Ottawa, Ontario, Canada), and lysates were cleared by centrifugation at 35,000 × g for 30 min at 4°C. The GST fusion proteins were captured using Protino Glutathione Agarose 4B (Macherey-Nagel, Düren, Germany), and the GST was cleaved at 4°C for 16 h using tobacco etch virus protease (Invitrogen, Life Technologies, Carlsbad, CA). The tobacco etch virus cleavage extended the FLNA constructs by two additional N-terminal amino acid residues, M and S. A HiLoad 26/60 Superdex 75 column (GE Healthcare, Chicago, IL) was utilized in the size exclusion chromatography of the desired fragments in 20 mM Tris (pH 8.0), 100 mM NaCl, and 1 mM dithiothreitol (DTT) using an ÄKTA prime system (GE Healthcare). Amicon ultracentrifugal devices (MilliporeSigma, Burlington, MA) were used for concentrating the proteins for downstream experiments. For NMR measurements, uniform 15N and 13C labeling of the filamin fragments was achieved using 1 g 15NH4Cl and 2 g D-glucose 13C (Cambridge Isotope Laboratories, Tewksbury, MA) per liter in M9 media. The proteins were expressed in BL21 Gold cells for 20 h at 25°C after induction with 1 mM isopropyl β-D-1-thiogalactopyranoside. The verification of homodispersity of each protein was done by analytical gel filtration (in Fig. S1) and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Six His-tagged PTPN12 fragments were also produced in BL21 and purified on nickel beads (Macherey-Nagel) using an imidazole 250 mM elution as previously described by Duval et al. (16). Purified proteins were dialyzed against phosphate buffer solution (8 mM Na2HPO4, 2 mM KH2PO4 (pH 7.8), 137 mM NaCl, 2.7 mM KCl) and quantified and analyzed by SDS-PAGE before use in SPR experiments.
FLNA6 homology model
The homology model of FLNA6 was built in two steps: 1) the sequence alignment of FLNA6 and template structure of FLNA5 (Protein Data Bank (PDB): 4M9P) (14) were constructed using the MALIGN tool in Bodil software (25) by employing a structure-based matrix (26) with a gap penalty of 40, and 2) the sequence-alignment-based model of FLNA6 was built using the nest tool in Jackal software (Honig Lab, New York, NY) (27).
NMR
NMR samples were prepared in 20 mM NaH2PO4, 50 mM NaCl, and 1 mM DTT buffer at pH 6.50, and then D2O was added to obtain 4% solutions. Protein concentrations were 0.5–1.1 mM. All NMR spectra were collected using a Bruker Avance III HD 800 MHz NMR spectrometer (Bruker, Billerica, MA), equipped with a cryogenically cooled TCI 1H, 13C, 15N triple-resonance probehead. The data were collected at 25°C. For the assignment of backbone chemical shifts, the following experiments were conducted: 1H, 15N heteronuclear single quantum correlation (HSQC), transverse relaxation optimized spectroscopy (TROSY)-HNCA, TROSY-HN(CO)CA, TROSY-HNCACB, and TROSY-HN(CO)CACB (for review, see (28)). All spectra were processed with TopSpin 3.5 and analyzed with NMRFAM-Sparky 1.4 (29). Time delays for 15N T1 relaxation data were 20, 100, 200, 400, 800, 1100, and 1400 ms. An exponentially decaying curve was fitted to the peak intensities as implemented in Dynamic Center software (Bruker). Chemical shift assignments of FLNA4–6 have been submitted to the Biological Magnetic Resonance Data Bank with accession code 27795.
HYDRONMR (30) was used to calculate the theoretical 15N T1 relaxation times at 303.13°K, using 0.008 poises for the solvent viscosity at 18.10 Tesla magnetic field. The relaxation data were calculated separately for the FLNA4–5 structure taken from the FLNA3–5 crystal structure (PDB: 4M9P) (14) and the FLNA6 homology model.
Limited proteolysis
Samples prepared in 20 mM Tris (pH 8.0), 100 mM NaCl, and 1 mM DTT were exposed to proteolytic digestion at 20°C by α-chymotrypsin (Sigma-Aldrich, St. Louis, MO) using a mass ratio of 1:1000. Protein fragments obtained from various incubation intervals were separated on 12% gels using SDS-PAGE. The entire experiment was conducted in triplicate, resulting in reproducible degradation patterns.
SAXS
A BM29 beamline (European Synchrotron Radiation Facility, Grenoble, France) was used to collect the SAXS data on a PILATUS 1M image plate using a sample to detector distance of 2.9 m and a wavelength of 1.0 Å (momentum transfer range 0.01 < q < 5 nm−1). Three different protein concentrations (1.0, 2.0, and 4.0 mg/mL) were used in the data acquisition. Before measurements, fresh DTT was added to 10 mM in the gel filtration buffer. The ATSAS software package (European Molecular Biology Laboratory, Hamburg, Germany) was utilized in the data processing (31). Guinier analysis performed using PRIMUS (32) and distance distribution functions calculated using DATGNOM (33) provided the estimates for the radius of gyration (Rg) and maximal dimensions (Dmax) of the particles. Apparent particle aggregation or repulsion was excluded in the Guinier analysis. The Porod volumes were estimated using the DATPOROD program in ATSAS (31). DAMMIF (34) on ATSAS online (https://www.embl-hamburg.de/biosaxs/atsas-online) was used for generating a total of 20 ab initio shape envelopes that were subsequently aligned against the most probable model, averaged, and ultimately filtered in DAMAVER (35). The resolution of the ab initio models were ultimately estimated using SASRES (36). The FLNA4–6 rigid body model was obtained using SASREF (37), and the final overlaying of the crystal structures and the ab initio models was achieved using SUPCOMB (38). The flexibility of the selected FLNA fragments was examined using the ensemble optimization method (EOM) (39, 40) on ATSAS online. In the wild-type (WT) EOM calculations, the FLNA4 and FLNA5 crystal structures and the FLNA6 model were used as rigid bodies. In the mutant protein EOM calculations, the FLNA4–5 domain pair was set as a random coil, whereas the FLNA6 model was provided as a structured domain. The WT fragments were analyzed using merged scattering data, whereas the mutated FLNA4–6 fragments data from a single protein concentration (4 mg/mL) was used. In addition, a control run for the FLNA4–6 WT fragment utilizing the mutant EOM parameters and WT SAXS data was performed. CORAL (41) was utilized to study the domain movements and the flexibility of the linker between the FLNA4–5 domain pair and the FLNA6 domain. In CORAL calculations, FLNA4–5 was kept in the fixed orientation, whereas the linker region and FLNA6 (aa 767–869) moved freely. The solution scattering of the selected atomic models obtained from CORAL and SASREF was evaluated using CRYSOL (42). The statistics from all of the SAXS analyses are shown in Tables S1–S3. The SAXS images were prepared using the PyMOL Molecular Graphics System, version 0.99 (Schrödinger, New York, NY) and GraphPad Prism 8 (GraphPad Software, San Diego, CA). The SAXS data have been submitted to Small Angle Scattering Biological Data Bank (SASBDB) with the accession codes SASDFD3 (FLNA4–6 WT), SASDFE3 (FLNA4–6 V711D, 4 mg/mL), SASDFF3 (FLNA4–6 V711D, 2 mg/mL), SASDFG3 (FLNA4–6 H743P, 4 mg/mL), and SASDFH3 (FLNA4–6 WT H743P, 2 mg/mL).
SPR experiments
The SPR experiments were conducted on the BIAcore 3000 system (GE Healthcare). The experiments were carried out at 25°C using HBS-EP (0.01 M HEPES (pH 7.4), 0.15 M NaCl, 3 mM EDTA, 0.005% v/v P20) as the running buffer. Purified PTPN12 fragments were immobilized on CM5 sensor chips (GE Healthcare) by amine coupling as recommended by the manufacturer for ∼200–300 resonance units. Samples of purified FLNA4–6 and the corresponding mutated fragments, V711D and H743P, were diluted in the running buffer and injected in single-cycle kinetics mode at five different concentrations (0.31, 0.62, 1.25, 2.50, and 5.00 μM) using a flow rate of 30 μL/min over the chip surface. Binding surfaces were regenerated to remove bound analyte by injecting 50 mM NaOH for 30 s. This regeneration condition removed analyte completely but retained the surface binding capacity of the PTPN12 functionalized chip. Kinetic constants were calculated by global fitting of the data to a 1:1 Langmuir binding model (single-cycle kinetics) after subtracting the control surface, using the BIAevaluation software, version 4.0.1 (GE Healthcare).
Results
Mutations at FLNA5 destroy the compact FLNA rod1 structure
To get information about how the MVP-causing mutations V711D and H743P located at FLNA5 effect FLNA structure, the mutations were inserted in the three-domain fragment consisting of domains from 4 to 6. This construct was used because it has flanking FLNA domains on both sides of the mutated FLNA5 domain, providing a reliable model of the full-length FLNA for in vitro structural studies. First, the analytical size exclusion analyses of WT and mutated FLNA4–6 fragments showed that the retention volumes of mutants are smaller than that of the WT fragment, indicating that mutations affect the shapes of FLNA4–6 (Fig. S1). SAXS was then employed to more closely study the effects of V711D and H743P mutations on FLNA4–6 structure (Table S1). The direct comparison of the forward scattering curves (Figs. 2 A and S2) and the corresponding Kratky, Porod, and distance distribution P(r) plots (43) (Fig. 2, B–D) of the WT and mutant proteins suggest structural unfolding of the V711D and H743P fragments. The EOM analyses of FLNA4–6 WT and mutant SAXS data provide further support for the structural misfolding of FLNA4–6 V711D and H743P fragments as both the Rg and Dmax distributions are much wider than those of the WT fragment (Fig. S3; Table S3). Also, for mutants, more structures, 14 (V711D) and 17 (H743P), are needed to fit the experimental data than for WT FLNA4-6 (four structures, Table S3).
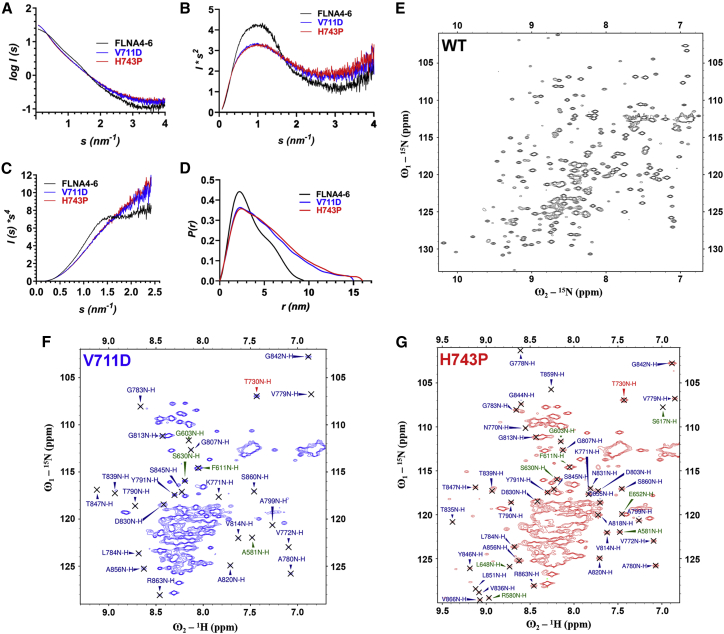
Figure 2.
FLNA-MVD causing mutations at FLNA5 destroyed the folding of both FLNA 5 and FLNA4. (A–D) The shape of the scattering curve (A) and Porod plot (C) of FLNA4–6 WT is typical for a folded protein, whereas the shape scattering curves (A) and Porod plots (C) of FLNA4–6 V711D and H743P show the mutant proteins were partially unfolded. The Kratky plot (B) further demonstrates that the FLNA4–6 WT is a multidomain protein with a flexible linker, whereas the mutated proteins are partially flexible. The distance distribution (P(r)) plot (D) reveals that the mutated proteins adopted significantly extended particle maximal dimensions in contrast to the WT. The SAXS plots were prepared using merged scattering data (of 2 and 4 mg/mL) from FLNA4–6 WT and the mutants in a single concentration (4 mg/mL). (E–G) Comparison of 1H 15N HSQC spectra of FLNA4–6 WT (E), V711D (F), and H743P (G) shows that both FLNA5 mutated fragments lack the FLNA5 (and most FLNA4) domain-specific NH crosspeaks in the region, typical for the structural protein, > 8.5 1H ppm. Instead, the vast majority of high-intensity crosspeaks were clustered in the region between 7.7 and 8.5 1H ppm, indicating that both mutants had a high proportion of unfolded polypeptide chain. In contrast, the WT spectrum showed a well-dispersed correlation map spanning from 7 to 10 1H ppm, with more uniform intensities between the crosspeaks. These data suggest that FLNA4–6 WT is comprised of three well-structured domains. The identity of the individual peaks in the mutant spectra (F and G) are shown, with FLNA4 peaks in green, FLNA5 peaks in red, and FLNA6 peaks in blue. To see this figure in color, go online.
It should, however, be kept in mind that SAXS only gives the average data of the entire FLNA4–6 fragment. Therefore, NMR spectroscopy was then employed to collect domain-level information about the effects of the V711D and H743P mutations on the structure of the FLNA4–6 fragment. First, the 15N-HSQC spectra for FLNA4–6 WT, V711D and H743P, were measured. The HSQC spectrum of the WT fragment (Fig. 2 E) was drastically different than those of the mutants, which were similar to each other (Fig. 2, F and G). The15N-HSQC spectrum of FLNA4–6 WT (Fig. 2 E) showed good dispersion of the crosspeaks, with rather similar intensities, as can be expected for a modular protein having three homologous, well-structured domains. In contrast, the 15N-1H correlation spectra of both mutants lack the vast majority of the NH crosspeaks in the structural fingerprint region (>8.5 ppm). Moreover, the high-intensity NH crosspeaks cluster in the middle of the 1HN chemical shift range (7.7–8.5 ppm 1H), indicating an improperly folded domain and the prevalence of structural disorder (Fig. 2, F and G).
To be able to analyze and compare all three spectra in more detail, the backbone assignment of FLNA4–6 WT was carried out using the triple-resonance HNCA, HN(CO)CA, HNCACB, and HN(CO)CACB experiments with the implementation of the TROSY (28). The close comparison of the 15N-HSQC spectra of FLNA4–6 WT and FLNA4–6 V711D and H743P mutants indicated that most of the crosspeaks corresponding to domain 5 and, interestingly, also to domain 4 were missing from the V711D and H743P 15N-HSQC spectra, whereas the crosspeaks corresponding to domain 6 could still be found in the mutant spectra (Fig. 2, E and G). In addition to the high-intensity crosspeaks, the 15N-HSQC spectra of the mutants also exhibit several broadened NH resonances, likely originating in domains 4 and 5. This suggests the presence of μ-ms timescale motions within FLNA4–5.
The almost complete unfolding of FLNA4–5 increased its susceptibility to proteolytic cleavage, as seen in Fig. 3 B. Controlled protease digestion assays showed that FLNA4–6 V711D and H743P were significantly more readily digested by chymotrypsin than FLNA4–6 WT. At the 60 min time point, the FLNA4–6 WT was largely intact, whereas practically no full-length species were left for mutant proteins. Simultaneously, as the full-length FLNA4–6 mutant fragments were digested by chymotrypsin, a new band corresponding to approximately a 10 kDa protein fragment formed. This could be the FLNA6 domain that, as a folded domain, is not prone to proteolysis, but we have no proof for that.
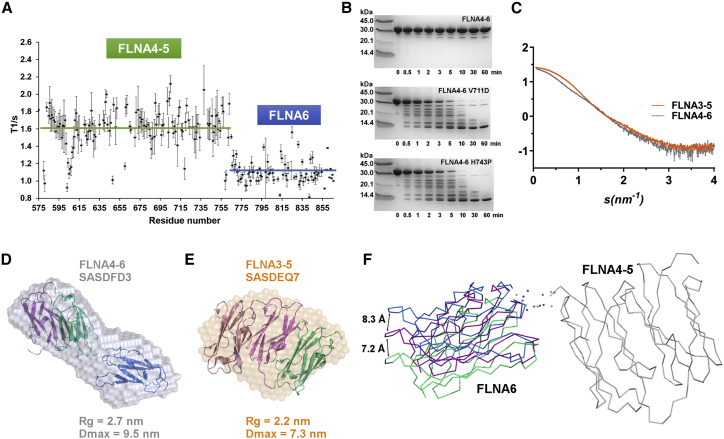
Figure 3.
FLNA6 is not part the compact structure formed by FLNA3–5. (A) The plot of 15N T1 relaxation times versus the FLNA4–6 amino acid sequence clearly indicates that FLNA4–5 moves together but independently from FLNA6. Horizontal lines indicate the average values calculated for FLNA4–5 and FLNA6. (B) The limited protease digestion indicated that both FLNA5 mutations destabilized the FLNA4–6 structure, as after 10 min, no FLNA4–6 mutant proteins were left, whereas WT stayed intact even after 60 min of proteolysis treatment. (C) The WT FLNA3–5 (orange) and FLNA4–6 (gray) fragments have distinct experimental SAXS profiles. The merged data of 2 and 4 mg/mL for both of the samples are presented. (D) The ab initio model of FLNA4–6 (surface presentation, gray, SASBDB ID SASDFD3) superimposed on the rigid body model of FLNA4–6 (with domain-specific coloring: FLNA4 = purple, FLNA5 = green, and FLNA6 = blue) obtained by SASREF. The normalized spatial discrepancy (NSD) of the alignment is 1.58. The χ2-value of the FLNA4–6 rigid body model against the experimental scattering data was 0.99 (estimated using CRYSOL) (42). (E) The ab initio model of FLNA3–5 (surface presentation = orange, SASBDB ID SASDEQ7) superimposed on the crystal structure of FLNA3–5 (PDB: 4M9P, with domain-specific coloring: FLNA3 = pink, FLNA4 = purple, and FLNA5 = green). The NSD of the alignment was 1.49. The χ2-value of the FLNA3–5 crystal structure against the experimental scattering data was 1.0 (estimated using CRYSOL) (42), as previously reported in (20). (F) The rigid body models of FLNA4–6 obtained using CORAL (41) demonstrate the linker flexibility (spheres) and the movement of the FLNA6 domain in respect to the domain pair of FLNA4–5 (gray). The domain pair FLNA4–5 is superimposed on FLNA4–6, and only the two most distant orientations (blue and green) and one middle orientation (purple) of FLNA6 are shown out of the 50 calculated models. Superimposition of the presented CORAL models with the SASREF model gave NSD values of 1.49 (blue), 1.54 (purple), and 1.66 (green). The black arrows indicate the approximate movements of the blue and green models from the middle orientation in Ångströms, measured using V814 as the landmark between the middle and the most distant orientations of FLNA6. The spheres representing the linker region between the FLNA4–5 domain pair and FLNA6 are dispersed, suggesting that the linker between the domains is mobile. To see this figure in color, go online.
Taken together, the data obtained from SAXS and NMR spectroscopy as well as from limited proteolysis indisputably showed that the FLNA-MVD causing V711D and H743P mutations almost completely destroyed the folding of the mutated FLNA5 as well as, interestingly, FLNA4.
FLNA6 is not part of the compact rod1 fragment and not affected by FLNA-MVD mutations
To date, the detailed atomic structure of FLNA4–6 remains undetermined. Domains 4 and 5 are known to form a compact structure, both in the crystal structure (Fig. 1 B) and in solution (14), but the orientation of FLNA6 with respect to FLNA5 has remained enigmatic. Both SAXS and NMR spectroscopy were employed to solve the overall structure of FLNA4–6 to get information about how FLNA5 mutation affects the overall rod1 structure. The comparison of FLNA3–5 and FLNA4–6 scattering curves shown in Fig. 3 C reveals that the effect of the domain 6 to FLNA4–5 structure is different than that of domain 3. The ab initio model calculated from SAXS data shows that the shape of FLNA4–6 (Fig. 3 D; Table S2) is elongated, whereas FLNA3–5 is more rounded (Fig. 3 E). The calculated SAXS-based rigid body model of the FLNA4–6 fragment reveals that domain 6 did not interact with FLNA4–5 (Fig. 3 D). The rigid body model of FLNA4–6 nicely fits with the ab initio envelope (Fig. 3 D), similar to the FLNA3–5 crystal structure (Fig. 3 E; (14)). Also, the molecular dimensions obtained from SAXS measurements revealed that FLNA4–6 is 2.4 nm longer than the compact FLNA3–5 fragment (14, 20) (Fig. 3, D and E; Table S1), indicating that FLNA6 is not as tightly packed as FLNA5.
To further investigate FLNA6 motion restrictions with respect to FLNA5, the 15N T1 relaxation times were measured using NMR spectroscopy. A plot of 15N T1 values versus amino acid sequence is shown in Fig. 3 A. The average T1 values are clearly higher for FLNA4 and FLNA5 in comparison to FLNA6. This indicates that rather than being “pearls on a string,” FLNA4 and 5 are tumbling as a larger structural unit with respect to FLNA6 and that tumbling of FLNA6 is minimally restricted with respect to FLNA4–5. To compare estimated 15N spin relaxation times for the separate FLNA4–5 and FLNA6 constructs, we simulated relaxation times for the FLNA4–5 crystal structure (PDB: 4M9P) (14) and homology model of FLNA6 using HYDRONMR software (30) (Fig. S4). It is evident that although expected 15N T1 values showed similar trends between the simulated and experimental values, meaning that elevated 15N T1 times were observed for the FLNA4 and 5 domains, the absolute 15N T1 values for the separate FLNA4–5 and FLNA6 domains were generally lower in simulated data. This indicates that molecular tumbling of FLNA6 is hindered by FLNA4–5 and vice versa in the FLNA4–6 construct, resulting in slower overall tumbling. The CORAL-based modeling of the SAXS data was then used to further investigate the motions of FLNA6 in respect to the FLNA4–5 domain pair. The results presented in Fig. 3 F show that FLNA6, connected by a short flexible linker to FLNA5, can adopt various orientations with respect to FLNA4–5. All these different orientations fit well to the experimental scattering data of FLNa4-6 (χ2-values 0.92–1.09, Table S2). The results from CORAL-based modeling of the SAXS data are in accordance with the results from EOM calculations, in which four structures were needed to fit the experimental data (Table S3) instead of one structure that would have represented the rigid structure.
Combining the results obtained from both SAXS and NMR studies suggested that FLNA6 does not interact with FLNA5, and its motion is only partially restricted by FLNA4–5 (Figs. 3, A, D, and F and S4). Accordingly, FLNA5 mutations only destroy the compact structure of FLNA4–5, whereas FLNA5–6 is not affected by FLNA5 mutations.
The effects of FLNA-MVD mutation on FLNA’s interactions
FLNA executes many of its functions via interactions with other proteins (9, 44, 45). Accordingly, the obvious mechanism for FLNA-MVD caused by the mutations would be the altered interactions with other proteins, followed by effects on downstream signaling. Most FLNA-binding partners have been mapped to rod2 domains, whereas only a few proteins have been shown to interact with rod1 domains. One of the rod1-interacting proteins is PTPN12. Recently, we have shown that the proline-rich domain (Pro4) of the C-terminal domain of PTPN12 binds FLNA domains 4–6, of which FLNA4 is the main binding site (20). Using SPR, we tested whether V711D and H743P mutations affect FLNA–PTPN12 interactions. Although an affinity constant of Kd 3.3 × 10−7 M was determined for the FLNA4–6 WT-PTPN12 interaction from the sensorgrams obtained using FLNA4–6 as an analyte on immobilized PTPN12 C-terminal fragment (aa 600–780) (20), the Kd was barely measurable for mutant recombinant FLNA-V711D and H743P (Fig. 4).
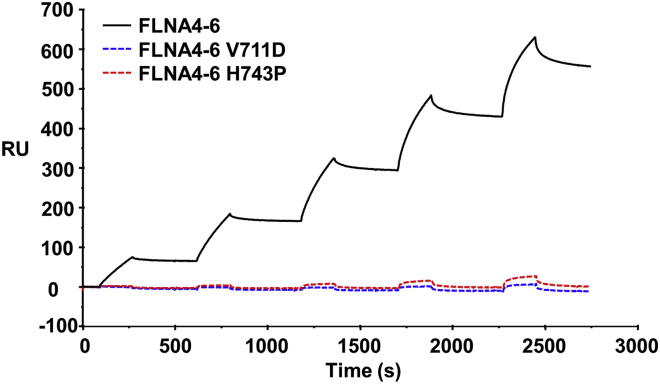
Figure 4.
FLNA-MVD causing mutations at FLNA5 abolished PTPN12 binding to FLNA. The PTPN12 C-terminal fragment (600–700) binds to FLNA4–6 WT with Kd 3.3 × 10−7 M (20) but binding to FLNA4–6 V711D and H743P were hardly detected in SPR experiments. To see this figure in color, go online.
Discussion
To date, the molecular bases of valvular diseases have been poorly understood, and this has restricted the development of new therapies to replace surgical treatment. In this study, we sought and found a molecular-level explanation of why the FLNA mutations V711D and H743P cause FLNA-MVD.
Our results clearly reveal that the FLNA mutations V711D and H743P both have structural consequences. An integrative approach utilizing NMR spectroscopy and SAXS measurements unambiguously indicated that the missense mutations destroyed the folding of the particular domain where the mutation was located. This is not surprising as the hydrophobic valine residue, which points toward the hydrophobic interior of the Ig domain of FLNa5 (Fig. 1 B), is mutated to a negatively charged and slightly larger aspartic acid residue, causing both electrostatic and steric conflicts that destroy domain folding. H743, in turn, is located in the middle of the β-strand (Fig. 1 B). Accordingly, the mutation to proline residue destroys the β-strand and consequently affects the stability of the neighboring β-strands and the entire Ig domain. Importantly, the FLNA5 mutations V711D and H743P were also shown to affect the stability of the neighboring N-terminal domain FLNA4 but not the neighboring C-terminal domain FLNA6. It was not surprising that the loss of the compact folding of FLNA5 affected the stability of FLNA4 as well because domain 4 had earlier been reported to be unstable when it is isolated but is stabilized by the neighboring domain 5 (14). Our results from both NMR 15N spin relaxation data and SAXS measurements, in turn, demonstrated that FLNA6 is not part of the compact rod1 substructure composed from domains 3–5. Accordingly, the destabilization of the adjacent FLNA5 domain does not influence the stability of FLNA6.
In cardiac valves, FLNA is submitted to intense hemodynamic stresses. Our recent study revealed that the P637Q mutation affects FLNA’s force resilience (20). With P637Q mutated FLNA, significantly lower forces were needed to detach FLNA4 and FLNA5 domains from each other than with WT FLNA. It can be speculated that V711D and H743P mutations also change FLNA’s ability to respond to forces because they destroy the compact and force-regulated rod1 substructure, making it inherently flexible and devoid of any force resilience ability. Several other disease-causing mutations have also been identified from FLNA. Interestingly, although disease-causing mutations are spread through FLNA, many are clustered in the compact FLNA regions of FLNA3–5 and FLNA16–17 (46, 47, 48, 49, 50). A skeletal dysplasia-causing mutation at FLNA16 has also been reported to change FLNA’s ability to respond to force (51). Accordingly, it seems that defects in FLNA’s force resilience might be involved in the pathogenesis of various FLNA-linked diseases. This could be expected because FLNA is known to be crucial for cellular force transmission (10). The connection of FLNA and mechanosensing is especially strong in the development and progression of cardiac diseases because of defects in mechanosensing known to cause various cardiac diseases (52, 53, 54). Moreover, FLNA is strongly expressed in endocardial cells during cardiac morphogenesis (12).
The almost complete unfolding of two FLNA domains, 4 and 5, also increased proteolytic digestion of FLNA. This might, of course, be one possible mechanism behind FLNA-MVD, although no in vivo data of the degradation of FLNA in FLNA-MVD has been reported. Similar FLNA domain unfolding due to disease-causing mutations have been reported, with FLNb17 mutations causing skeletal dysplasia called Larsen syndrome (51).
Our results also revealed that FLNA5 mutations abolished FLNA’s interactions with PTPN12. The main binding site of PTPN12 has been mapped to domain 4 (20). The P637Q mutation that is located at FLNA4 has recently been reported to prevent the PTPN12 interaction. FLNA5-PTPN12 interaction is, however, very weak (20). As the FLNA5 mutations V711D and H743P not only destroy the folding of domain 5 but also FLNA4, it is not surprising that these mutations also abolish PTPN12 binding to FLNA. Other proteins, such as protein kinase Syk (55) and DPP9 (56), have also been mapped to bind FLNA5. The loss of FLNA5 folding due to FLNA-MVD mutations obviously abolished their binding to FLNA as well. Whether Syk or DPP9 have roles in FLNA-MVD pathogeneses is not known.
Taken together, the underlying mechanism behind FLNA-MVD seems to be linked to the unfolding of FLNA4 and FLNA5, which might cause the proteolytic digestion of cells, abolish PTPN12 interaction, and potentially also affect FLNA’s force resilience. Which one of these is the most essential molecular mechanism is not known. The importance of FLNA misfolding in the FLNA-MVD pathogeneses provides a tempting idea to use molecular chaperones (57) as a therapeutic treatment of FLNA-MVD.
Conclusions
Cardiac valve diseases are common, affecting 3% of the population. Currently, no treatments other than cardiac surgery are available, but surgery is expensive and has high levels of risk. However, the lack of structural level information of the proteins involved has restricted the development of new therapies. The results presented here with regard to structural and functional consequences of FLNA5 mutations, together with recently reported FLNA4 mutations, suggest that both FLNA force resilience and interaction with PTPN12 are important for FLNA-MVD pathogenesis. Although our results do not provide a complete molecular-level explanation for FLNA-MVD, they provide a crucial step toward understanding the underlying molecular mechanism behind valvular dystrophy and a possible objective for the development of drug-based therapeutics.
Author Contributions
T.J.K.H. Validation, Formal analysis, Investigation, Writing—original draft, Writing—Review & Editing, Visualization. R.C. Formal analysis, Investigation. S.L. Formal analysis, Investigation. M.H. Supervision. J.M. Writing—original draft, Writing—Review & Editing, Supervision, Funding Acquisition. P.P. Validation, Formal analysis, Investigation, Writing—original draft, Writing—Review & Editing, Supervision, Funding Acquisition. U.P. Validation, Formal analysis, Investigation, Writing—original draft, Writing—Review & Editing, Supervision, Project Administration, Funding Acquisition.
Acknowledgments
European Synchrotron Radiation Facility is thanked for providing the beamline BM29 access. We thank Mike Maillasson 1-Plateforme IMPACT Biogenouest-SFR François Bonamy-Université de Nantes for his technical expertise in SPR experiments.
This work was supported by Academy of Finland (283481 to U.P. and 288235 to P.P.), the Fédération Française de Cardiologie (2011, Paris, France to J.M.), and INSERM Translational Research Grant (2012–2016, Paris, France). Dr. R.C. is supported by a grant from Institut de France-Fondation Lefoulon-Delalande (Paris, France) and a “Connect Talent” research chair from Region Pays de la Loire and Nantes Metropole (France).
Editor: Jill Trewhella.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.08.032.
Supporting Material
References
- 1.Levine R.A., Hagége A.A., Yacoub M.H., Leducq Mitral Transatlantic Network Mitral valve disease--morphology and mechanisms. Nat. Rev. Cardiol. 2015;12:689–710. doi: 10.1038/nrcardio.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kyndt F., Gueffet J.P., Schott J.J. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation. 2007;115:40–49. doi: 10.1161/CIRCULATIONAHA.106.622621. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein J.A., Bernstein D., Hudgins L. Familial cardiac valvulopathy due to filamin A mutation. Am. J. Med. Genet. A. 2011;155A:2236–2241. doi: 10.1002/ajmg.a.34132. [DOI] [PubMed] [Google Scholar]
- 4.Le Tourneau T., Le Scouarnec S., Schott J.J. New insights into mitral valve dystrophy: a Filamin-A genotype-phenotype and outcome study. Eur. Heart J. 2018;39:1269–1277. doi: 10.1093/eurheartj/ehx505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Flier A., Sonnenberg A. Structural and functional aspects of filamins. Biochim. Biophys. Acta. 2001;1538:99–117. doi: 10.1016/s0167-4889(01)00072-6. [DOI] [PubMed] [Google Scholar]
- 6.Feng Y., Chen M.H., Walsh C.A. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc. Natl. Acad. Sci. USA. 2006;103:19836–19841. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stossel T.P., Condeelis J., Shapiro S.S. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 8.Sauls K., de Vlaming A., Norris R.A. Developmental basis for filamin-A-associated myxomatous mitral valve disease. Cardiovasc. Res. 2012;96:109–119. doi: 10.1093/cvr/cvs238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou A.X., Hartwig J.H., Akyürek L.M. Filamins in cell signaling, transcription and organ development. Trends Cell Biol. 2010;20:113–123. doi: 10.1016/j.tcb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Ehrlicher A.J., Nakamura F., Stossel T.P. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature. 2011;478:260–263. doi: 10.1038/nature10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baldassarre M., Razinia Z., Calderwood D.A. Filamins regulate cell spreading and initiation of cell migration. PLoS One. 2009;4:e7830. doi: 10.1371/journal.pone.0007830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norris R.A., Moreno-Rodriguez R., Markwald R.R. Expression of the familial cardiac valvular dystrophy gene, filamin-A, during heart morphogenesis. Dev. Dyn. 2010;239:2118–2127. doi: 10.1002/dvdy.22346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruskamo S., Gilbert R., Pentikäinen U. The C-terminal rod 2 fragment of filamin A forms a compact structure that can be extended. Biochem. J. 2012;446:261–269. doi: 10.1042/BJ20120361. [DOI] [PubMed] [Google Scholar]
- 14.Sethi R., Seppälä J., Ylänne J. A novel structural unit in the N-terminal region of filamins. J. Biol. Chem. 2014;289:8588–8598. doi: 10.1074/jbc.M113.537456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sethi R., Ylänne J. Small-angle X-ray scattering reveals compact domain-domain interactions in the N-terminal region of filamin C. PLoS One. 2014;9:e107457. doi: 10.1371/journal.pone.0107457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duval D., Lardeux A., Merot J. Valvular dystrophy associated filamin A mutations reveal a new role of its first repeats in small-GTPase regulation. Biochim. Biophys. Acta. 2014;1843:234–344. doi: 10.1016/j.bbamcr.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duval D., Labbé P., Mérot J. MVP-associated filamin a mutations affect FlnA-PTPN12 (PTP-PEST) interactions. J. Cardiovasc. Dev. Dis. 2015;2:233–247. doi: 10.3390/jcdd2030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Y., Lu Z. Regulation of tumor cell migration by protein tyrosine phosphatase (PTP)-proline-, glutamate-, serine-,and threonine-rich sequence (PEST) Chin. J. Cancer. 2013;32:75–83. doi: 10.5732/cjc.012.10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Playford M.P., Lyons P.D., Schaller M.D. Identification of a filamin docking site on PTP-PEST. J. Biol. Chem. 2006;281:34104–34112. doi: 10.1074/jbc.M606277200. [DOI] [PubMed] [Google Scholar]
- 20.Haataja T.J.K., Bernardi R.C., Pentikäinen U. Non-syndromic mitral valve dysplasia mutation changes the force resilience and interaction of human filamin A. Structure. 2019;27:102–112.e4. doi: 10.1016/j.str.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Östman A., Hellberg C., Böhmer F.D. Protein-tyrosine phosphatases and cancer. Nat. Rev. Cancer. 2006;6:307–320. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura F., Stossel T.P., Hartwig J.H. The filamins: organizers of cell structure and function. Cell Adhes. Migr. 2011;5:160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellanger J.M., Astier C., Debant A. The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat. Cell Biol. 2000;2:888–892. doi: 10.1038/35046533. [DOI] [PubMed] [Google Scholar]
- 24.Gileadi O., Burgess-Brown N.A., Pantic N.H. High throughput production of recombinant human proteins for crystallography. Methods. Mol. Biol. 2008;426:221–246. doi: 10.1007/978-1-60327-058-8_14. [DOI] [PubMed] [Google Scholar]
- 25.Lehtonen J.V., Still D.J., Johnson M.S. BODIL: a molecular modeling environment for structure-function analysis and drug design. J. Comput. Aided Mol. Des. 2004;18:401–419. doi: 10.1007/s10822-004-3752-4. [DOI] [PubMed] [Google Scholar]
- 26.Johnson M.S., Overington J.P. A structural basis for sequence comparisons. An evaluation of scoring methodologies. J. Mol. Biol. 1993;233:716–738. doi: 10.1006/jmbi.1993.1548. [DOI] [PubMed] [Google Scholar]
- 27.Petrey D., Xiang Z., Honig B. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins. 2003;53(Suppl 6):430–435. doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- 28.Permi P., Annila A. Coherence transfer in proteins. Prog. Nucl. Magn. Reson. Spectrosc. 2004;44:97–137. [Google Scholar]
- 29.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuler B. Perspective: chain dynamics of unfolded and intrinsically disordered proteins from nanosecond fluorescence correlation spectroscopy combined with single-molecule FRET. J. Chem. Phys. 2018;149:010901. doi: 10.1063/1.5037683. [DOI] [PubMed] [Google Scholar]
- 31.Franke D., Petoukhov M.V., Svergun D.I. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Cryst. 2017;50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konarev P.V., Volkov V.V., Svergun D.I. PRIMUS : a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 33.Petoukhov M.V., Konarev P.V., Svergun D.I. ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. J. Appl. Cryst. 2007;40:223–228. [Google Scholar]
- 34.Franke D., Svergun D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volkov V.V., Svergun D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuukkanen A.T., Kleywegt G.J., Svergun D.I. Resolution of ab initio shapes determined from small-angle scattering. IUCrJ. 2016;3:440–447. doi: 10.1107/S2052252516016018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petoukhov M.V., Svergun D.I. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozin M.B., Svergun D.I. Automated matching of high- and low-resolution structural models research papers Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 2001;34:33–41. [Google Scholar]
- 39.Tria G., Mertens H.D., Svergun D.I. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. 2015;2:207–217. doi: 10.1107/S205225251500202X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernadó P., Mylonas E., Svergun D.I. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 41.Petoukhov M.V., Franke D., Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Svergun D., Barberato C., Koch M.H. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 43.Rambo R.P., Tainer J.A. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura F., Osborn T.M., Stossel T.P. Structural basis of filamin A functions. J. Cell Biol. 2007;179:1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Razinia Z., Mäkelä T., Calderwood D.A. Filamins in mechanosensing and signaling. Annu. Rev. Biophys. 2012;41:227–246. doi: 10.1146/annurev-biophys-050511-102252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robertson S.P., Jenkins Z.A., Krakow D. Frontometaphyseal dysplasia: mutations in FLNA and phenotypic diversity. Am. J. Med. Genet. A. 2006;140:1726–1736. doi: 10.1002/ajmg.a.31322. [DOI] [PubMed] [Google Scholar]
- 47.Robertson S.P., Twigg S.R., Wilkie A.O., OPD-spectrum Disorders Clinical Collaborative Group Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat. Genet. 2003;33:487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- 48.Robertson S.P. Otopalatodigital syndrome spectrum disorders: otopalatodigital syndrome types 1 and 2, frontometaphyseal dysplasia and Melnick-Needles syndrome. Eur. J. Hum. Genet. 2007;15:3–9. doi: 10.1038/sj.ejhg.5201654. [DOI] [PubMed] [Google Scholar]
- 49.Daniel P.B., Morgan T., Robertson S.P. Disease-associated mutations in the actin-binding domain of filamin B cause cytoplasmic focal accumulations correlating with disease severity. Hum. Mutat. 2012;33:665–673. doi: 10.1002/humu.22012. [DOI] [PubMed] [Google Scholar]
- 50.Bicknell L.S., Farrington-Rock C., Robertson S.P. A molecular and clinical study of Larsen syndrome caused by mutations in FLNB. J. Med. Genet. 2007;44:89–98. doi: 10.1136/jmg.2006.043687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seppälä J., Bernardi R.C., Pentikäinen U. Skeletal dysplasia mutations effect on human filamins’ structure and mechanosensing. Sci. Rep. 2017;7:4218. doi: 10.1038/s41598-017-04441-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krüger M., Linke W.A. Titin-based mechanical signalling in normal and failing myocardium. J. Mol. Cell. Cardiol. 2009;46:490–498. doi: 10.1016/j.yjmcc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 53.Herum K.M., Lunde I.G., Christensen G. The soft- and hard-heartedness of cardiac fibroblasts: mechanotransduction signaling pathways in fibrosis of the heart. J. Clin. Med. 2017;6:1–31. doi: 10.3390/jcm6050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyon R.C., Zanella F., Sheikh F. Mechanotransduction in cardiac hypertrophy and failure. Circ. Res. 2015;116:1462–1476. doi: 10.1161/CIRCRESAHA.116.304937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Falet H., Pollitt A.Y., Hartwig J.H. A novel interaction between FlnA and Syk regulates platelet ITAM-mediated receptor signaling and function. J. Exp. Med. 2010;207:1967–1979. doi: 10.1084/jem.20100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Justa-schuch D., Silva-garcia M., Geiss-friedlander R. DPP9 is a novel component of the N-end rule pathway targeting the tyrosine kinase Syk. Elife. 2016;5:e16370. doi: 10.7554/eLife.16370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki Y. Emerging novel concept of chaperone therapies for protein misfolding diseases. Proc. Jpn. Acad. Ser B Phys. Biol. Sci. 2014;90:145–162. doi: 10.2183/pjab.90.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.