Abstract
Hemoglobin A1c (HbA1c) is widely used to diagnose diabetes and assess glycemic control in individuals with diabetes. However, nonglycemic determinants, including genetic variation, may influence how accurately HbA1c reflects underlying glycemia. Analyzing the NHLBI Trans-Omics for Precision Medicine (TOPMed) sequence data in 10,338 individuals from five studies and four ancestries (6,158 Europeans, 3,123 African-Americans, 650 Hispanics, and 407 East Asians), we confirmed five regions associated with HbA1c (GCK in Europeans and African-Americans, HK1 in Europeans and Hispanics, FN3K and/or FN3KRP in Europeans, and G6PD in African-Americans and Hispanics) and we identified an African-ancestry-specific low-frequency variant (rs1039215 in HBG2 and HBE1, minor allele frequency (MAF) = 0.03). The most associated G6PD variant (rs1050828-T, p.Val98Met, MAF = 12% in African-Americans, MAF = 2% in Hispanics) lowered HbA1c (−0.88% in hemizygous males, −0.34% in heterozygous females) and explained 23% of HbA1c variance in African-Americans and 4% in Hispanics. Additionally, we identified a rare distinct G6PD coding variant (rs76723693, p.Leu353Pro, MAF = 0.5%; −0.98% in hemizygous males, −0.46% in heterozygous females) and detected significant association with HbA1c when aggregating rare missense variants in G6PD. We observed similar magnitude and direction of effects for rs1039215 (HBG2) and rs76723693 (G6PD) in the two largest TOPMed African American cohorts, and we replicated the rs76723693 association in the UK Biobank African-ancestry participants. These variants in G6PD and HBG2 were monomorphic in the European and Asian samples. African or Hispanic ancestry individuals carrying G6PD variants may be underdiagnosed for diabetes when screened with HbA1c. Thus, assessment of these variants should be considered for incorporation into precision medicine approaches for diabetes diagnosis.
Keywords: hemoglobin A1c, whole-genome sequence association analyses, The Trans-Omics for Precision Medicine (TOPMed) program, multi-ancestry sample
Introduction
Hemoglobin A1c (HbA1c) is a convenient indirect measure of long-term exposure to blood glucose concentrations. HbA1c estimates the proportion of glycated hemoglobin, an irreversible chemical modification of the hemoglobin molecule by blood glucose, in the blood.1 As HbA1c reflects average ambient glycemia over the previous two to three months, the life of an erythrocyte, it is a commonly used as a test to diagnose diabetes (MIM: 125853 and MIM: 222100) and estimate glycemic control in individuals with diabetes. However, non-glycemic variation in HbA1c due to differences in erythrocyte turnover can influence how accurately HbA1c reflects underlying glycemia.2
We previously conducted a trans-ethnic genome-wide association study (GWAS) meta-analysis of HbA1c in 159,940 individuals from four ancestries (European, African, East Asian, and South Asian). We identified 60 common (minor allele frequency [MAF] greater than 5%) genetic variants associated with HbA1c, of which 19 were classified as “glycemic” and 22 as “erythrocytic” based on the probable biological mechanism through which they appeared to influence HbA1c levels.3 Genetic variants affecting HbA1c via erythrocyte biological pathways may lead to diagnostic misclassification of ambient glycemia and thus of diabetes status. HbA1c GWAS have so far focused on genetic variants imputed to HapMap,4 and therefore genetic discovery efforts have been focused on common variants. Low-frequency (0.5% < MAF < 5%) and rare (MAF < 0.5%) genetic variants and their associated impact on the diagnostic accuracy of HbA1c have not been systematically examined but are suspected to occur.5, 6 Further, previous GWAS have shown that the combined effect of HbA1c-related common variants causes differences in HbA1c that were three times greater in individuals of African ancestry compared with those of European ancestry (0.8 [%-units] versus 0.25 [%-units]).3 This relatively large difference in HbA1c was mainly driven by a single African-ancestry-specific missense variant (rs1050828-T, p.Val98Met) in the Glucose-6-phosphatase Dehydrogenase gene (G6PD [MIM: 305900]), which causes G6PD deficiency (MIM: 300908).7, 8 As the genetic architecture of HbA1c appears to differ by ancestry, as does type 2 diabetes risk, it is imperative to understand the genetic basis of HbA1c in different ancestral groups to ensure that large-effect ancestry-specific variants, similar to the G6PD variant, are uncovered.9
We sought to identify common, low frequency, and rare genetic variants (single-nucleotide variants [SNVs] and structural variations) associated with HbA1c through association analyses in diabetes-free individuals from four ancestries by using whole-genome sequencing (WGS) data from the National Institutes of Health (NIH) National Heart, Lung, and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) program. We hypothesized that variants that are in the low frequency spectrum and that have relatively large effects will be detected even with modest sample sizes. To uncover additional distinct erythrocytic variants associated with HbA1c, we performed gene-based tests and fine-mapping by using sequential conditional association analyses of erythrocytic loci that reached genome-wide significance in this study. We then sought replication of genome-wide and sub-genome-wide TOPMed signals in participants from the UK Biobank who had European, African, or Asian ancestry.
Material and Methods
Populations and Participants
We included in our analyses 10,338 TOPMed participants without diabetes from five cohorts: the Old Order Amish study (n = 151), the Atherosclerosis Risk in Communities Study (ARIC, n = 2,415), the Framingham Heart Study (FHS, n = 2,236), the Jackson Heart Study (JHS, n = 2,356), and the Multi-Ethnic Study of Atherosclerosis (MESA, n = 3,180). These participants represent four ancestry groups: Europeans (EA, n = 6,158), African-Americans (AA, n = 3,123), Hispanics or Latinos (HA, n = 650), and East Asians (AS, n = 407) (Table S1). Descriptions of each cohort are available in the Supplemental Data. Diabetes was defined as fasting glucose (FG) ≥ 7 mmol/L after ≥ 8 h, HbA1c ≥ 6.5%-units, 2 h glucose by an oral glucose tolerance test ≥ 11.1 mmol/L, non-fasting glucose ≥ 11.1 mmol/L, physician-diagnosed diabetes, self-reported diabetes, or use of an antidiabetic medication. Measures of FG and 2 h glucose made in whole blood were corrected to plasma levels using the correction factor of 1.13. Individual studies applied further sample exclusions where applicable, including pregnancy and type 1 diabetes status.
Measurement of HbA1c and Erythrocytic Traits
The National Glycohemoglobin Standardization Program (NGSP) certified assays10 used to measure HbA1c in each cohort are indicated in Table S2. HbA1c was expressed in NGSP %-units. Measurements for red blood cell (RBC) count ( × 1012/L), hemoglobin (HB; g/dL), hematocrit (HCT; %), mean corpuscular volume (MCV; fL), mean corpuscular hemoglobin (MCH; pg), mean corpuscular hemoglobin concentration (MCHC; g/dL), and red blood cell distribution width (RDW; %) were obtained from complete blood count panels performed using standard assays.
Whole-Genome Sequencing
The NHLBI TOPMed program provided WGS, performed at an average depth of 38 × by several sequencing centers (New York Genome Center, Broad Institute of MIT and Harvard, University of Washington Northwest Genomics Center, Illumina Genomic Services, Macrogen Corporation, and Baylor Human Genome Sequencing Center) using DNA from blood. Details regarding the laboratory methods, data processing, and quality control are described on the TOPMed website (see URL in the Web Resources section) and in documents included in each TOPMed accession released on the database of Genotypes and Phenotypes (dbGaP). Processing of whole genome sequences was harmonized across genomic centers using a standard pipeline.11
We used TOPMed freeze 5b that comprised 54,508 samples. We performed variant discovery and genotype calling jointly by using the GotCloud pipeline across TOPMed parent studies for all samples. A support vector machine quality filter was trained using known variants (positive training set) and Mendelian-inconsistent variants (negative training set). The TOPMed data coordinating center performed additional quality control checks for sample identity issues including pedigree errors, sex discrepancies, and genotyping concordance. After site level filtering, TOPMed freeze 5b consisted of ∼438 million SNVs and ∼33 million short insertion-deletion variants. Read mapping was done using the 1000 Genomes Project reference sequence versions for human genome build GRCh38.
The study was approved by the appropriate institutional review boards (IRBs) and informed consent was obtained from all participants.
Statistical Analyses in TOPMed
We first pooled the data across all of the studies, and then we performed pooled WGS association analysis of HbA1c in each ancestry separately using GENetic EStimation and Inference in Structured samples (GENESIS, see URL in the Web Resources section)12 on the Analysis Commons.13 We used linear mixed effect models to test the association of HbA1c with the genetic variants individually while adjusting for sex and age at HbA1c measurement and study, and allowing for heterogeneous variance across study groups. We accounted for relatedness within each ancestry by using an empirical kinship matrix (Genetic Relatedness Matrix [GRM]) calculated globally with Mixed Model Analysis for Pedigrees and Populations (MMAP, see URL in the Web Resources section)14, 15 and using the pooled sample of all TOPMed freeze 5b participants (54k participants), and variants with an MAF ≥ 0.001 (totaling ∼26.5M variants), without linkage disequilibrium (LD) pruning. We then meta-analyzed ancestry-specific results.
We excluded from our single-variant analyses variants with a minor allele count (MAC) of less than 20 across the combined samples. We used a significance threshold of p < 2 × 10−8 to report an association as genome-wide significant for common, low-frequency, and rare genetic variants, which was slightly more stringent than the widely adopted p value threshold of 5 × 10−8 in GWAS, based on estimations for genome-wide significance for WGS studies in UK10K.16, 17 For the X chromosome, genotypes were coded as 0 and 2 for males and 0, 1, and 2 for females, and sex-stratified analyses (analyses conducted separately in males and females) were also performed. We performed additional analyses in AA, adjusted on the sickle cell trait (SCT) variant rs334, as well as haplotype analyses with this variant, using the R package haplo.stats.
Because loci that influence HbA1c through erythrocytic mechanisms are expected to cause nonglycemic variation in HbA1c, we sought to classify HbA1c-associated loci as “glycemic” or “erythrocytic” using mediation analyses on FG, HB, MCV, MCH, or MCHC in TOPMed. If the HbA1c-variant association effect size decreased by more than 25% when we added FG to the regression model, the variant was classified as “glycemic.” If a variant was not glycemic, and if the effect size decreased by more than 25% upon adding HB, MCV, MCH, or MCHC to the regression model, the variant was classified as “erythrocytic.” If association effect sizes were unchanged by or increased after mediation adjustment, then the variant was considered “unclassified.”
If HbA1c-associated variants remained unclassified by the mediation analyses, we used association analysis results with FG to classify them as glycemic (p < 0.005), and association analysis results with erythrocytic traits (HB, MCV, MCH, MCHC, RBC, HCT, or RDW) to classify them as erythrocytic (p < 0.005). Association results for FG were obtained from a TOPMed freeze 5b pooled association analysis performed using a linear mixed effect model in GENESIS, adjusted on age, age squared, sex, body mass index (BMI), and self-reported ancestry (n = 26,883). Association results for erythrocytic traits were obtained from several sources. We first used TOPMed freeze 5b single-variant association analyses performed using a score test in GENESIS and adjusted for age, sex, and study with a GRM while allowing for heterogeneous variance across study groups (n = 25,080). The number of individuals included per study in each analysis is available in Table S3. Analyses were performed at the University of Washington using the TOPMed pipeline. We also used association results from published GWAS in European (UK Biobank and INTERVAL studies; n = 173,480),18 Hispanic (Hispanic Community Health Study and Study of Latinos [HCHS/SOL]; n = 12,502),19 and African American participants (Continental Origins and Genetic Epidemiology [COGENT] Network [n ∼ 16,500]20 and the Candidate gene Association Resource (CARe) Project [n = 7,112]),21 and exome genotyping in 130,273 multi-ethnic individuals.22
For loci classified as erythrocytic, we performed sequential conditional analyses to determine the number of distinct signals in each region. The regions were defined based on LD plots. We used a Bonferroni correction for the number of genetic variants in the region with MAC ≥ 20 to define a signal as distinct. We also performed gene-based and burden tests in GENESIS using a different selection of rare genetic variants (MAF ≤ 1%) based on functional annotations (missense, high confidence loss-of-function, or synonymous variants).
Results from each analysis were combined across ancestries or between males and females by meta-analysis using METAL.23 The heterogeneity test between males and females was calculated using the following formula:
This assessed the difference in effect sizes between males (βmales) and females (βfemales) while accounting for correlation (r) between male and female statistics due to relatedness and was calculated outside of METAL. LD calculations in the TOPMed data and regional plots were done, within ancestry group, using the Omics Analysis, Search and Information System (OASIS, see URL in the Web Resources section). Functional annotations were performed using the WGS Annotator (WGSA).24
Statistical Analyses in the UK Biobank
We used the UK Biobank (UKBB, see URL in the Web Resources section), a prospective cohort study with deep genetic and phenotypic data collected on approximately 500,000 individuals aged between 40 and 69 at recruitment from across the United Kingdom, as an independent sample for external replication of our findings. The centralized analysis of the genetic data, including genotype quality, population structure and relatedness of the genetic data, and efficient phasing and genotype imputation, has been described extensively elsewhere.25 Two similar arrays were used for genotyping (Applied Biosystems UK Biobank Lung Exome Variant Evaluation and UK Biobank Axiom Arrays) and pre-phasing was performed using markers present on both arrays. Phasing on the autosomes was carried out using SHAPEIT3 and 1000 Genomes phase 3 panel to help with the phasing of non-European ancestry samples. Imputations were carried out using the IMPUTE4 program with the Haplotype Reference Consortium (HRC) reference panel or with a merged UK10K and 1000 Genomes phase 3 reference panel. For chromosome X, haplotype estimation and genotype imputation were carried out separately on the pseudo-autosomal and non-pseudo autosomal regions.
We identified UKBB African-ancestry participants using the following three self-reported ethnicities: “Caribbean,” “African,” and “Any other Black background.” We identified UKBB European-ancestry participants using the following three self-reported ethnicities: “White,” “British,” and “Irish.” We identified UKBB Asian-ancestry participants using the “Chinese” self-reported ancestry. We excluded participants with diabetes defined by the use of antidiabetic medication, self-reported physician diagnosis, FG ≥ 7 mmol/L, or non-fasting glucose ≥ 11.1 mmol/L or HbA1c ≥ 6.5%-units. Because African-ancestry UKBB participants were admixed, we generated principal components (PCs) using smartPCA23, 24 (see URL in the Web Resources section) based on 72,300 common genetic variants in low LD selected with PLINK (see URL in the Web Resources section).25 For each PC, ethnic outliers lying more than 6 SD away from the mean were excluded. Additionally, we excluded samples with high heterozygosity and high missing rate, sex aneuploidy, or different genomic and stated sex. We used the R package Scalable and Accurate Implementation of Generalized mixed model (SAIGE, see URL in the Web Resources section)26 and linear mixed effect models to evaluate the association of TOPMed genome-wide and sub-genome-wide signals with HbA1c, adjusting for age and sex, and using a GRM. We also used UKBB exome sequence data available in 50K participants to check the concordance of genotypes with imputed data for rare variants.
Results
HbA1c-Associated Regions Using WGS
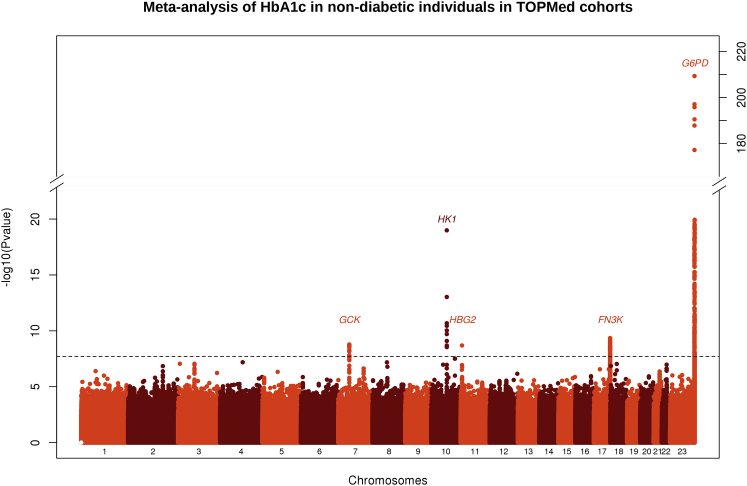
We included in our analyses 10,338 TOPMed participants representing four ancestry groups: EA (n = 6,158), AA (n = 3,123), HA (n = 650), and AS (n = 407; Table S1). TOPMed studies were composed of middle- to older-aged participants of EA, AA, HA, or AS ancestry with comparable mean HbA1c, mean fasting glucose (FG), and mean hemoglobin (HB, Table S4). A total of 13,079,661 variants (EA), 21,443,543 variants (AA), 9,567,498 variants (HA), and 6,567,324 variants (AS) passed filters and were included in the analyses. QQ-plots and Manhattan plots of WGS associations with HbA1c from ancestry-specific analyses and the meta-analysis are provided in Figure 1 and Figures S1 and S2. Using a significance threshold of p < 2 × 10−8 to report an association as genome-wide significant, we detected five regions associated with HbA1c, including one locus that had not been identified in the trans-ethnic GWAS meta-analysis of HbA1c3 (a low-frequency AA-specific variant, rs1039215 in HBG2 [MIM: 142250] and HBE1 [MIM: 142100], MAF = 0.03). Regional plot for the HBG2 and HBE1 locus is provided in Figure S3. The four other SNVs were located in regions previously identified as associated with HbA1c in trans-ethnic meta-analyses3: rs2971670 in GCK (MIM: 138079) on chromosome 7 (Pmeta = 1.7 × 10−9, mainly associated in EA and AA, r2 = 0.59, D′ = 1 with rs4607517, the index SNV in published GWAS), rs17476364 in HK1 (MIM: 142600) on chromosome 10 (Pmeta = 3.1 × 10−21, mainly associated in EA and HA, r2 = 0.10, D′ = 1 with rs10823343, r2 = 0.18, D′ = 0.50 with rs4745982, index SNVs in published GWAS), rs113373052 in FN3K (MIM: 608425) on chromosome 17 (Pmeta = 4.5 × 10−10, associated in EA, r2 = 0.92, D′ = 0.99, with rs1046896, index SNV in published GWAS), and rs1050828 in G6PD on chromosome X (Pmeta = 5.1 × 10−210, associated in AA and HA, monomorphic in the other ancestries).3 The top SNV or structural variant in each region detected at the genome-wide threshold (p < 2 × 10−8) is indicated in Table 1, and the top one detected at the sub-genome-wide threshold (p < 5 × 10−7) is indicated in Table S5. Genetic association results (summary statistics) are available in the Type 2 Diabetes Knowledge Portal (see URL in the Web Resources section).
Figure 1.
Manhattan Plot of the Meta-Analysis of HbA1c in Non-Diabetic Individuals in TOPMed Cohorts
The –log10 (p value) for each single-nucleotide variant on the y axis is plotted against the build 38 genomic position on the x axis (chromosomal coordinate). The dashed horizontal line indicates the genome-wide significance threshold of p = 2 × 10−8. The y axis was truncated for ease of interpretation.
Table 1.
Most Associated Single Nucleotide Variants in the Regions Detected at the Genome-Wide Level (p < 2 × 10−8) by Ancestry and in the Meta-Analysis of HbA1c in Non-Diabetic Individuals in TOPMed Cohorts
|
- |
- |
- |
- |
Meta-Analysis |
Europeans (n = 6,158) |
African-Americans (n = 3,123) |
Hispanics (n = 650) |
Asians (n = 407) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MarkerIDa | rsID | Closest Gene | Ref/Alt Allele | Beta | SE | p Value | AAFb | Beta | SE | p Value | AAFb | Beta | SE | p Value | AAFb | Beta | SE | p Value | AAFb | Beta | SE | p Value |
| 7:44186502 | rs2971670 | GCK | C/T | 0.04 | 0.01 | 1.7E-09 | 0.18 | 0.03 | 0.01 | 1.6E-05 | 0.18 | 0.06 | 0.01 | 1.6E-04 | 0.21 | 0.05 | 0.03 | 0.04 | 0.19 | 0.04 | 0.03 | 0.20 |
| 10:69334748 | rs17476364 | HK1 | T/C | −0.09 | 0.01 | 3.1E-21 | 0.10 | −0.08 | 0.01 | 3.2E-18 | 0.02 | −0.09 | 0.04 | 0.04 | 0.06 | −0.15 | 0.04 | 5.4E-04 | - | - | - | - |
| 11:5464062 | rs1039215 | HBG2 and HBE1 | A/G | −0.21 | 0.04 | 2.0E-09 | - | - | - | - | 0.03 | −0.21 | 0.04 | 2.0E-09 | - | - | - | - | - | - | - | - |
| 17:82739582 | rs113373052 | FN3K | C/T | 0.03 | 0.01 | 4.5E-10 | 0.32 | 0.03 | 0.01 | 4.0E-08 | 0.29 | 0.02 | 0.01 | 0.06 | 0.39 | 0.03 | 0.02 | 0.15 | 0.49 | 0.05 | 0.02 | 0.04 |
| X:154536002 | rs1050828 | G6PD | C/T | −0.41 | 0.01 | 5.1E-210 | - | - | - | - | 0.12 | −0.41 | 0.01 | 8.4E-205 | 0.02 | −0.37 | 0.07 | 8.3E-07 | - | - | - | - |
MarkerID is defined as Chromosome: Position. Positions are indicated on Build 38.
AAF: effect (alternate) allele frequency
Classification of HbA1c-Associated Loci Based on Their Biological Pathways
Because loci that influence HbA1c through erythrocytic pathways are expected to cause nonglycemic variation in HbA1c, we sought to classify HbA1c-associated loci as glycemic or erythrocytic through the use of mediation analyses in TOPMed and association analyses with glycemic and erythrocytic traits in TOPMed or using results from the literature. Mediation analyses and look-up in WGS analysis of FG in TOPMed classified the GCK variants as glycemic (Tables S6–S8). Association analyses with erythrocytic traits in published GWAS and in TOPMed showed that the rs17476364-C allele, in the HK1 gene, was positively associated with HB, MCV, MCH, MCHC, red blood cell count (RBC), HCT, and RDW in Europeans18 and that the rs1050828-T allele, in the G6PD gene, was positively associated with MCH in African-Americans and MCV in African-Americans and Hispanics or Latinos and negatively associated with HCT and HB in African-Americans and RBC and RDW in African-Americans and Hispanics or Latinos19, 20, 21 (Tables S6, S8, and S9). Results of the mediation and association analyses in TOPMed are available in Tables S6 and S8 for the genome-wide variants and in Tables S7 and S10 for the sub-genome variants. Association results with erythrocytic traits from published GWAS are available in Table S9. Among the 26 variants meeting the sub-genome-wide threshold, four were common and 22 were low frequency or rare. Two of the common HbA1c-associated variants were reported previously:3 the glycemic variant at G6PC2 (MIM: 608058) and the erythrocytic variant at TMPRSS6 (MIM: 609862). Among the genome-wide significant loci, HK1 (EA) and G6PD (AA and HA) were classified as erythrocytic, as was the low-frequency variant rs1039215 in HGB2 and HBE1, which was negatively associated with HB, HCT, MCV, and MCH. The significant FN3K variant was unclassified.
Characterization of Distinct Signals at Erythrocytic HbA1c-Associated Loci
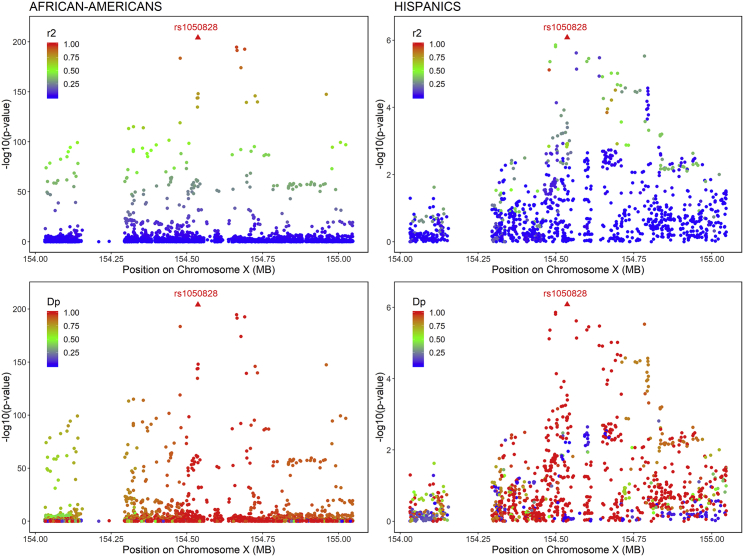
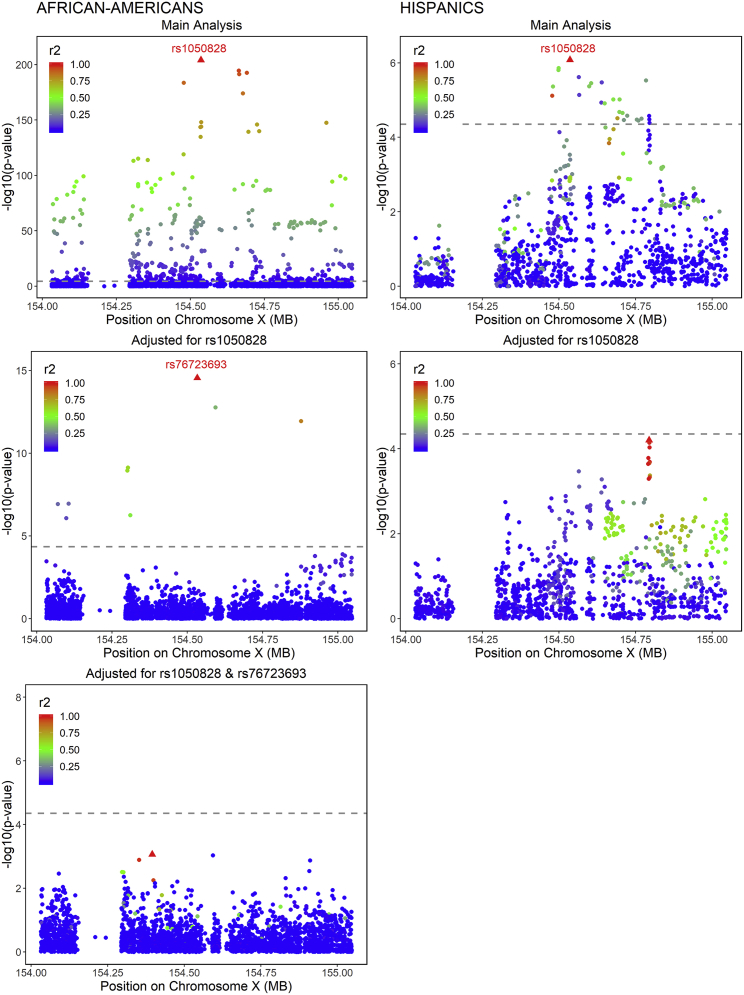
For loci classified as erythrocytic (HK1, HBG2 and HBE1 and G6PD), we performed sequential conditional analyses to detect distinct signals in addition to the top SNV. The regions were defined, based on LD plots, as ± 60kb region around HK1, +/− 250kb region around HBG2 and HBE1, and ± 500kb region around G6PD (Figures S3–S5). We did not detect secondary associations in the HK1 and HBG2 and HBE1 regions at a threshold of 4.3 × 10−5 and 8.5 × 10−6 respectively. In the G6PD region, the top SNV (rs1050828, p.Val98Met, T-allele frequency 12% in AA, 2% in HA, and 0% in EA and AS) was associated with lower HbA1c in both AA and HA. This SNV accounted for 23% of HbA1c variance in AA and 4% in HA. The LD in the G6PD region is complex, with a strong haplotype effect in AA and HA (Figure 2). By performing conditional analyses on the top SNV (rs1050828), we were able to detect an additional rare signal (G-allele frequency 0.5%) in AA (rs76723693, p.Leu353Pro, Bcond = −0.50, Pcond = 2.8 × 10−15) that was distinct from rs1050828 (r2 = 0.0006, D′ = 1, Figure 3). The threshold to detect an additional signal was fixed to 1.7 × 10−5 in AA and 4.5 × 10−5 in HA. The three other SNVs located in the same region that were sub-genome-wide significant in the main analysis (rs143745197, rs184539426, and rs189305788) were not significant after adjusting for both rs1050828 and rs76723693. We detected significant or suggestive associations by using gene-based (p = 9.7 × 10−11) and burden tests (p = 4.7 × 10−5) when aggregating 15 missense rare variants with MAF ≤ 1% in G6PD (Table S11). Because association from SKAT was more significant than the association of rs76723693 from single-SNV association analysis, it suggested that several rare missense variants in G6PD were associated with HbA1c. We performed additional association and conditional analyses in G6PD for missense variants with 10 < MAC < 20. In addition to rs76723693, one rare missense variant (rs5030872, MAF = 0.002) was suggestively associated with HbA1c (B = −0.64, p = 3.4 × 10−6) and became significantly associated with HbA1c when we adjusted for both rs1050828 and rs76723693 (Bcond = −0.71, Pcond = 1.3 × 10−9).
Figure 2.
Regional HbA1c Association Plots in the G6PD Region in Non-Diabetic African-Americans and Hispanics in TOPMed Cohorts
Association plots are displayed separately in African-Americans (left side) and in Hispanics (right side). Single-nucleotide variants are plotted with their p values (-log10 values, y axis) as a function of build 38 genomic position on chromosome X (x axis). The local linkage disequilibrium (LD) structure (r2—top or D′—bottom) between the top associated single-nucleotide variant, rs1050828 (red triangle), and correlated proxies is indicated in color according to a blue-to-red scale from r2/D′ = 0 to 1, and this local LD structure was calculated separately in non-diabetic African-American and Hispanic TOPMed cohorts.
Figure 3.
Sequential Conditional Analyses in the G6PD Region in Non-Diabetic African-Americans and Hispanics in TOPMed Cohorts
Association plots are displayed separately in African-Americans (left side) and in Hispanics (right side). Single-nucleotide variants are plotted with their p values (-log10 values, y axis) as a function of build 38 genomic position on chromosome X (x axis). The local linkage disequilibrium (LD) structure (r2) between the top associated single-nucleotide variant (red triangle) and correlated proxies is indicated in color according to a blue to red scale from r2 = 0 to 1 and was calculated separately in non-diabetic African-American and in Hispanic TOPMed cohorts. Significance thresholds to declare an association signal as distinct are indicated by a dashed grey line (p = 1.7 × 10−5 in African-Americans and p = 4.5 × 10−5 in Hispanics, based on the number of single-nucleotide variants in the region with a minor allele count greater than 20).
The HBG2 and HBE1 variant rs1039215 (p = 2.0 × 10−9) is located at 327 kb from the SCT variant rs334 (p = 2.9 × 10−8). A recent paper reported rs1039215 to be in LD (r2 > 0.2) with the SCT variant (rs334, r2 = 0.24, D′ = −0.83) and correlated with HBG2 gene expression in whole blood.27 To determine whether rs1039215 was distinct from rs334, we calculated LD and performed conditional analyses. In TOPMed, rs1039215 was in modest LD with rs334 (r2 = 0.30, D′ = 0.88, Figure S3), and conditioning on rs334 attenuated its association (decrease in p value from 2.0 × 10−9 to 2.0 × 10−3, Table S12). These results suggest that the association of rs1039215 with HbA1c may be explained by the SCT rs334 variant. Nevertheless, the haplotype rs334-A and rs1039215-G was more significantly associated with HbA1c than were the haplotypes rs334-A and rs1039215-A and rs334-T and rs1039215-G (Table S13). Thus, the association of the variant rs1039215 in HBG2 and HBE1 could be partially distinct from the known association at the SCT variant. In our AA sample, 12 individuals each carried two copies of the rs1050828-T allele and at least one copy of the rs1039215-G allele and had a mean HbA1c of 4.60 (0.41) versus 5.65 (0.39) for the 2,372 individuals who carried no risk allele at the two variants (Table S14).
HbA1c-Lowering Effects of G6PD Variants in AA and HA
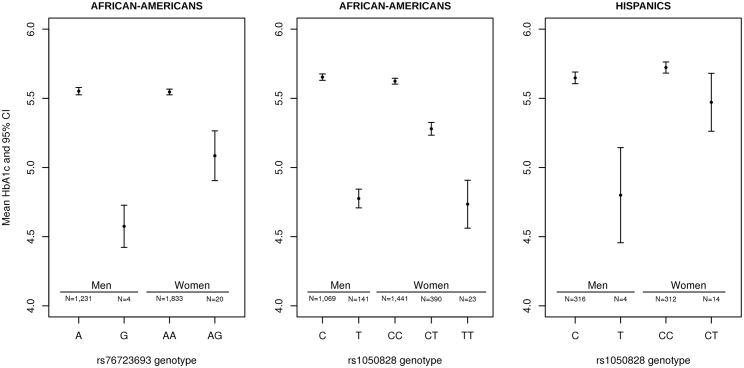
In sex-stratified analyses in AA, the G6PD rs76723693 variant was associated with a decrease in HbA1c of 0.98%-units (95% CI 0.51–1.44) per allele in hemizygous men and 0.46%-units (95% CI 0.26–0.66) in heterozygous women, whereas the G6PD rs1050828 variant was associated with a decrease in HbA1c of 0.88%-units (95% CI 0.81–0.95) per allele in hemizygous men and 0.34%-units (95% CI 0.30–0.39) in heterozygous women (Table 2 and Figure 4). We detected heterogeneity in effect sizes between males and females for rs1050828 (Phet = 0.008) but not for rs76723693 (Phet = 0.19). In HA, the G6PD rs1050828 variant was associated with a decrease in HbA1c of 0.84%-units (95% CI 0.47–1.22) per allele in hemizygous men and 0.25%-units (95% CI 0.06–0.45) in heterozygous women (Table 3 and Figure 4). Upon adjusting on the SCT variant, rs334, we found that the association of these two SNVs did not attenuate (Table S12).
Table 2.
Combined and Sex-Stratified Conditional Association Analyses of HbA1c for the Two Distinct Single Nucleotide Variants in the G6PD Region in Non-Diabetic African-Americans in TOPMed Cohorts
| - |
- |
- |
- |
Pooled Analysis (n = 3,123) |
Conditional Analysis on rs1050828 |
Males (n = 1,269) |
Females (n = 1,854) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MarkerIDa | rsID | Ref/Alt Allele | AAFb | Beta | SE | p Value | Beta | SE | p Value | Beta | SE | p Value | Beta | SE | p Value | Heterogeneity p Valuec |
| X:154533025 | rs76723693 | A/G | 0.005 | −0.44 | 0.07 | 2.2E-09 | −0.50 | 0.06 | 2.8E-15 | −0.52 | 0.12 | 6.8E-6 | −0.33 | 0.10 | 6.2E-04 | 0.19 |
| X:154536002 | rs1050828 | C/T | 0.12 | −0.41 | 0.01 | 8.4E-205 | - | - | - | −0.44 | 0.02 | 2.3E-148 | −0.37 | 0.02 | 2.7E-69 | 0.008 |
MarkerID is defined as Chromosome: Position. Positions are indicated on Build 38.
AAF: Alternate allele frequency
Heterogeneity p Value: test of difference in effect sizes between males and females, accounting for the correlation between males and females statistics
Figure 4.
Mean HbA1c Levels According to Genotypes at Both G6PD Variants (rs76723693 and rs1050828) and Stratified by Sex
Plots are displayed separately in African-Americans (AA, left and center) and in Hispanics (HA, right). Due to the limited sample size of African-American women carrying two copies of the rare G allele at rs76723693, they are not represented on this plot. In AA, the G6PD rs76723693 variant was associated with a decrease in HbA1c of 0.98%-units (95% confidence interval [CI] 0.51–1.44) per allele in hemizygous men and 0.46%-units (95% CI 0.26–0.66) in heterozygous women; whereas the G6PD rs1050828 variant was associated with a decrease in HbA1c of 0.88%-units (95% CI 0.81–0.95) per allele in hemizygous men and 0.34%-units (95% CI 0.30–0.39) in heterozygous women In HA, the G6PD rs1050828 variant was associated with a decrease in HbA1c of 0.84%-units (95% CI 0.47–1.22) per allele in hemizygous men and 0.25%-units (95% CI 0.06–0.45) in heterozygous women.
Table 3.
Analysis of HbA1c Associations with rs1050828, X:154536002, Alternate Allele T, in Non-Diabetic African-Americans and Hispanics in TOPMed Cohorts
|
- |
African-Americans |
Hispanics |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | N | AAFa | Beta | SE | p Value | % Of Variance Explained | n | AAFa | Beta | SE | p Value | % of Variance Explained |
| All | 3,123 | 0.12 | −0.41 | 0.01 | 8.4E-205 | 0.23 | 650 | 0.02 | −0.37 | 0.07 | 8.3E-07 | 0.04 |
| Males | 1,269 | 0.12 | −0.44 | 0.02 | 2.3E-148 | 0.35 | 323 | 0.01 | −0.45 | 0.11 | 3.5E-05 | 0.05 |
| Females | 1,854 | 0.12 | −0.37 | 0.02 | 2.7E-69 | 0.14 | 327 | 0.02 | −0.28 | 0.10 | 4.5E-03 | 0.02 |
AAF: alternate allele frequency
Population Reclassification of Diabetes Diagnosis Based on G6PD Variants
Despite the lower diagnostic sensitivity of HbA1c28, 29 compared to FG or 2 h glucose in an oral glucose tolerance test, diabetes screening using only HbA1c is not uncommon in clinical practice due to its lower intra-individual variability and the convenience of performing the test in the non-fasting state. To assess the potential public health impact of G6PD variants in AA, we designed a hypothetical scenario, using National Health and Nutrition Examination Survey (NHANES, see URL in the Web Resources section) 2015–2016, and estimated the number of African Americans and Hispanics whose diagnosis of diabetes would have been missed if they had been screened by a single HbA1c measurement without confirmatory testing by direct glucose measurements. We assumed that the effects of the two G6PD variants rs1050828 and rs76723693 were not accounted for, and the standard diagnostic threshold of ≥6.5% was uniformly applied to the entire population. We restricted the NHANES analytic sample to adults aged ≥18 years and who self-identified as “Non-Hispanic Black” or “Mexican American/Other Hispanic” respectively. We defined the proportions of individuals carrying the G6PD variants by their allele frequencies and assumed that the “true” HbA1c values of these individuals were higher by their genetic effect sizes from the association analyses.
In non-Hispanic Black individuals (n = 1,227), an estimated 2.32% with HbA1c < 6.5%-units may be considered to have diabetes upon accounting for the effect size and observed allele frequency of the common G6PD variant, rs1050828. An additional 0.13% with HbA1c < 6.5%-units may be considered to have diabetes upon accounting for the effect of the rare G6PD variant, rs76723693. In Mexican American/Other Hispanic individuals (n = 1,768), an additional 0.26% with HbA1c < 6.5%-units may be considered to have diabetes due to the effect of rs1050828. According to the 2016 United States Census Bureau, approximately 30.14 and 39.33 million adults identified themselves as African American and Hispanic adults, respectively; this suggests that 740,000 African American adults, of whom 40,000 would have diabetes attributed to the rare variant, rs76723693, and 100,000 Hispanic adults with diabetes would remain undiagnosed when screened by a single HbA1c measurement if this genetic information was not taken into account (Tables S15 and S16).
Replication of Results in the UKBB
We selected a total of 5,311 African-ancestry, 398,122 European-ancestry, and 1,331 Asian-ancestry non-diabetic UKBB participants for replication. In the African-ancestry sample, 2,247 participants were males, mean age (SD) was 51.2 (7.8), and mean HbA1c (SD) was 5.5 (0.5). In the European-ancestry sample, 179,055 participants were males, mean age (SD) was 56.6 (8.0), and mean HbA1c (SD) was 5.3 (0.3). In the Asian-ancestry sample, 490 participants were males, mean age (SD) was 52.1 (7.6), and the mean HbA1c (SD) was 5.5 (0.4) (see Supplemental Data).
The four variants of interest (rs334 [SCT], rs1039215 [HBG2], rs76723693, and rs1050828 [G6PD]) had a good quality of imputation (info score > 0.50, Table 4). We observed significant and consistent associations of the two G6PD variants with HbA1c (rs1050828-T, B = −0.38, p < 2.2 × 10−16 and rs76723693-G, B = −0.20, p = 0.008, Table 4). The association of the rare G6PD rs76723693-G variant was even more significant when both G6PD variants were in the model (B = −0.25, p = 5.1 × 10−5). We observed a high concordance between exome sequence data and imputed genotypes for the rare G6PD rs76723693 SNV. Only seven individuals among 49,741 had a discordance of genotypes between imputed and exome sequence data, using dosage ranges of [0–0.2], [0.9–1.1], and [1.8–2.0] to classify AA, GA, and GG genotypes respectively. The additional G6PD variant with 10 < MAC < 20 in TOPMed, rs5030872, was not present in the UKBB. Thus, GWAS, even with large sample sizes, are limited to genetic variants present in imputation reference panels. We did not detect significant associations of HBG2 and SCT variants with HbA1c in the UKBB. Association results for the TOPMed genome-wide and sub-genome-wide associations in UKBB are provided in Tables 4 and S17. In UKBB African-ancestry participants, in addition to G6PD, we replicated the known HbA1c loci GCK and FN3K. We also found nominal associations with HbA1c (p < 0.05) for rs77974976 in CTTNBP21 (B = −0.12, p = 0.02), rs143745197 in ATP2B3 (B = −0.21, p = 2.4 × 10−6), and rs189305788 in F8 (B = −0.18 p = 0.01). The SNV rs143745197 was in LD with our main G6PD signal (B = −0.02, p = 0.64 after conditioning on rs1050828) and the SNV rs189305788 was in LD with our additional distinct G6PD signal (B = −0.03, p = 0.83 after conditioning on rs76723693). We did not replicate the sub-genome-wide signal in XPNPEP1 (B = −0.01, p = 0.95). However, the genetic variant rs551601853 was rarer in the UKBB compared to TOPMed (MAC = 6 in UKBB versus MAC = 20 in TOPMed). In UKBB European-ancestry participants, we replicated the known HbA1c loci: GCK, HK1, FN3K, G6PC2, TMC8, and TMPRSS6. We did not replicate the Asian-specific signal in EMCN.
Table 4.
Association Results of the Main TOPMed Signals in the UK Biobank
| - |
- |
- |
- |
- |
African Ancestry |
European Ancestry |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MarkerIDa | rsID | Closest Gene | Alternate Allele | Imputation Info Score | Alternate Allele Frequency | Beta | SE | p Value | Alternate Allele Frequency | Beta | SE | p Value |
| 7:44186502 | rs2971670 | GCK | T | 0.999 | 0.18 | 0.06 | 0.01 | 2.2E-07 | 0.18 | 0.04 | 0.001 | 7.8E-233 |
| 10:69334748 | rs17476364 | HK1 | C | 0.997 | 0.01 | −0.08 | 0.04 | 5.7E-02 | 0.11 | −0.12 | <1E-04 | <1E-250 |
| 11:5464062 | rs1039215 | HBG2 and HBE1 | G | 0.939 | 0.99 | −0.02 | 0.05 | 6.0E-01 | – | – | – | – |
| 11:5227002 | rs334 | SCT | A | 0.892 | 0.02 | −0.002 | 0.04 | 9.6E-01 | – | – | – | – |
| 17:82739582 | rs113373052 | FN3K | T | 0.998 | 0.27 | 0.03 | 0.01 | 8.7E-03 | 0.31 | 0.03 | 0.001 | 2.2E-214 |
| X:154533025 | rs76723693 | G6PD | G | 0.638 | 0.004 | −0.20 | 0.08 | 7.6E-03 | – | – | – | – |
| X:154536002 | rs1050828 | G6PD | T | 0.920 | 0.16 | −0.38 | 0.01 | 7.3E-310 | – | – | – | – |
MarkerID is defined as Chromosome:Position. Positions are indicated on Build 38.
Discussion
Through association analysis and fine-mapping of HbA1c-associated loci in 10,338 individuals from four different ancestries, we demonstrated how deep large-scale WGS can be used to identify common, low frequency, and rare genetic variants that influence nonglycemic variation in HbA1c in multi-ethnic populations. While fewer loci were uncovered in this WGS study compared to array genotyping in larger studies, WGS did enhance our understanding of the allelic heterogeneity at known HbA1c-associated loci. Moreover, the multi-ethnic panel provided more resolution on the range of alleles and haplotype effects within these loci. We confirmed four known regions associated with HbA1c (the glycemic GCK, erythrocytic HK1 and G6PD, and unclassified FN3K and/or FN3KRP regions) and identified two AA-specific low frequency or rare erythrocytic variants in G6PD (rs76723693) and HBG2 and HBE1 (rs1039215); these variants, to our knowledge, have not been reported in previous trans-ethnic GWAS meta-analyses of HbA1c. The magnitude and direction of effect of the association of rs76723693 and rs1039215 with HbA1c was similar in the two largest TOPMed AA cohorts (JHS and MESA, Table S18). The association of rs76723693 with HbA1c was replicated in UKBB African-ancestry participants. We detected significant association through the use of a gene-based test when aggregating rare missense variants in G6PD; this result indicates that other rare missense variants in G6PD, in addition to rs76723693, are associated with HbA1c. We showed that the association of the HBG2 and HBE1 variant, rs1039215, with HbA1c was partially distinct from the known SCT rs334 variant in HBB (MIM: 141900), and this suggests that genetic variation at hemoglobin genes other than HBB may also influence HbA1c. Individuals carrying both the G6PD variant (rs1050828) and the HBG2 and HBE1 variant (rs1039215) had even lower HbA1c than did those carrying only one of the two variants. Hemizygous males and homozygous females for the G6PD rs1050828-T allele who carried one or more copies of the HBG2 rs1039215-G allele had a mean HbA1c that was 1.05%-unit lower than the mean HbA1c of individuals who carried none of these alleles.
The HbA1c variant in AA, rs1039215 on chromosome 11, lies in an intron of the hemoglobin subunit gamma 2 gene HBG2 and the hemoglobin subunit epsilon 1 gene HBE1. HBG2, in addition to HBG1, encodes the gamma chain of hemoglobin, which combines with two alpha chains to form fetal hemoglobin. Fetal hemoglobin is known to interfere with the measurement of HbA1c by some laboratory assays,30, 31, 32 and so persistence of fetal hemoglobin may be a mechanism through which rs1039215 influences HbA1c measurements. While a small degree of analytic interference with SCT has been reported for the Tosoh 2.2 and G7 assays used by JHS, MESA, and ARIC to measure HbA1c (see NGSP website URL in the Web Resources section),33, 34 no interference has been reported for the Bio-Rad Variant II Turbo assay used by UKB35 (Table S2). Thus, assay interference may explain the lack of association of rs334 with HbA1c in UKBB. Alternatively, as our haplotype analysis indicated that the haplotype containing both rs334-A in HBB and rs1039215-G in HBG2 was likely the main driver of this signal in the region, the true causal variant could be another variant within this haplotype that partially explains the reported association of rs334 with HbA1c.
Both variants identified in G6PD in AA are missense and pathogenic variants for G6PD deficiency (as reported in ClinVar [VCV000037123 and VCV000010388]), an X-linked genetic disorder characterized by a defect in glucose-6-phosphate dehydrogenase enzymatic action; this defect causes erythrocytes to break down prematurely. Thus, the mechanism through which these G6PD variants lower HbA1c is most likely shortening of the erythrocytic lifespan (Table S19). Assuming random X inactivation, we would expect the difference between hemizygous males and males who do not carry the rs1050828-T variant to be two times larger than the difference between heterozygous females and females who do not carry the rs1050828-T variant; however, in this study, we observed a slightly lower than expected effect in females. We posit that this heterogeneity of genetic effects based on sex may be due to X-linked dosage compensation and not actual sex differences.36 While these G6PD variants lower HbA1c in AA, with the same direction of effect as the sickle cell trait variant,5 we note that the lowering effect of these genetic variants on HbA1c does not explain the higher mean HbA1c in AA individuals compared to EA individuals with similar glycemia that has been reported in observational studies.37, 38
We found that rs1050828, the common G6PD variant in AA, was also associated with HbA1c in HA (sample composed of 51% Mexican, 14% Dominican, 14% Puerto Rican, 11% South American, and 10% others) suggesting that carriers of this variant are likely to be underdiagnosed if a single HbA1c measurement is used to screen two of the largest racial and ethnic minorities in the United States for diabetes. If G6PD variants were ignored, we estimate that approximately 840,000 African American and Hispanic adults with diabetes would remain undiagnosed. Because Hispanic populations are ancestrally diverse, and that diversity includes a range of African ancestry (∼7% of HA in our study have > 50% AA ancestry and ∼25% have > 15% AA ancestry), G6PD variants were expected to occur at some lower frequency in HA compared to AA. Given that there is no universal screening for G6PD deficiency in North America, asymptomatic carriers are unlikely to know their G6PD status.7 While misclassification of diabetes status at the population level was largely driven by the common G6PD variant rs1050828 and not the rare rs76723693 variant (found in 0.5% of the AA population), in this era of precision medicine, there is a growing need for clinical practice to account for the impact of rare genetic variants with clinically meaningful effects on routinely performed diagnostic tests even if the proportion of carriers in the population is small. We also predict that other G6PD deficiency variants, particularly common in African, Mediterranean, and Asian populations,39 will impact HbA1c’s utility as a diagnostic measure. Until genetic information is universally accessible to adjust diagnostic thresholds of imperfect biomarkers, our findings support current clinical practice guidelines from the American Diabetes Association which recommends the use of both FG and HbA1c in combination for diabetes diagnosis, especially given that the diagnostic level of HbA1c is conservative relative to FG.40
Strengths of this investigation include the analysis of extremely high-quality deep sequence data from a large, multi-ancestry sample including five studies and four ancestries to investigate the association of genetic variation across the full allelic spectrum with HbA1c, a diagnostic test that is central to the care of individuals with diabetes. As previous GWAS on HbA1c have included a smaller sample of Hispanic ancestry, this study represents a larger genetic discovery effort for ethnic minorities in the United States. Our study demonstrates that both common and rare genetic variation differ across ancestries and inform the unique genetic architecture of HbA1c. We recognize limitations. We acknowledge that even with a high imputation score, the imputation accuracy of rare variants in the UKBB may not be as optimal as that of common variants. Our hypothetical scenario using NHANES data to assess the population impact of G6PD genetics on diabetes screening does not take into account the full complement of clinical and genetic information contributing to nonglycemic variation in HbA1c, some of which may raise, and not lower, HbA1c, thus reducing the risk of under-detection.41 For instance, structural variants (e.g., copy number variation) were not included in our analyses. Nevertheless, our findings highlight the potential for underdiagnosis in carriers of these variants, which disproportionately affects certain ethnic minorities, if HbA1c is used as the only diagnostic test for diabetes. Our sample sizes, and thus our power, are limited compared to GWAS, particularly for Hispanic and Asian populations. For instance, we did not detect an association signal for rs76723693 in HA because only one individual in our HA population carried a copy of the rare variant. Including larger samples of individuals of non-European ancestries will improve the characterization of rare variants with clinically meaningful effects on HbA1c. We acknowledge that the genome-wide threshold used in this paper may not represent the true number of independent genetic variants (SNVs as well as insertion-deletion variants) tested using our sequence data.
In this study, we identified common and rare genetic variants that cause nonglycemic differences in HbA1c with important clinical and public health implications. Because HbA1c is commonly used in diabetes screening and management worldwide, disregarding the effects of these rare and common genetic variants that tend to occur in high diabetes-risk minority groups could result in delayed diagnosis or under-detection of diabetes in carriers of these variants. To avoid such disparities in care, an assessment of these variants should be considered for incorporation into precision medicine approaches for diabetes diagnosis.
Declaration of Interests
The authors declare no competing interests. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
Acknowledgments
Whole-genome sequencing: Whole-genome sequencing (WGS) for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). Acknowledgments and funding information for each study are available in the Supplemental Data. Centralized read mapping and genotype calling, along with variant quality metrics and filtering, were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1). Phenotype harmonization, data management, sample-identity QC, and general study coordination were provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed. The contributions of the investigators of the NHLBI TOPMed Consortium are gratefully acknowledged. Full authorship list is available in the Supplemental Data.
Funding: L.M.R. is funded by NHLBI grant T32 HL129982. A.K.M. is supported by National Institutes of Health (NIH) National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants K01 DK107836 and R03 DK118305. P.S.dV. is supported by American Heart Association grant 18CDA34110116. J.B.M. and the project are supported by NIH NIDDK grants K24 DK080140, U01 DK078616, and U01 DK105554 Opportunity Pool OP6. The Analysis Commons was funded by R01HL131136. H.M.H. was supported by NHLBI training grants T32 HL007055 and T32 HL129982 and American Diabetes Association grant 1-19-PDF-045. J.R.O. is supported by the TOPMed grant U01 HL137181.
Published: September 26, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.08.010.
Contributor Information
Chloé Sarnowski, Email: chloesar@bu.edu.
Aaron Leong, Email: ASLEONG@partners.org.
Web Resources
Analysis Commons, http://analysiscommons.com
Database of Genotypes and Phenotypes (dbGaP), https://www.ncbi.nlm.nih.gov/gap/
GENetic EStimation and Inference in Structured samples (GENESIS), http://bioconductor.org/packages/release/bioc/html/GENESIS.html
HbA1c Assay Interferences information on the National Glycohemoglobin Standardization Program (NGSP) website, http://www.ngsp.org/interf.asp
Mixed Model Analysis for Pedigrees and Populations (MMAP), https://mmap.github.io/
National Health and Nutrition Examination Survey (NHANES): https://wwwn.cdc.gov/nchs/nhanes/continuousnhanes/default.aspx?BeginYear=2015
NHLBI TOPMed Consortium, https://www.nhlbiwgs.org/topmed-banner-authorship
Omics Analysis, Search and Information System (OASIS), https://omicsoasis.github.io
Online Mendelian Inheritance in Man (OMIM), https://www.omim.org
Scalable and Accurate Implementation of Generalized mixed model (SAIGE), https://github.com/weizhouUMICH/SAIGE
TOPMed laboratory methods, data processing, and quality control details, https://www.nhlbiwgs.org/methods
TOPMed Pipeline, https://github.com/UW-GAC/analysis_pipeline
TOPMed website, https://www.nhlbiwgs.org
Trans-Omics for Precision Medicine (TOPMed) Consortium full authorship list, https://www.nhlbiwgs.org/topmed-banner-authorship
Type 2 Diabetes Knowledge Portal, http://www.type2diabetesgenetics.org
UK Biobank, https://www.ukbiobank.ac.uk/
Supplemental Data
References
- 1.Mortensen H.B., Christophersen C. Glucosylation of human haemoglobin a in red blood cells studied in vitro. Kinetics of the formation and dissociation of haemoglobin A1c. Clin. Chim. Acta. 1983;134:317–326. doi: 10.1016/0009-8981(83)90370-4. [DOI] [PubMed] [Google Scholar]
- 2.Leong A., Meigs J.B. Type 2 diabetes prevention: Implications of hemoglobin A1c genetics. Rev. Diabet. Stud. 2015;12:351–362. doi: 10.1900/RDS.2015.12.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wheeler E., Leong A., Liu C.T., Hivert M.F., Strawbridge R.J., Podmore C., Li M., Yao J., Sim X., Hong J., EPIC-CVD Consortium. EPIC-InterAct Consortium. Lifelines Cohort Study Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med. 2017;14:e1002383. doi: 10.1371/journal.pmed.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacy M.E., Wellenius G.A., Sumner A.E., Correa A., Carnethon M.R., Liem R.I., Wilson J.G., Sacks D.B., Jacobs D.R., Jr., Carson A.P. Association of sickle cell trait with Hemoglobin A1c in African Americans. JAMA. 2017;317:507–515. doi: 10.1001/jama.2016.21035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strickland S.W., Campbell S.T., Little R.R., Bruns D.E., Bazydlo L.A.L. Recognition of rare hemoglobin variants by hemoglobin A1c measurement procedures. Clin. Chim. Acta. 2018;476:67–74. doi: 10.1016/j.cca.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 7.Leong A. Is there a need for neonatal screening of glucose-6-phosphate dehydrogenase deficiency in Canada? McGill J. Med. 2007;10:31–34. [PMC free article] [PubMed] [Google Scholar]
- 8.Motulsky A.G., Stamatoyannopoulos G. Clinical implications of glucose-6-phosphate dehydrogenase deficiency. Ann. Intern. Med. 1966;65:1329–1334. doi: 10.7326/0003-4819-65-6-1329. [DOI] [PubMed] [Google Scholar]
- 9.Paterson A.D. HbA1c for type 2 diabetes diagnosis in Africans and African Americans: Personalized medicine NOW! PLoS Med. 2017;14:e1002384. doi: 10.1371/journal.pmed.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Little R.R., Rohlfing C.L., Sacks D.B., National Glycohemoglobin Standardization Program (NGSP) Steering Committee Status of hemoglobin A1c measurement and goals for improvement: from chaos to order for improving diabetes care. Clin. Chem. 2011;57:205–214. doi: 10.1373/clinchem.2010.148841. [DOI] [PubMed] [Google Scholar]
- 11.Regier A.A., Farjoun Y., Larson D.E., Krasheninina O., Kang H.M., Howrigan D.P., Chen B.J., Kher M., Banks E., Ames D.C. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun. 2018;9:4038. doi: 10.1038/s41467-018-06159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng X., Gogarten S.M., Lawrence M., Stilp A., Conomos M.P., Weir B.S., Laurie C., Levine D. SeqArray-a storage-efficient high-performance data format for WGS variant calls. Bioinformatics. 2017;33:2251–2257. doi: 10.1093/bioinformatics/btx145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brody J.A., Morrison A.C., Bis J.C., O’Connell J.R., Brown M.R., Huffman J.E., Ames D.C., Carroll A., Conomos M.P., Gabriel S., NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium. TOPMed Hematology and Hemostasis Working Group. CHARGE Analysis and Bioinformatics Working Group Analysis commons, a team approach to discovery in a big-data environment for genetic epidemiology. Nat. Genet. 2017;49:1560–1563. doi: 10.1038/ng.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amin N., van Duijn C.M., Aulchenko Y.S. A genomic background based method for association analysis in related individuals. PLoS ONE. 2007;2:e1274. doi: 10.1371/journal.pone.0001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leutenegger A.L., Prum B., Génin E., Verny C., Lemainque A., Clerget-Darpoux F., Thompson E.A. Estimation of the inbreeding coefficient through use of genomic data. Am. J. Hum. Genet. 2003;73:516–523. doi: 10.1086/378207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanai M., Tanaka T., Okada Y. Empirical estimation of genome-wide significance thresholds based on the 1000 Genomes Project data set. J. Hum. Genet. 2016;61:861–866. doi: 10.1038/jhg.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu C., Tachmazidou I., Walter K., Ciampi A., Zeggini E., Greenwood C.M., UK10K Consortium Estimating genome-wide significance for whole-genome sequencing studies. Genet. Epidemiol. 2014;38:281–290. doi: 10.1002/gepi.21797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodonsky C.J., Jain D., Schick U.M., Morrison J.V., Brown L., McHugh C.P., Schurmann C., Chen D.D., Liu Y.M., Auer P.L. Genome-wide association study of red blood cell traits in Hispanics/Latinos: The Hispanic Community Health Study/Study of Latinos. PLoS Genet. 2017;13:e1006760. doi: 10.1371/journal.pgen.1006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z., Tang H., Qayyum R., Schick U.M., Nalls M.A., Handsaker R., Li J., Lu Y., Yanek L.R., Keating B., BioBank Japan Project. CHARGE Consortium Genome-wide association analysis of red blood cell traits in African Americans: the COGENT Network. Hum. Mol. Genet. 2013;22:2529–2538. doi: 10.1093/hmg/ddt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo K.S., Wilson J.G., Lange L.A., Folsom A.R., Galarneau G., Ganesh S.K., Grant S.F., Keating B.J., McCarroll S.A., Mohler E.R., 3rd Genetic association analysis highlights new loci that modulate hematological trait variation in Caucasians and African Americans. Hum. Genet. 2011;129:307–317. doi: 10.1007/s00439-010-0925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chami N., Chen M.H., Slater A.J., Eicher J.D., Evangelou E., Tajuddin S.M., Love-Gregory L., Kacprowski T., Schick U.M., Nomura A. Exome Genotyping Identifies Pleiotropic Variants Associated with Red Blood Cell Traits. Am. J. Hum. Genet. 2016;99:8–21. doi: 10.1016/j.ajhg.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., White S., Peng B., Johnson A.D., Brody J.A., Li A.H., Huang Z., Carroll A., Wei P., Gibbs R. WGSA: An annotation pipeline for human genome sequencing studies. J. Med. Genet. 2016;53:111–112. doi: 10.1136/jmedgenet-2015-103423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Gabrielsen M.E., Wolford B.N., LeFaive J., VandeHaar P., Gagliano S.A., Gifford A. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shriner D., Rotimi C.N. Whole-genome-sequence-based haplotypes reveal single origin of the sickle allele during the Holocene wet phase. Am. J. Hum. Genet. 2018;102:547–556. doi: 10.1016/j.ajhg.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cowie C.C., Rust K.F., Byrd-Holt D.D., Gregg E.W., Ford E.S., Geiss L.S., Bainbridge K.E., Fradkin J.E. Prevalence of diabetes and high risk for diabetes using A1C criteria in the U.S. population in 1988-2006. Diabetes Care. 2010;33:562–568. doi: 10.2337/dc09-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selvin E., Steffes M.W., Gregg E., Brancati F.L., Coresh J. Performance of A1C for the classification and prediction of diabetes. Diabetes Care. 2011;34:84–89. doi: 10.2337/dc10-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Little R.R., Roberts W.L. A review of variant hemoglobins interfering with hemoglobin A1c measurement. J. Diabetes Sci. Technol. 2009;3:446–451. doi: 10.1177/193229680900300307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohlfing C.L., Connolly S.M., England J.D., Hanson S.E., Moellering C.M., Bachelder J.R., Little R.R. The effect of elevated fetal hemoglobin on hemoglobin A1c results: five common hemoglobin A1c methods compared with the IFCC reference method. Am. J. Clin. Pathol. 2008;129:811–814. doi: 10.1309/YFVTUD0GHJF7D16H. [DOI] [PubMed] [Google Scholar]
- 32.Sarnowski C., Hivert M.F. Impact of genetic determinants of HbA1c on Type 2 diabetes risk and diagnosis. curr. diab. rep. 2018;18:52. doi: 10.1007/s11892-018-1022-4. [DOI] [PubMed] [Google Scholar]
- 33.Roberts W.L., Safar-Pour S., De B.K., Rohlfing C.L., Weykamp C.W., Little R.R. Effects of hemoglobin C and S traits on glycohemoglobin measurements by eleven methods. Clin. Chem. 2005;51:776–778. doi: 10.1373/clinchem.2004.047142. [DOI] [PubMed] [Google Scholar]
- 34.Rohlfing C., Hanson S., Little R.R. Measurement of hemoglobin A1c in patients with sickle cell trait. JAMA. 2017;317:2237. doi: 10.1001/jama.2017.4643. [DOI] [PubMed] [Google Scholar]
- 35.Mongia S.K., Little R.R., Rohlfing C.L., Hanson S., Roberts R.F., Owen W.E., D’Costa M.A., Reyes C.A., Luzzi V.I., Roberts W.L. Effects of hemoglobin C and S traits on the results of 14 commercial glycated hemoglobin assays. Am. J. Clin. Pathol. 2008;130:136–140. doi: 10.1309/1YU0D34VJKNUCGT1. [DOI] [PubMed] [Google Scholar]
- 36.Sidorenko J., Kassam I., Kemper K., Zeng J., Lloyd-Jones L., Montgomery G.W., Gibson G., Metspalu A., Esko T., Yang J. The effect of X-linked dosage compensation on complex trait variation. bioRxiv. 2018 doi: 10.1038/s41467-019-10598-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergenstal R.M., Gal R.L., Connor C.G., Gubitosi-Klug R., Kruger D., Olson B.A., Willi S.M., Aleppo G., Weinstock R.S., Wood J., T1D Exchange Racial Differences Study Group Racial differences in the relationship of glucose concentrations and Hemoglobin A1c levels. Ann. Intern. Med. 2017;167:95–102. doi: 10.7326/M16-2596. [DOI] [PubMed] [Google Scholar]
- 38.Ziemer D.C., Kolm P., Weintraub W.S., Vaccarino V., Rhee M.K., Twombly J.G., Narayan K.M., Koch D.D., Phillips L.S. Glucose-independent, black-white differences in hemoglobin A1c levels: a cross-sectional analysis of 2 studies. Ann. Intern. Med. 2010;152:770–777. doi: 10.7326/0003-4819-152-12-201006150-00004. [DOI] [PubMed] [Google Scholar]
- 39.Gómez-Manzo S., Marcial-Quino J., Vanoye-Carlo A., Serrano-Posada H., Ortega-Cuellar D., González-Valdez A., Castillo-Rodríguez R.A., Hernández-Ochoa B., Sierra-Palacios E., Rodríguez-Bustamante E., Arreguin-Espinosa R. Glucose-6-Phosphate Dehydrogenase: update and analysis of new mutations around the world. Int. J. Mol. Sci. 2016;17:E2069. doi: 10.3390/ijms17122069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.American Diabetes Association 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2019. Diabetes Care. 2019;42(Suppl 1):S13–S28. doi: 10.2337/dc19-S002. [DOI] [PubMed] [Google Scholar]
- 41.Jun, G., Sedlazeck, F.J., Chen, H., Yu, B., Qi, Q., Krasheninina, O., Carroll, A., Liu, X., Mansfield, A., Zarate, S., et al. (2018). PgmNr 3186/W: Identification of novel structural variations affecting common and complex disease risks with >16,000 whole genome sequences from ARIC and HCHS/SOL. https://www.ashg.org/2018meeting/pdf/ASHG-2018-poster-abstracts.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.