Abstract
Large-scale multi-ethnic cohorts offer unprecedented opportunities to elucidate the genetic factors influencing complex traits related to health and disease among minority populations. At the same time, the genetic diversity in these cohorts presents new challenges for analysis and interpretation. We consider the utility of race and/or ethnicity categories in genome-wide association studies (GWASs) of multi-ethnic cohorts. We demonstrate that race/ethnicity information enhances the ability to understand population-specific genetic architecture. To address the practical issue that self-identified racial/ethnic information may be incomplete, we propose a machine learning algorithm that produces a surrogate variable, termed HARE. We use height as a model trait to demonstrate the utility of HARE and ethnicity-specific GWASs.
Keywords: trans-ethnic GWAS, genetic ancestry, multi-ethnic cohort, biobank, ethnicity-specific trait loci, self-reported race/ethnicity, stratified analysis
Introduction
Genome-wide association studies (GWASs) have become a powerful approach for exploring the genetic basis of complex phenotypes. While earlier studies focused on populations of predominantly European descent, recent efforts have aimed to substantially expand racial and ethnic diversity. The Million Veterans Program1 (MVP) represents a multi-ethnic cohort, which has enrolled more than 750,000 veteran volunteers, completed genotyping in more than 350,000 participants to date, and includes a wealth of phenotypes and health outcomes. Questions have arisen while performing GWASs in a multi-ethnic cohort regarding the definition and the use of an individual’s ancestry. Dense genotype data have enabled accurate estimation of individual ancestry,2, 3, 4 which has been shaped by reproductive isolation and admixture through human history. At the same time, many studies also obtain racial/ethnic information on participants through questionnaires or electronic health records (EHR). In this paper, we will refer to this latter information as self-identified race/ethnicity (SIRE) to distinguish from genetically inferred ancestry (GIA). A primary goal of multi-ethnic GWASs is to characterize ethnicity-specific trait loci or heterogeneous genetic effect across populations. An example of ethnicity-specific locus is CD36 (MIM: 173510) for high-density lipid cholesterol (HDL), for which the putative causal variant (rs2366858) is only polymorphic in populations of African descent.5 A well-known example of heterogeneous genetic effect is the APOE (MIM: 107741) e4 allele, which is polymorphic in many populations but confers greater risk of Alzheimer disease in Asians compared to other populations.6, 7 The mechanisms underlying such heterogeneity are not well understood and may include unaccounted-for causal variants nearby or interaction with environmental or genetic factors that vary across populations. With the goal of effectively characterizing ethnicity-specific trait loci and interpreting heterogeneous genetic effects, we investigate the analytic issues related to ancestry, race, and ethnicity in multi-ethnic GWASs.
To date, most GWASs stratify on SIRE and adjust GIA within SIRE as covariates. The stratification by SIRE often implicitly occurs at the recruitment or genotyping stages, which focus on populations described by a single SIRE, such as Hispanics, Europeans/European Americans, African Americans/Afro-Caribbean, or East Asians, among others. Within each race/ethnicity, GIA is adjusted as covariates to account for genetic structure within a SIRE.4, 8 Results from these ethnicity-specific studies are combined through meta-analysis within an ethnicity or through trans-ethnic analysis across ethnicities.9, 10 In contrast, in recent Biobank-based multi-ethnic cohort studies, participants are recruited, phenotyped, and genotyped according to a uniform protocol. For such studies, two analytic strategies can be considered. One approach, which we will refer to as mega-analysis, performs association mapping on the entire cohort, adjusting for population structure in the entire cohort using GIA. While simple to implement, results from such an analysis are difficult to interpret: a significant trait locus may be relevant in one racial/ethnic group, a few groups, or all groups. When the representation of ethnicities is unbalanced, the association results are likely driven by the group with the largest sample size. Furthermore, we show through simulation and analyses of real data that, compared to stratified analysis, mega-analysis often loses statistical power when the causal variant is minority specific or its allelic effect varies between populations.
The alternative approach performs stratified analyses for each racial/ethnic group. In addition to the interpretability of association findings, this approach enables meaningful comparison between studies and meta-analysis across studies. However, the question remains how strata should be defined in a multi-ethnic cohort, in which participants are enrolled without restrictions based on race or ethnicity. We reason that SIRE and GIA have complementary strengths. In epidemiologic studies, there is a long history of stratifying on SIRE. This is because SIRE acts as a surrogate to an array of social, cultural, behavioral, and environmental variables, many of which are correlated with trait variation or disease risk.11, 12, 13 Hence, stratifying on SIRE has the potential benefits of reducing heterogeneity of these non-genetic variables and decoupling the correlation between genetic and non-genetic factors. For genetic association studies, the SIRE categories recapitulate the continental-level genetic ancestry structure;14, 15, 16 therefore, population-specific trait variants are likely to be enriched in one or a few SIRE groups. However, SIRE can be incomplete and of varying accuracy depending on the source. In MVP, SIRE is derived from direct responses to survey questionnaires and from text mining of the Department of Veterans Affairs EHR. This leaves 3.67% of the participants without any SIRE information; additionally, inconsistency occurs when consolidating multiple sources. The missing and imperfect SIRE is expected in most multi-ethnic EHR-based biobank cohorts. In contrast, GIA—in the form of principal components or admixture proportions—can be estimated for every GWAS participant. Previous population genetic studies have demonstrated that GIA and self-identified racial/ethnic information have a high correlation, but one does not unambiguously determine the other. Specifically, in admixed groups such as African Americans and Hispanics, genetic ancestries vary continuously among individuals along axes that represent admixture proportions; defining strata based on GIA requires thresholds that are often ad hoc.17 Conversely, the distribution of ancestry proportions may partially overlap between different racial/ethnic groups and cannot be separated based on GIA alone.18, 19
Motivated by these practical challenges, we propose a supervised learning algorithm that defines a categorical stratification variable in a multi-ethnic GWAS. The variable, termed HARE (harmonized ancestry and race/ethnicity), uses GIA to refine SIRE for genetic association studies in three ways: identify individuals whose SIRE is likely inaccurate, reconcile conflicts among multiple SIRE sources, and impute missing racial/ethnic information when the predictive confidence is high. We describe the relationship between HARE, racial/ethnic, and genetic ancestry in MVP, a representative US-based multi-ethnic cohort. Using HARE as the stratifying variable, we investigate the effectiveness of detecting ethnicity-specific trait loci through simulation as well as analysis of height as a model trait in the MVP.
Material and Methods
HARE
The goal of HARE is to define strata for ethnicity-specific GWAS analyses. The computation of HARE consisted of two components: first, in the “training” step, a support vector machine (SVM) model was built, which learned the correspondence between GIA and SIRE; second, in the “assignment” step, HARE was determined based on SIRE, GIA, and the output from the SVM. The assignment follows the decision tree of Figure 1.
Figure 1.
Decision Tree for HARE Assignment
For each individual, denote the support vector machine predicted probabilities, arranged in decreasing order from the most likely stratum, L1, to the least likely stratum, LK. If the individual’s SIRE is not missing, PSIRE denotes the support vector machine predicted probability corresponding to SIRE; otherwise PSIRE is undefined. For analyses reported in this study, t1 = 40 and t2 = 20.
Training of SVM
The SVM used GIA, the top 30 PCs in our analysis, as predictors and SIRE as response. Because SIRE is a multi-class categorical variable, we first trained a one-versus-one classifier with a radial basis function kernel for every pair of categories. These binary classifiers were then combined using a pairwise coupling model to produce a multi-class probability vector for each individual.20 The individual classifiers had two tuning parameters: the inverse variance of the kernel, , controls the radius of influence exerted by a single training sample, while the regularization constant, C, encourages sparse models. These parameters were optimized by searching a two-dimensional grid and using a 5-fold cross-validation. More details are given in the caption of Figure S1. In the MVP analysis, SIRE took four values: Hispanic, non-Hispanic Asian, non-Hispanic black, and non-Hispanic white, as described below.
Assignment of HARE
Given an individual’s genetic PCs, the multi-class SVM outputs a probability vector, , representing the predicted membership probability for each of the K distinct categories. Let L1 denote the stratum corresponding to the highest predicted probability, L2 be the stratum corresponding to the second highest predicted probability, and so on, such that . For individuals whose SIRE is non-missing and consistent across records, let PSIRE denote the predicted probability corresponding to SIRE. For each individual, HARE is assigned according to the decision tree in Figure 1, or equivalently, as:
Note that when SIRE is non-missing and strongly contradicts GIA, we set HARE as missing rather than re-assigning the individual according to the predicted stratum L1.
HARE may be unassigned (missing) for some individuals. We set t1 = 40 and t2 = 20; lower t1 and higher t2 will result in more individuals with missing HARE, through removing more outliers and assigning fewer individuals, respectively.
User-Selected Parameters
All results presented in this paper used the SVM trained on top 30 PCs. Comparing the assignment using 30 PCs versus 20 PCs revealed a discordance rate of 1.3%. This level of consistency is not surprising, because as higher PCs tend to describe finer-level populations structure, they are less informative for the four major HARE groups. On the other hand, if a PC were entirely uninformative, it will be ignored during the SVM training. Naturally, including many unnecessary PCs will increase computation burden. Therefore, we recommend using an upper limit of the PCs that are relevant; specifically, for major race/ethnicity strata, 30 PCs suffice. The two thresholds, t1 and t2, control the stringencies with which the outliers are removed and individuals without SIRE are assigned a HARE. Varying these parameter values in a wide range from 0 to 100, we found that the HARE assignment was quite stable (Figure S2). This analysis also provides practical guidance in choosing the thresholds. In our study, we chose the values of 40 and 20, respectively, based on the visual inspection that the slope of the curves is fairly shallow at these values.
SIRE and GIA in the Million Veteran Program (MVP)
The MVP, launched in 2011 by the Department of Veteran Affairs Office of Research and Development, was a nation-wide research program aiming to acquire new biological insights and to elucidate the genetic basis of diseases, with the ultimate goal of further refining precision medicine to Veteran Affairs health care.1 MVP participants consented to a blood draw and to have their DNA extracted for genomic profiling and linked to their full electronic health record within the VA. Both MVP Biobank and this analysis were approved by the VA institutional review boards.
Unless otherwise noted, analyses presented in this paper included 351,820 MVP participants, who were genotyped using a customized Affymetrix Axiom Biobank array of 723,305 variants. For GIA, the top 30 PCs were computed using program FlashPCA21 on an extended genotype dataset that included all MVP participants and an additional 2,504 individuals from the 1000 Genomes Phase 3 data.22 To aid interpretation, we also estimated individual ancestry proportions using the program ADMIXTURE23 with K = 5 and augmented the MVP participants with individuals from 1000 Genomes Phase 3 data that approximated European (GBR), African (YRI/LWK), East Asian (CHB), South Asian (GIH/PJL), and Native American (PEL) ancestral populations. We note that this admixture analysis was designed to qualitatively complement the PCA analysis: as the 1000 Genomes individuals included in this analysis did not fully represent ancestry diversity in MVP, various model assumptions in ADMIXTURE were violated; therefore, we caution quantitative interpretation of the estimated admixture proportions.
SIRE in MVP was derived based on information collected from the VA Corporate Data Warehouse (CDW) and the MVP Baseline Survey (MVP-BS). Overall, ∼60% of participants had consistent SIRE, while the remaining participants either had no SIRE or had multiple and inconsistent responses among two or more SIRE determinations in CDW and the MVP-BS. Because our goal was to define ethnicity-specific strata for subsequent GWAS analyses, we focused on defining four groups—Hispanics, non-Hispanic Asian, non-Hispanic black, and non-Hispanic white—which have moderately large sample sizes for adequately powered genetic association analysis. For this reason, we set the SIRE of individuals whose responses were not in one of these four categories as “missing,” which included American Indian, Alaska Native, Native Hawaiian, Other Pacific Islanders, and multi-race/ethnicity responses.
To train the SVM model described above, we constructed a training dataset that included 201,931 individuals whose SIRE was unambiguous and was one of the four groups. The top 30 PCs were used as predictors. To reduce the influence of a few outliers on the SVM model, we repeated the SVM training step once after removing 1,547 individuals, for whom the predicted most likely group is not the same as SIRE. Thus, the final SVM model used to compute the predicted probability vectors was based on 200,384 individuals. The assignment of HARE followed the decision tree in Figure 1. Because our training dataset did not include American Indian, Alaska Native, Native Hawaiians, and Pacific Islanders, the HARE for individuals reporting SIRE entirely from one of these populations were set to missing. Altogether, 6,257 individuals had missing HARE.
As an assessment of its statistical accuracy, we applied the SVM trained on the 201,931 individuals, described above, to a non-overlapping set of 27,974 MVP participants, for whom SIRE was available and genotyping was completed on the same Affymetrix array at a later date. PCs of these individuals were calculated by projecting onto the axes determined based on the main cohort.21 Thus, these individuals were not used in any step during the training of the SVM model. We then assigned strata assuming either their SIRE is known or unknown, and we compared these assignments with the actual SIRE.
Simulation
We performed simulation studies to characterize the statistical power for detecting minority-specific trait variants using HARE-stratified analysis as compared to that of a mega-analysis approach. In brief, we first selected a minority-specific causal variant as described below. A quantitative phenotype was then simulated using program GCTA24 and the MVP genotype data, according to the genotype at the causal variant and assuming that it explains a specific proportion of the phenotypic variance, h2. The causal variant was then removed from the dataset, and SNPs within a ± 100K base pair (bp) window of the causal variant were scanned for association using PLINK.25 Thus, the genotype data used for the association analysis represented realistic LD patterns both within ethnicity and between ethnicities. For each causal variant, a total of 100 phenotypes were simulated for each specific h2, and power was defined as the proportion of simulations, in which at least one tag SNP near the causal variant was associated with the phenotype at p < 5 × 10−8. This process was repeated for ten different values of h2 ranging from 0.0001 to 0.01. To eliminate population stratification, in HARE-stratified analysis, the top 10 PCs calculated within a HARE stratum were adjusted as covariates. For the mega-analysis, the top 20 PCs computed on the entire cohort were adjusted as covariates.
To select causal variants, we considered rare and common causal variants separately because the LD pattern around these causal variants are likely to differ. For rare causal variants, we randomly selected 125 unlinked SNPs such that the minor allele frequency (MAF) was less than 1% in one HARE minority strata while absent in all other HARE strata; these included 105 variants that were polymorphic only in non-Hispanic black and 20 that were polymorphic only in Hispanics. Requiring a causal variant to have an MAF > 10% in one minority population while monomorphic in all other strata yielded very few SNPs. Therefore, we relaxed the population-specific criterion and instead looked for relatively common variants that preferentially occur in one stratum. Specifically, we selected (1) 103 variants with MAF > 0.1 in non-Hispanic black, MAF ≤ 5 × 10−4 in non-Hispanic white and MAF ≤ 1 × 10−2 in Hispanics and (2) 3 variants with MAF > 0.1 in Hispanics, MAF ≤ 5 × 10−4 in non-Hispanic white, and MAF ≤ 2 × 10−3 in non-Hispanic black.
GWAS for Height in MVP data
Of 351,820 MVP participants, 342,883 had height measurements after excluding extreme outliers (height < 48 or > 99 inches) and amputees. We then took the average of measurements that were made within 3 years from an individual’s enrollment date, excluding measures more than 3 inches from the individual’s average height. A multi-ethnic GWAS using both stratified and mega-analysis were performed within each HARE stratum and in the entire cohort, respectively, using the same strategy to control for population stratification as described in the Simulation section above. We also performed fixed-effects, inverse-variance weighted meta-analysis combining four HARE groups using PLINK.25 Significant SNPs within 1 Mb were considered as the same locus. For validation, we compared GWAS results in MVP with UKB GWAS26 for height in 452K individuals of European ancestry, and to WHI, which included 8,149 African American women.27
Results
HARE in the MVP
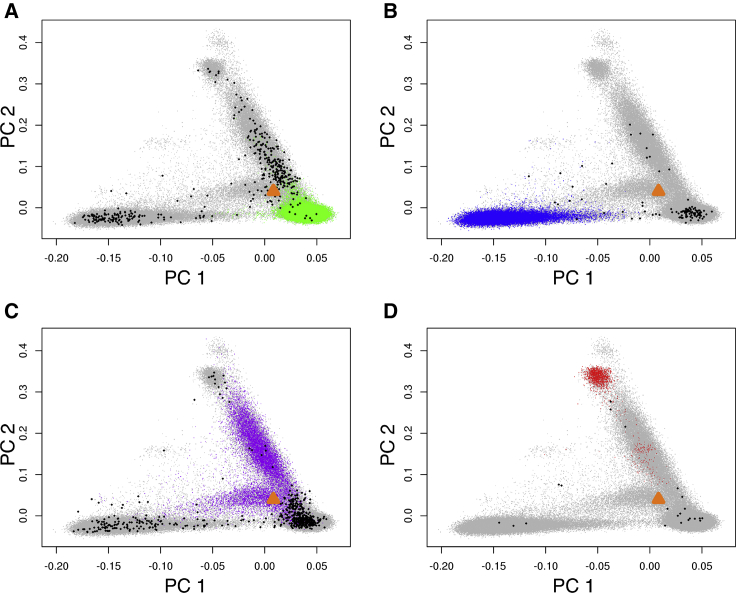
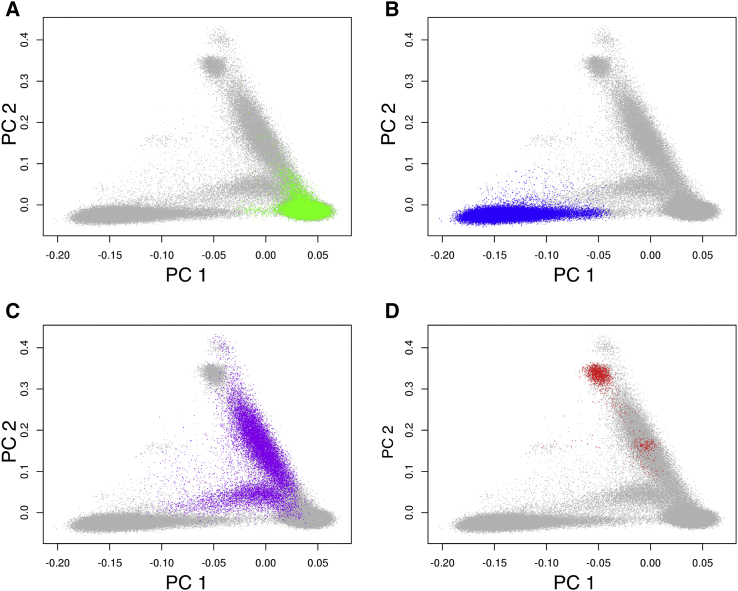
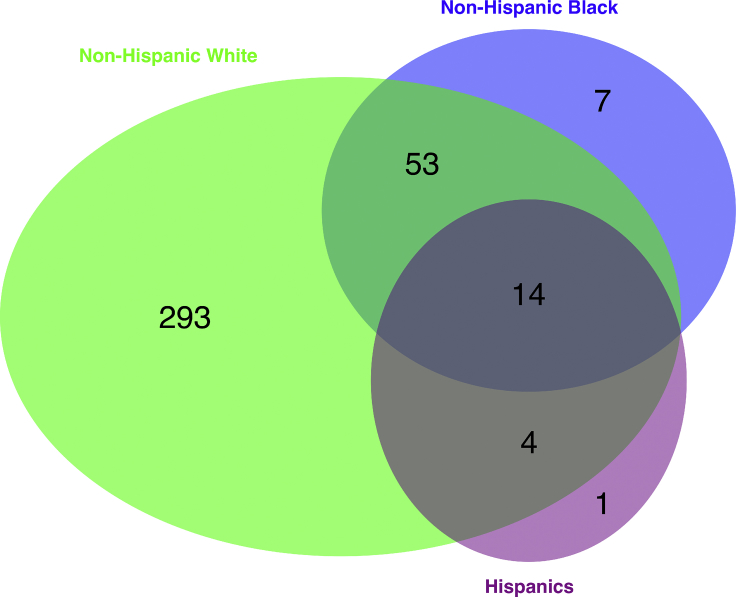
Of 351,820 individuals, all but 6,257 (1.78%) were assigned to one of the four non-overlapping HARE groups: Hispanics, non-Hispanic white, non-Hispanic black, and non-Hispanic Asian (Table 1). Figures 2 and 3 compare GIA and HARE; the interpretation of the genetic PCs is assisted using a model-based admixture analysis, which included Europeans, Africans, Native Americans, East Asians, and South Asians as the ancestral individuals (see Material and Methods). The first two PCs, computed using the genotypes of the entire MVP cohort, represented the variation of African (PC1) and Native American/Asian (PC2) ancestry. As expected, the ancestries of individuals in the non-Hispanic black group varied along PC1 that described the difference among European ancestry and African ancestry (Figures 2B, 3B, and S3B). Likewise, Hispanic individuals showed varying proportions of European, African, and Native American ancestry (Figures 2C, 3C, and S3C). The non-Hispanic Asian group consisted of two components, corresponding to the East and South Asian populations, respectively, according to the admixture analysis (Figures 2D, 3D, and S3D). Interestingly, European admixture (greater than 20%) were inferred in 12% (n = 364) of the individuals in the HARE non-Hispanic Asian group. Among this group, 46% (n = 166) individuals had “Asian” as the only SIRE information; an additional 25% (n = 91) indicated both Asian and European ancestries. This likely reflected recent admixture between Asian Americans and European Americans. Although it would have been feasible to train the support vector machine to learn East Asian and South Asian as two separate HARE categories, we chose to group them into one stratum because the statistical power of subsequent genetic association analysis would likely be low in this group due to relatively small sample size (n = 3,054).
Table 1.
Comparison between HARE and SIRE among 351,820 MVP Participants
| HARE |
SIRE |
|||||
|---|---|---|---|---|---|---|
| Non-Hispanic White | Non-Hispanic Black | Hispanics | Non-Hispanic Asian | Missing | Total | |
| Non-Hispanic White | 163,267 | 0 | 0 | 0 | 85,240 | 248,507 |
| Non-Hispanic Black | 0 | 25,830 | 0 | 0 | 42,325 | 68,155 |
| Hispanics | 0 | 0 | 10,306 | 0 | 15,541 | 25,847 |
| Non-Hispanic Asian | 0 | 0 | 0 | 1,449 | 1,605 | 3,054 |
| Missing | 400 | 110 | 546 | 23 | 5,178 | 6,257 |
| Total | 163,667 | 25,940 | 10,852 | 1,472 | 149,889 | 351,820 |
Figure 2.
The First Two Principal Components of Genetically Inferred Ancestry and HARE Assignments for Individuals, whose SIRE Is Non-missing and Consistent across Records
Colored points represent individuals whose HARE agrees with SIRE. Black points highlight individuals whose genetically inferred ancestry strongly disagrees with SIRE; subsequently HARE for these individuals is set to missing. All other MVP participants are denoted in gray. The gold triangle indicates a hypothetical individual whose HARE could be non-Hispanic European, Hispanic, or missing, depending on her SIRE. Shown are non-Hispanic white (A), non-Hispanic black (B), Hispanic (C), and non-Hispanic Asian (D).
Figure 3.
The First Two Principal Components of Genetically Inferred Ancestry and HARE Assignments for Individuals, whose SIRE Is Missing or Inconsistent across Records
Colored points represent individuals, whose HARE is assigned to one of the strata. Shown are non-Hispanic white (A), non-Hispanic black (B), Hispanic (C), and non-Hispanic Asian (D).
Among nearly 202,000 individuals with SIRE, 1,079 (0.53%) had GIA strongly indicating a different racial/ethnic group. These individuals are highlighted in Figure 2 (black points) and appear as genetic outliers compared to others with the same SIRE (colored points). As it was not possible to resolve the source of the discrepancy, we set HARE of these individuals to missing. Among the nearly 150,000 individuals whose SIRE was missing or not used in the training procedure, 144,711 (96.55%) were assigned into one of four HARE groups. It should be emphasized that the assignment of HARE depended on both the individual’s GIA and whether the individual has known SIRE. Consider a hypothetical individual with PC coordinate marked by the triangle on Figure 2. The ancestry of this individual was estimated to derive 62.8%, 12.9%, 15.7%, 2.4%, and 6.1% from European, African, Native American, East Asian, and South Asian ancestral populations, respectively. The predicted most likely stratum by the support vector machine model was European, followed by Hispanic. Suppose the individual’s SIRE was European, HARE would be non-Hispanic white because the prediction agrees with SIRE . If, on the other hand, the SIRE of this individual were Hispanic, the probability ratio between European and Hispanic would not be large enough to reject SIRE ; hence the HARE of this individual would be assigned as Hispanic. Finally, if the individual’s SIRE were missing, HARE would remain missing because the most likely stratum (European) and the second most likely stratum (Hispanic) would be too close to call . A consequence of the asymmetric decision is that a comparison between Figures 2 and 3 indicates that the HARE clusters representing the four major groups are slightly more diffused among individuals with SIRE (Figure 2) compared to the corresponding clusters among individuals with missing SIRE (Figure 3).
To assess its statistical accuracy, we applied the support vector machine model to an independent “test cohort,” consisting of 27,974 veteran participants, whose SIRE was known and whose genotype became available in a second stage. Genotypes of these individuals were not used in any part of model training. Because the SIRE of these individuals was known, their HARE followed the left branch of the decision tree in Figure 1. In total, 0.4% (117/27,974) outliers (strong disagreement between genetic ancestry and SIRE) were detected, comparable to the outlier proportion observed in the main cohort (1,079/201,931) (Tables 1 and S1). Alternatively, assuming the SIRE information were unknown, we defined pseudo-HARE for these individuals following the right branch of the decision tree. Considering SIRE as the gold standard strata assignment, the off diagonal in Table S2 provides an estimate of prediction error (0.46%); however, 116 of the 130 “mis-classification” were among the outliers detected in Table S1. Hence, we estimated a statistical “error” rate between 0.05% and 0.46%.
Detecting Population-Specific Trait Variants
To gain an intuition about the difference between mega-analysis GWASs and stratified GWASs for identifying ethnicity-specific trait variant, we first considered a simple model that permitted analytic comparison. Let SNP G be the true functional variant influencing a trait, Y, where G is only polymorphic in a minority population. A common scenario is that G is not genotyped, but instead association is tested between the phenotype and a nearby tagging SNP, M. The stratified approach tests the association at M in the minority population using the linear model, . Alternatively, mega-analysis tests the model, , where Z adjusts for population structure. By analytically deriving the distribution of the test statistics for each model, we identified factors that determined the relative efficiency of the two approaches (Supplemental Material and Methods). As expected, the advantage of the stratified analysis over mega-analysis was more apparent when the LD between M and G was high or the minor allele frequency of M in the non-minority cohort was high, because the presence of both alleles at M added noise. In addition, we showed that the stratified analysis tended to outperform mega-analysis when the heritability attributable to the causal variant, h2, was low or the representation of the specific minority population, as a fraction of the entire cohort, was low. These observations are important: first, the genetic architectures of complex traits and diseases are likely to be highly polygenic, and thus the contribution of any single variant to the overall phenotypic variation is expected to be low; second, in population cohorts that are recruited without over-sampling a specific race/ethnicity, the representation of minority populations is expected to be low. In other words, the analytic consideration suggested that mega-analysis may lose substantial power in identifying ethnicity-specific trait variants in realistic settings.
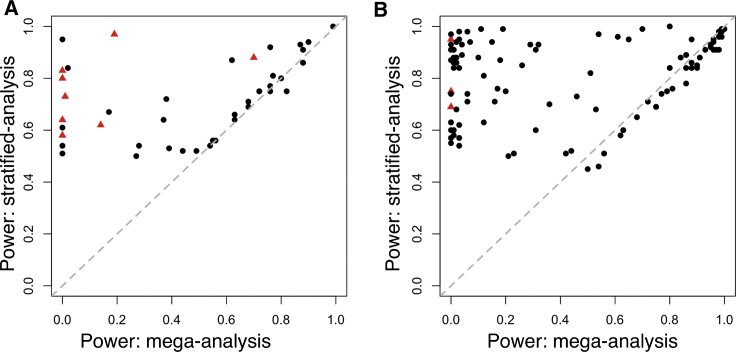
In reality, the power of detecting an ethnicity-specific locus depends on the linkage disequilibrium (LD) between the causal variant and its best tagging-SNP in the neighborhood, and the covariates, Z, take on continuous values representing multi-dimensional ancestry variation. To compare mega-analysis and ethnicity-specific analysis in a more realistic setting, we performed simulation using real genotype data, which preserved realistic LD patterns and tag SNP allele frequencies near the causal variants. Because the power to detect the un-genotyped causal variant depended on the LD pattern around the causal variant, which varied widely across the sampled causal variants, power also varied broadly at any given level of h2 (Figure S4). Figure 4 compares the power of the stratified and mega-analysis by choosing the smallest h2 value, at which at least one of the two models reached a power of 50%. For rare variants (MAF ≤ 0.01), 36 variants were detected at a higher power using stratified analysis, 3 variants were detected at a slightly higher power under the mega-analysis, and 3 variants had equal power (in 100 simulations). The remaining variants had an observed detection rate of 0 or 1, for both approaches and in all the values of h2 considered (0.0001–0.01). Thus, consistent with the analytic derivation, for rare causal variants, minority-stratified analysis almost always outperforms mega-analysis, the gain can be substantial, and the trend is consistent for the range of h2 considered (Figures S4). Furthermore, even for causal variants that were relatively common in minority populations and not completely monomorphic in Europeans, stratified analysis was never much worse, but could be considerably more powerful, compared to the mega-analysis (Figure 4B).
Figure 4.
Simulation Results Comparing Statistical Power for Detecting Minority-Specific Causal Variants using Mega-analysis (x axis) versus Stratified Analysis (y axis)
Black dots indicate causal SNPs predominantly occurring in non-Hispanic blacks; red triangles indicate causal SNPs predominantly occurring in Hispanics. Shown are (A) rare variants with MAF ≤ 0.01; (B) common variants with MAF ≥ 0.1. Causal SNPs detected by both methods with power of 0 or 1 are omitted. Comparison of power for fixed levels of genetic variance explained by the causal variants can be found in Figure S4.
GWAS for Height in MVP
A total of 372 distinct loci reached genome-wide significance for height in one of the HARE groups; as expected, the number of significant loci was positively related to the sample size28 (Figure 5). Of these, 21 loci were found in exactly one HARE group and would have been missed in the mega-analysis of the entire MVP cohort (Table S3). Nineteen of these loci were found in the non-Hispanic white group; at a Bonferroni corrected level of 0.05/19 = 0.002, all loci were replicated by the UK Biobank (UKB).26, 29 It was more challenging to validate the remaining two loci that were only significant in MVP non-Hispanic black and not in the mega-analysis due to the unavailability of large-scale minority cohorts. The index SNP (rs1560489) at the locus on chromosome 4 was replicated in an African American cohort of the Women’s Health Initiative SNP Health Association Resource (WHI-SHARe, p = 1.07 × 10−3, N = 8,149),27 whereas the locus on chromosome 2 did not show evidence of association in that study (p = 0.23). Curiously, the index SNP (rs6719889) at this locus reached a suggestive association in the UKB with the allelic effect in the same direction (p = 8.37 × 10−6); as the region has not been previously implicated for human height, it is a candidate region for future studies. Meta-analysis across the four HARE strata yielded 63 additional loci, bringing the total to 416 genome-wide significant loci; the corresponding number of genome-wide significant loci, using mega-analysis, is 410. Taken together, these results demonstrate that HARE-stratified analysis yielded ethnicity-informative association findings, which could be meaningfully meta-analyzed with other multi-ethnic cohorts; this kind of meta-analysis could not be done if each multi-ethnic cohort simply performed mega-analysis because the ethnicity representation may vary substantially across cohorts.
Figure 5.
Number of Genome-wide Significant Height Loci in Each HARE Group
Due to the relatively small sample size of non-Hispanics Asians, no genomic region reached genome-wide significance in this group, and therefore this HARE group is not included. SNPs with p < 5 × 10−8 are considered significant; SNPs within 1 Mb are grouped into a single locus.
Discussion
HARE aims to maximize discoveries and enhance interpretation in multi-ethnic GWAS cohorts. By leveraging self-identified race/ethnicity and genetically inferred ancestry, HARE offers a working definition for partitioning a multi-ethnic cohort into non-overlapping strata, which, in our application to MVP, approximate race/ethnicity that is used to characterize existing GWASs. This definition enables most, if not all, individuals to be included in the GWAS analyses, regardless of whether self-identified race/ethnicity is available. Simulation and height analyses demonstrate that, compared to mega-analysis, HARE-stratified analysis provides added benefits of identifying trait loci that occur predominantly in one stratum. Additionally, HARE-stratified analysis enables discovering and characterization of trait-associated variants with allelic heterogeneity across ethnicities. Taking into consideration such heterogeneity is important for estimating the genetic risk of diseases for minority individuals.30, 31, 32 Furthermore, GWAS results based on a HARE stratum can be incorporated into meta-analyses with other studies of similar ancestry background, thereby improving the power for detecting minority-specific trait variants with low-frequency or moderate allelic effects.
HARE combines genetic ancestry and race/ethnicity information and is motivated by the empirically observed correlation between continental level genetic ancestry and major race/ethnicity. Yet HARE differs from both GIA and SIRE, and it is not intended to replace either. For individuals with unambiguous SIRE, HARE is identical as SIRE with few exceptions. These exceptions may arise simply due to data entry error or it may reflect the distinction between an individual’s social identity and his/her genetic ancestry. In the MVP cohort as well as previous studies, such occurrence is generally rare; in our implementation, HARE becomes missing. Effectively this is equivalent to the current practice of excluding individuals as genetic outliers from ethnicity-specific GWASs. For an individual whose SIRE is ambiguous, either because there is no data or there are inconsistent responses from multiple sources, we use genetic information to identify the stratum that most resembles the individual with respect to genetic ancestry. This, however, should not be considered as predicting an individual’s SIRE, as the ambiguity and the missingness of SIRE may not be random. Furthermore, for studies that examine the effects of social identities in a trait or disease, we recommend restricting the analyses to individuals with SIRE. For this reason, we emphasize that future studies should continue to obtain SIRE information whenever possible.
Likewise, HARE differs from the often-adopted population genetic structure approaches used to study human demographic history, which model a number of ancestral, reproductively isolated, populations; genomes of a contemporary individual can be attributed entirely to one of the ancestral populations, or as a mixture from several ancestral populations.3, 16, 23, 33 In contrast, HARE strata do not correspond to these ancestral populations. Moreover, each HARE stratum is not genetically homogeneous: the within-HARE genetic structure reflects variation in admixture proportions and/or geographic cline. Therefore, GWASs within each HARE stratum need to account for genetic structure by adjusting PCs, ancestry proportions, or genetic relationship matrix.
Our study focuses on stratified analysis by major race/ethnicity (as defined by the US Census), currently the most commonly adopted stratifying unit used in multi-ethnic GWASs. It is well appreciated that finer-scale structure exists within each race/ethnicity; researchers may wish to focus on strata defined within a race/ethnic group. For example, Conomos et al. aims to perform association studies within the Hispanics by defining strata corresponding to Cuban, Dominican, Puerto Rican, Mexican, Central American, or South American.13 By training a support vector machine classifier based on individuals self-identified as members of these groups, the analytic framework we describe here can be used to assign Hispanic individuals without self-identified information. While from a method point of view this is possible, we offer two pieces of practical advice: first, there is a trade-off between the homogeneity and sample size within each stratum. Therefore, when the primary role of the stratum variable is to assist genetic association studies, it is important to focus on strata that are likely to achieve adequate sample sizes. Second, in a multi-ethnic Biobank cohort, we recommend characterizing population structure in a hierarchical fashion by first defining major race/ethnic strata (HARE). Subsequently each HARE stratum can be further characterized to reflect finer-scale structure, by using PCs computed within the HARE. This approach will not only reduce the computational cost but will also lead to more interpretable stratum definition and reduce statistical uncertainties. We have developed HARE in the context of MVP, in which the proportion of closely related individuals is low and has little influence on the top PCs. If a cohort consists of high proportions of relatives, modified algorithms can be adopted to characterize population structure while reducing the effects of related individuals.34
Lastly, we note that the observations of heterogeneous association across HARE strata, or other population units, does not automatically inform the mechanisms underlying the heterogeneity, which may or may not be genetic. To date, the mechanism that underlies the heterogenous allelic risk of ApoE e4 allele on Alzheimer disease across ethnicities—biological or otherwise—remains poorly understood. This observation also argues against approaches that stratify on the ancestry of genomic segments, because the modifier of the allelic effect may reside elsewhere in the genome or it may not be genetic at all. Despite such uncertainty, the knowledge of heterogeneous allelic effect allows more precise and individualized utilization of genetic information in disease prediction, prevention, or intervention. To probe into the cause of the heterogeneity, much more comprehensive genetic, environmental, and lifestyle risk factors need to be measured and investigated.
Declaration of Interests
S.L.D. has received research grants from the following for-profit organizations in the last three years: AbbVie Inc., Anolinx LLC, Astellas Pharma Inc., AstraZeneca Pharmaceuticals LP, Boehringer Ingelheim International GmbH, Celgene Corporation, Eli Lilly and Company, Genentech Inc., Genomic Health, Inc., Gilead Sciences Inc., GlaxoSmithKline PLC, Innocrin Pharmaceuticals Inc., Janssen Pharmaceuticals, Inc., Kantar Health, Myriad Genetic Laboratories, Inc., Novartis International AG, and PAREXEL International Corporation through the University of Utah or Western Institute for Biomedical Research. S.M.D. has received research grant from RenalytixAI and CytoVas through the University of Pennsylvania.
Acknowledgments
This research is supported by funding from the Department of Veterans Affairs Office of Research and Development, Million Veteran Program Grant I01-BX003362 (PIs P.S.T. and K.-M.C.), I01-BX003340 (PIs P.W.F.W. and K.C.), and I01-BX002641 (PI P.S.T.). Additionally, H.F. and H.T. are supported by US National Institutes of Health grants R01 GM073059 and R35 GM127063 and Y.V.S. is supported by R01 NR013520. This publication does not represent the views of the Department of Veterans Affairs or the United States Government. A list of MVP investigators can be found in Supplemental Acknowledgments.
Published: September 26, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.08.012.
Contributor Information
Yan V. Sun, Email: yvsun@emory.edu.
Hua Tang, Email: huatang@stanford.edu.
the VA Million Veteran Program:
J. Michael Gaziano, Rachel Ramoni, Jim Breeling, Kyong-Mi Chang, Grant Huang, Sumitra Muralidhar, Christopher J. O’Donnell, Philip S. Tsao, Sumitra Muralidhar, Jennifer Moser, Stacey B. Whitbourne, Jessica V. Brewer, John Concato, Stuart Warren, Dean P. Argyres, Brady Stephens, Mary T. Brophy, Donald E. Humphries, Nhan Do, Shahpoor Shayan, Xuan-Mai T. Nguyen, Saiju Pyarajan, Kelly Cho, Elizabeth Hauser, Yan Sun, Hongyu Zhao, Peter Wilson, Rachel McArdle, Louis Dellitalia, John Harley, Jeffrey Whittle, Jean Beckham, John Wells, Salvador Gutierrez, Gretchen Gibson, Laurence Kaminsky, Gerardo Villareal, Scott Kinlay, Junzhe Xu, Mark Hamner, Kathlyn Sue Haddock, Sujata Bhushan, Pran Iruvanti, Michael Godschalk, Zuhair Ballas, Malcolm Buford, Stephen Mastorides, Jon Klein, Nora Ratcliffe, Hermes Florez, Alan Swann, Maureen Murdoch, Peruvemba Sriram, Shing Shing Yeh, Ronald Washburn, Darshana Jhala, Samuel Aguayo, David Cohen, Satish Sharma, John Callaghan, Kris Ann Oursler, Mary Whooley, Sunil Ahuja, Amparo Gutierrez, Ronald Schifman, Jennifer Greco, Michael Rauchman, Richard Servatius, Mary Oehlert, Agnes Wallbom, Ronald Fernando, Timothy Morgan, Todd Stapley, Scott Sherman, Gwenevere Anderson, Elif Sonel, Edward Boyko, Laurence Meyer, Samir Gupta, Joseph Fayad, Adriana Hung, Jack Lichy, Robin Hurley, Brooks Robey, and Robert Striker
Web Resources
Supplemental Data
References
- 1.Gaziano J.M., Concato J., Brophy M., Fiore L., Pyarajan S., Breeling J., Whitbourne S., Deen J., Shannon C., Humphries D. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 2.Falush D., Stephens M., Pritchard J.K. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang H., Peng J., Wang P., Risch N.J. Estimation of individual admixture: analytical and study design considerations. Genet. Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 4.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 5.Coram M.A., Duan Q., Hoffmann T.J., Thornton T., Knowles J.W., Johnson N.A., Ochs-Balcom H.M., Donlon T.A., Martin L.W., Eaton C.B. Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am. J. Hum. Genet. 2013;92:904–916. doi: 10.1016/j.ajhg.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrer L.A., Cupples L.A., Haines J.L., Hyman B., Kukull W.A., Mayeux R., Myers R.H., Pericak-Vance M.A., Risch N., van Duijn C.M., APOE and Alzheimer Disease Meta Analysis Consortium Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 7.Liu M., Bian C., Zhang J., Wen F. Apolipoprotein E gene polymorphism and Alzheimer’s disease in Chinese population: a meta-analysis. Sci. Rep. 2014;4:4383. doi: 10.1038/srep04383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris A.P. Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol. 2011;35:809–822. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Rooij F.J.A., Qayyum R., Smith A.V., Zhou Y., Trompet S., Tanaka T., Keller M.F., Chang L.C., Schmidt H., Yang M.L., BioBank Japan Project Genome-wide Trans-ethnic Meta-analysis Identifies Seven Genetic Loci Influencing Erythrocyte Traits and a Role for RBPMS in Erythropoiesis. Am. J. Hum. Genet. 2017;100:51–63. doi: 10.1016/j.ajhg.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burchard E.G., Ziv E., Coyle N., Gomez S.L., Tang H., Karter A.J., Mountain J.L., Pérez-Stable E.J., Sheppard D., Risch N. The importance of race and ethnic background in biomedical research and clinical practice. N. Engl. J. Med. 2003;348:1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- 12.Risch N., Burchard E., Ziv E., Tang H. Categorization of humans in biomedical research: genes, race and disease. Genome Biol. 2002;3:t2007. doi: 10.1186/gb-2002-3-7-comment2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conomos M.P., Laurie C.A., Stilp A.M., Gogarten S.M., McHugh C.P., Nelson S.C., Sofer T., Fernández-Rhodes L., Justice A.E., Graff M. Genetic Diversity and Association Studies in US Hispanic/Latino Populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet. 2016;98:165–184. doi: 10.1016/j.ajhg.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg N.A., Pritchard J.K., Weber J.L., Cann H.M., Kidd K.K., Zhivotovsky L.A., Feldman M.W. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 15.Tang H., Quertermous T., Rodriguez B., Kardia S.L., Zhu X., Brown A., Pankow J.S., Province M.A., Hunt S.C., Boerwinkle E. Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. Am. J. Hum. Genet. 2005;76:268–275. doi: 10.1086/427888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Myers R.M. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 17.Klarin D., Damrauer S.M., Cho K., Sun Y.V., Teslovich T.M., Honerlaw J., Gagnon D.R., DuVall S.L., Li J., Peloso G.M., Global Lipids Genetics Consortium. Myocardial Infarction Genetics (MIGen) Consortium. Geisinger-Regeneron DiscovEHR Collaboration. VA Million Veteran Program Genetics of blood lipids among ∼300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 2018;50:1514–1523. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banda Y., Kvale M.N., Hoffmann T.J., Hesselson S.E., Ranatunga D., Tang H., Sabatti C., Croen L.A., Dispensa B.P., Henderson M. Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the genetic epidemiology research on adult health and aging (GERA) cohort. Genetics. 2015;200:1285–1295. doi: 10.1534/genetics.115.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryc K., Durand E.Y., Macpherson J.M., Reich D., Mountain J.L. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet. 2015;96:37–53. doi: 10.1016/j.ajhg.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu T.-F., Lin C.-J., Weng R.C. Probability Estimates for Multi-class Classification by Pairwise Coupling. J. Mach. Learn. Res. 2004;5:975–1005. [Google Scholar]
- 21.Abraham G., Qiu Y., Inouye M. FlashPCA2: principal component analysis of Biobank-scale genotype datasets. Bioinformatics. 2017;33:2776–2778. doi: 10.1093/bioinformatics/btx299. [DOI] [PubMed] [Google Scholar]
- 22.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H., 1000 Genomes Project Consortium An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carty C.L., Johnson N.A., Hutter C.M., Reiner A.P., Peters U., Tang H., Kooperberg C. Genome-wide association study of body height in African Americans: the Women’s Health Initiative SNP Health Association Resource (SHARe) Hum. Mol. Genet. 2012;21:711–720. doi: 10.1093/hmg/ddr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wasserstein R.L., Lazar N.A. The ASA Statement on p -Values: Context, Process, and Purpose. Am. Stat. 2016;70:129–133. [Google Scholar]
- 29.Canela-Xandri O., Rawlik K., Tenesa A. An atlas of genetic associations in UK Biobank. Nat. Genet. 2018;50:1593–1599. doi: 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coram M.A., Fang H., Candille S.I., Assimes T.L., Tang H. Leveraging Multi-ethnic Evidence for Risk Assessment of Quantitative Traits in Minority Populations. Am. J. Hum. Genet. 2017;101:218–226. doi: 10.1016/j.ajhg.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scutari M., Mackay I., Balding D. Using Genetic Distance to Infer the Accuracy of Genomic Prediction. PLoS Genet. 2016;12:e1006288. doi: 10.1371/journal.pgen.1006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grinde K.E., Qi Q., Thornton T.A., Liu S., Shadyab A.H., Chan K.H.K., Reiner A.P., Sofer T. Generalizing polygenic risk scores from Europeans to Hispanics/Latinos. Genet. Epidemiol. 2019;43:50–62. doi: 10.1002/gepi.22166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pritchard J.K., Stephens M., Rosenberg N.A., Donnelly P. Association mapping in structured populations. Am. J. Hum. Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conomos M.P., Miller M.B., Thornton T.A. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol. 2015;39:276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.