Abstract
Purpose
Concurrent chemoradiation therapy (CRT) is the principal treatment modality for locally advanced lung cancer. Cell death due to CRT leads to the release of cell-free DNA (cfDNA) and circulating tumor DNA (ctDNA) into the bloodstream, but the kinetics and characteristics of this process are poorly understood. We hypothesized that there could be clinically meaningful changes in cfDNA and ctDNA during a course of CRT for lung cancer.
Methods and materials
Multiple samples of plasma were obtained from 24 patients treated with CRT for locally advanced lung cancer to a mean dose of 66 Gy (range, 58-74 Gy) at the following intervals: before CRT, at weeks 2 and 5 during CRT, and 6 weeks after treatment. cfDNA was quantified, and a novel next generation sequencing (NGS) technique using enhanced tagged/targeted-amplicon sequencing was performed to analyze ctDNA.
Results
Patients for whom specific mutations in ctDNA were undetectable at the baseline time point had improved survival, and potentially etiologic driver mutations could be tracked throughout the course of CRT via NGS in multiple patients. We quantified the levels of cfDNA from patients before CRT, at week 2, week 5, and at 6 weeks after treatment. No differences were observed at weeks 2 and 5 of therapy, but we noted a significant increase in cfDNA in the posttreatment follow-up samples compared with samples collected before CRT (P = .05).
Conclusions
Dynamic changes in both cfDNA and ctDNA were observed throughout the course of CRT in patients with locally advanced lung cancer. Specific mutations with therapeutic implications can be identified and tracked using NGS methodologies. Further work is required to characterize the changes in cfDNA and ctDNA over time in patients treated with CRT and to assess the predictive and prognostic potential of this powerful technology.
Introduction
Every year, more than 65,000 people receive a diagnosis of stage III non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). The nonoperative standard of care for stage III NSCLC/SCLC is chemoradiation therapy (CRT), often followed by adjuvant immunotherapy. Despite aggressive treatment, the median survival is approximately 25 months for NSCLC and 23 months for SCLC.1, 2 Identifying predictive and prognostic factors before, during, or immediately after CRT in stage III disease could allow for adaptation of treatment to optimize patient outcomes or to detect minimal residual disease after therapy. In metastatic NSCLC, patient-specific molecular subtyping of biopsied tissue, circulating tumor cells, or circulating DNA can guide therapy decisions but has had little impact in the stage III setting.3, 4
Both cell-free DNA (cfDNA) and circulating tumor DNA (ctDNA) are promising biomarkers for lung cancers treated with radiation therapy.5, 6, 7, 8 cfDNA is created when cells undergo cell death and release fragmented DNA into the bloodstream. ctDNA is a subset of cfDNA that can be distinguished from cfDNA of noncancerous origin by the presence of cancer-specific mutations. We hypothesized that there would be quantitative and qualitative changes in cfDNA and ctDNA during a course of CRT for locally advanced lung cancer and that these changes may have prognostic potential.
Methods and Materials
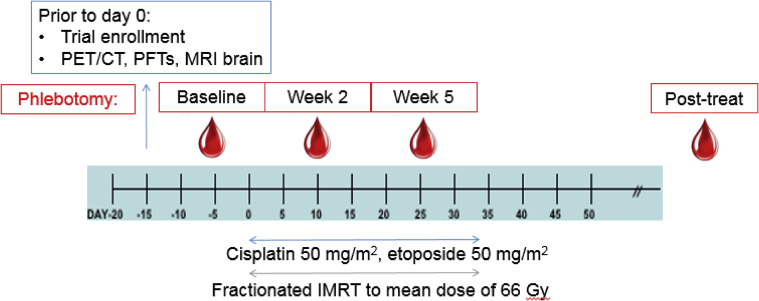
We obtained patient plasma from 24 patients treated with cisplatin/etoposide and concurrent radiation therapy for locally advanced lung cancer on an institutional review board–approved trial (21 with NSCLC; 3 with SCLC). Patients were treated with accelerated fractionation (6 fractions/week) to a mean dose of 66 Gy (range, 58-74 Gy) as described previously.9 Unless patients refused phlebotomy, plasma was obtained at 4 time points: before therapy, at weeks 2 and 5 during CRT, and approximately 6 weeks after treatment (Fig 1). Samples were single spun at acquisition. After thawing on ice, single aliquots of plasma were spun for 15 minutes at 15,000 × g at 4°C and subsequently processed via the Maxwell RSC ccfDNA Plasma Kit. cfDNA was quantified with fluorimetry using the QuantiFluor ONE dsDNA system (Promega Corp, Madison, WI). cfDNA concentration was normalized by plasma volume.
Figure 1.
Eligible patients were initially staged and found to have locally advanced but nonmetastatic lung cancer. Blood was obtained from eligible patients before therapy (baseline), during weeks 2 and 5 of chemoradiation therapy, and at approximately 6 weeks after treatment. Patients were treated with cisplatin/etoposide and concurrent radiation therapy with accelerated fractionation.
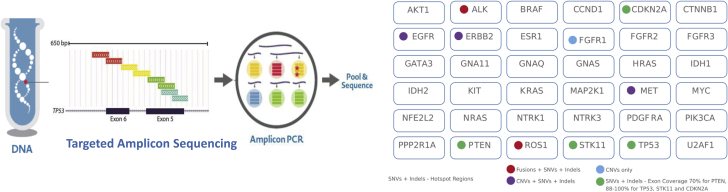
A next generation sequencing (NGS) technique known as enhanced tagged/targeted-amplicon sequencing (eTAm-Seq) was subsequently employed. eTAm-Seq sequencing is an amplicon sequencing method that can detect and quantify cancer mutations in plasma, with identification of >99% point mutations at >0.5% allelic fraction and a reportable limit of 0.06%.8, 10 All genes in the panel are implicated in the pathogenesis of lung cancer (Fig 2).
Figure 2.
Targeted amplicon sequencing was performed on cfDNA to analyze the genes indicated, all of which are implicated in the pathogenesis of lung cancer. Exon tiling to assess the majority of the noted gene, hotspot analysis of relevant areas of genes, and copy-number variant analysis were performed as indicated. Abbreviations: cfDNA = cell-free DNA; indels = in-frame deletions; SNV = single nucleotide variation.
Overall survival (OS) was calculated from diagnosis using the Kaplan-Meier method, and hazard ratio estimates were obtained using the proportional hazards model. Changes from baseline in cfDNA concentration were assessed using signed rank tests. P values < .20 were considered significant.
Results
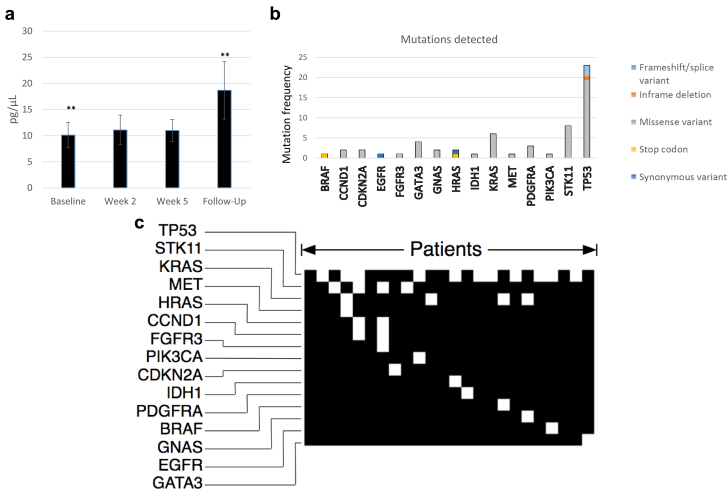
Ninety samples of plasma were examined from a cohort of patients undergoing CRT for locally advanced lung cancer at the time points shown (Fig 1). The median time between the pretreatment baseline and posttreatment follow-up sample was 92 days. The mean normalized concentration of cfDNA isolated from pretreatment baseline (n = 24), week 2 (n = 24), week 5 (n = 22), and posttreatment follow-up samples (n = 20) was 10.1 pg/μL (σ = 8.9), 11.1 pg/μL (σ = 10.0), 11.1 pg/μL (σ = 7.2), and 18.7 pg/μL (σ = 18.5), respectively. There was a significant increase in cfDNA between the baseline and posttreatment follow-up samples (P = .05; Fig 3a). Using the reverse Kaplan-Meier method, the median clinical follow-up in the entire sample was 60 months (80% confidence interval [CI], 40-61 months), and median OS was 28 months (80% CI, 20-52 months). There was no clear correlation of cfDNA concentration with OS or disease-free survival.
Figure 3.
(a) Mean cell-free DNA concentration during various phlebotomy time points before, during, and after chemoradiation therapy. ** Significance in the change in cell-free DNA concentration from the baseline to the 6-week posttreatment follow-up samples. (b) Detected mutated genes are indicated in the frequency histogram; type of mutation is superimposed in color. (c) Qualitative plot of mutations in circulating tumor DNA. Each column represents a unique patient.
Of the 90 evaluable samples, NGS detected mutations in 58 samples. Twenty-one of the 24 patients had detectable ctDNA at any point during the study. The median mutant allele fraction was 0.75% (range, 0.09%-50.74%). Two patients had germline-inherited mutations in genes GATA3 and CDKN2A based on allele fractions of approximately 50%. The majority of mutations identified were missense variants (88%), with stop codons (5.2%), frameshift/spice variants (1.7%), in-frame deletions (1.7%), and synonymous variants (3.4%) representing the minority (Fig 3b).
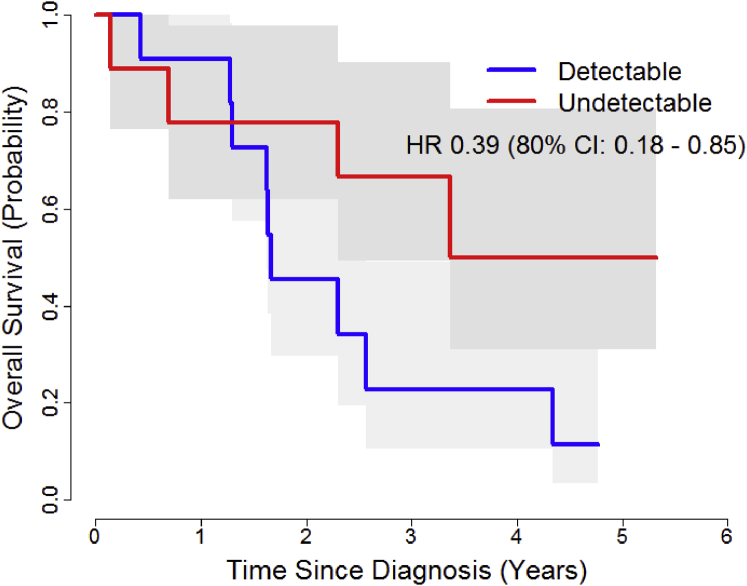
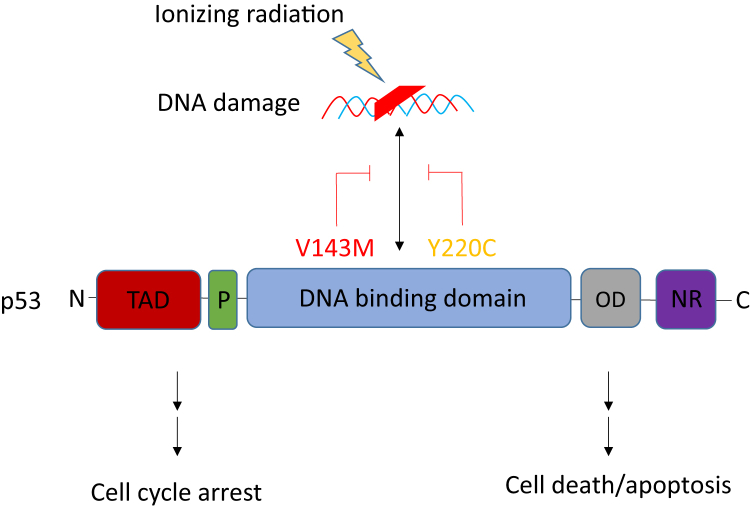
Patients with undetectable baseline ctDNA had better OS (hazard ratio, 0.39; 80% CI, 0.18-0.85; P = .12). Median OS was 4.3 years in patients with no detectable mutations (n = 9) and 1.66 years in patients for whom mutations were detected (n = 11; P = .1; Fig 4). The 2-year survival probability was 0.78 and 0.45, respectively (80% CI for the difference. 0.06-0.59). Interestingly, the evolution of mutations in individual patients during therapy could also be observed. For example, a 56-year-old man with T2N3M0 NSCLC who was treated to 58 Gy with cisplatin/etoposide had no detectable mutations at the baseline or week 2 time points, but a point mutation in the DNA binding domain of p53 at amino acid 143 appeared during week 5 of CRT. At the subsequent follow-up blood draw, an additional mutation appeared in the DNA binding domain of p53 at amino acid 220 (Fig 5). Both sites affect p53's function as a tumor suppressor gene, abrogating its ability to sense damage to ionizing radiation and thereby leading to apoptosis.11, 12 The clinical result in this patient was recurrent disease with local failure at 9 months post CRT.
Figure 4.
Overall survival curves in patients for whom mutations were detectable versus undetectable at the baseline blood collection timepoint.
Figure 5.
p53 was the most frequently detected mutated gene. The domain structure of p53 is shown, as well as 2 mutations observed in a patient with a particularly poor outcome. Mutations in the DNA binding domain of p53 abrogate its ability to sense DNA damage and thereby reduce the probability of both cell cycle arrest and apoptosis. Abbreviations: C = C-terminus; D = oligimerization domain; N = N-terminus; NR=negative regulatory domain; P = proline-rich domain; TAD = transactivation domain.
Discussion
Our study provides insight into the dynamic changes of both cfDNA and ctDNA in patient plasma before, during, and after CRT for locally advanced lung cancer. cfDNA and ctDNA represent dynamic, “real-time” biomarkers due to a half-life of 90 minutes, are easy to obtain via simple phlebotomy, and can be characterized by multiple technologies. We observed that the concentration of cfDNA increased after CRT (Fig 3a). The fact that cfDNA concentration is higher after cancer therapy is not surprising in the context of expected cell death after CRT, and other groups have explored this phenomenon in lung cancer and other tumor types.13, 14, 15, 16 Further work is required to determine whether the kinetics of cfDNA, a nonspecific marker of overall DNA in the circulation, has either predictive or prognostic significance in the context of CRT. Our study did not find a clear correlation between cfDNA and clinical outcome.
Fluctuations in normal tissue injury (rather than tumor cell death) may contribute to variations in cfDNA; thus, an assessment of ctDNA represents a more specific assay. Using NGS, we characterized a number of unique mutations in ctDNA that could be subsequently tracked during CRT (Fig. 3b, 3c), the most frequent of which were in the p53 gene. The detection and subsequent monitoring of ctDNA in localized malignancies is a novel concept, with limited prior studies of patients treated with other modalities and in other cancers.7, 8 Our study is unique in that only patients treated exclusively with CRT for locally advanced lung cancer were included. The observation that patients for whom mutations were undetectable before therapy (at baseline) had better OS suggests that there may be prognostic potential for ctDNA in this patient population.
Beyond the quantitative aspect of ctDNA to assess tumor burden, the qualitative impact of specific mutations on treatment outcome is intriguing. For instance, one poorly performing patient (with a survival of 10 months) in the baseline mutation cohort had a mutation in KRAS (G12V), a well-established harbinger of early recurrence and poor survival.17 Missense mutations in other genes, such as STK11/LKB1, were also observed in poorly performing patients with baseline mutations, although this gene has been somewhat less well characterized in the literature. We also observed the evolution of mutations that may predict for treatment failure during CRT, including DNA-binding domain mutations in p53, which may diminish p53's ability to sense DNA damage and thereby lead to cell cycle arrest and cell death (Fig. 5).11, 12 The relevance of these findings requires further validation in a larger cohort.
Conclusions
Despite a prospective design and robust clinical follow-up, the number of patients in this study was relatively small, and the study was hampered by occasional phlebotomy refusal and/or nonevaluable samples. The latter issue may be attributed to technical factors, most notably the prolonged time during which blood was frozen, limited volume of plasma available, and the potential contamination of some samples with genomic DNA owing to single-spin centrifugation of the samples. However, the assessment of cfDNA concentration and ctDNA via NGS clearly represents a powerful tool, and further clinical trials using the analysis of cfDNA should be undertaken in the radiation oncology clinic.
Footnotes
Platform presentation at the American Society for Radiation Oncology, San Diego, CA, 2017.
Sources of support: Funded in part by a grant from the North Carolina Lung Cancer Initiative.
Disclosures: Dr Torok reports grants from the Lung Cancer Initiative during the conduct of the study. Dr Nixon reports consulting/advisory positions with Novartis, Pfizer, and Cerulean Pharma, as well as research funding from TRACON Pharma, Amgen, Novartis, Incyte, Seattle Genetics, Genentech/Roche, and Acceleron Pharma. No other conflicts of interest were reported.
References
- 1.Bradley J.D., Paulus R., Komaki R. Standard-dose versus high-dose conformal radiotherapy with concurrent and consolidation carboplatin plus paclitaxel with or without cetuximab for patients with stage IIIA or IIIB non-small-cell lung cancer (RTOG 0617): A randomised, two-by-two factorial phase 3 study. Lancet Oncol. 2015;16:187–199. doi: 10.1016/S1470-2045(14)71207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonia S.J., Villegas A., Daniel D. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N Engl J Med. 2017;377:1919–1929. doi: 10.1056/NEJMoa1709937. [DOI] [PubMed] [Google Scholar]
- 3.Neal J.W., Gainor J.F., Shaw A.T. Developing biomarker-specific end points in lung cancer clinical trials. Nat Rev Clin Oncol. 2015;12:135–146. doi: 10.1038/nrclinonc.2014.222. [DOI] [PubMed] [Google Scholar]
- 4.Wan J.C.M., Massie C., Garcia-Corbacho J. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhuri A.A., Binkley M.S., Osmundson E.C., Alizadeh A.A., Diehn M. Predicting radiotherapy responses and treatment outcomes through analysis of circulating tumor DNA. Semin Radiat Oncol. 2015;25:305–312. doi: 10.1016/j.semradonc.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deig C.R., Mendonca M.S., Lautenschlaeger T. Blood-based nucleic acid biomarkers as a potential tool to determine radiation therapy response in non-small cell lung cancer. Radiat Res. 2017;187:333–338. doi: 10.1667/RR14613.1. [DOI] [PubMed] [Google Scholar]
- 7.Newman A.M., Bratman S.V., To J. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nature Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forshew T., Murtaza M., Parkinson C. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra168. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 9.Kelsey C.R., Das S., Gu L., Dunphy F.R., III, Ready N.E., Marks L.B. Phase 1 dose escalation study of accelerated radiation therapy with concurrent chemotherapy for locally advanced lung cancer. Int J Radiat Oncol Biol Phys. 2015;93:997–1004. doi: 10.1016/j.ijrobp.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Plagnol V., Woodhouse S., Howarth K. Analytical validation of a next generation sequencing liquid biopsy assay for high sensitivity broad molecular profiling. PLoS One. 2018;13 doi: 10.1371/journal.pone.0193802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato S., Han S.Y., Liu W. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brady C.A., Attardi L.D. p53 at a glance. J Cell Sci. 2010;123:2527–2532. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sozzi G., Conte D., Leon M. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol. 2003;21:3902–3908. doi: 10.1200/JCO.2003.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Mehra N., Dolling D., Sumanasuriya S. Plasma cell-free DNA concentration and outcomes from taxane therapy in metastatic castration-resistant prostate cancer from two phase III trials (FIRSTANA and PROSELICA) Eur Urol. 2018;74:283–291. doi: 10.1016/j.eururo.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyun M.H., Sung J.S., Kang E.J. Quantification of circulating cell-free DNA to predict patient survival in non-small-cell lung cancer. Oncotarget. 2017;8:94417–94430. doi: 10.18632/oncotarget.21769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B.T., Drilon A., Johnson M.L. A prospective study of total plasma cell-free DNA as a predictive biomarker for response to systemic therapy in patients with advanced non-small-cell lung cancers. Ann Oncol. 2016;27:154–159. doi: 10.1093/annonc/mdv498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renaud S., Falcoz P.E., Schaeffer M. Prognostic value of the KRAS G12V mutation in 841 surgically resected Caucasian lung adenocarcinoma cases. Br J Cancer. 2015;113:1206–1215. doi: 10.1038/bjc.2015.327. [DOI] [PMC free article] [PubMed] [Google Scholar]