Abstract
The second Newborn Sequencing in Genomic Medicine and Public Health study was a randomized, controlled trial of the effectiveness of rapid whole-genome or -exome sequencing (rWGS or rWES, respectively) in seriously ill infants with diseases of unknown etiology. Here we report comparisons of analytic and diagnostic performance. Of 1,248 ill inpatient infants, 578 (46%) had diseases of unknown etiology. 213 infants (37% of those eligible) were enrolled within 96 h of admission. 24 infants (11%) were very ill and received ultra-rapid whole-genome sequencing (urWGS). The remaining infants were randomized, 95 to rWES and 94 to rWGS. The analytic performance of rWGS was superior to rWES, including variants likely to affect protein function, and ClinVar pathogenic/likely pathogenic variants (p < 0.0001). The diagnostic performance of rWGS and rWES were similar (18 diagnoses in 94 infants [19%] versus 19 diagnoses in 95 infants [20%], respectively), as was time to result (median 11.0 versus 11.2 days, respectively). However, the proportion diagnosed by urWGS (11 of 24 [46%]) was higher than rWES/rWGS (p = 0.004) and time to result was less (median 4.6 days, p < 0.0001). The incremental diagnostic yield of reflexing to trio after negative proband analysis was 0.7% (1 of 147). In conclusion, rapid genomic sequencing can be performed as a first-tier diagnostic test in inpatient infants. urWGS had the shortest time to result, which was important in unstable infants, and those in whom a genetic diagnosis was likely to impact immediate management. Further comparison of urWGS and rWES is warranted because genomic technologies and knowledge of variant pathogenicity are evolving rapidly.
Keywords: infant, intensive care unit, whole-genome sequencing, whole-exome sequencing, genetic disease, diagnosis, ultra-rapid whole-genome sequencing, genomic medicine, precision medicine
Introduction
Genetic diseases are a leading cause of infant mortality, particularly among ∼15% of US infants admitted to neonatal, pediatric, and cardiovascular ICUs (NICU, PICU, CVICU, respectively) (see March of Dimes Data Book in Web Resources).1, 2, 3, 4, 5, 6 Disease progression can be extremely rapid in infants, necessitating early etiologic diagnosis in order to inform interventions that can lessen suffering, morbidity, and mortality.7, 8 Timely diagnosis requires genome-scale testing since more than 14,000 simple genetic diseases have been described and their presentations often overlap in seriously ill infants (see Web Resources). Examples include seizures, respiratory and cardiac failure, hypotonia, hypoglycemia, and jaundice. Furthermore, simple genetic disease presentations are frequently formes frustes of classical descriptions.7, 8 Over the last 8 years, methods have been developed for increasingly rapid diagnosis of genetic diseases by rWGS and rWES.9, 10, 11, 12 rWES examines ∼2% of the genome, representing almost all known exons and immediate flanking intronic regions typically within 10–20 base pairs of the exons. rWGS, in contrast, examines all exons and introns, and ∼90% of the genome.
Since 2012, the evidence has steadily increased that rapid genomic sequencing, with trio testing when possible, results in a meaningful improvement in the net health outcome in seriously ill infants with suspected genetic disorders of unknown etiology. In previous studies that have examined the utility of rWGS and rWES as a diagnostic test in infants with suspected genetic diseases, rates of diagnosis of simple genetic diseases have ranged from 42% to 57%, changes in medical management from 30% to 72%, and altered outcomes from 24% to 34%.7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26 This evidence has led to calls for implementation in national healthcare systems as the new standard of care.27, 28, 29 The National Health Service of the United Kingdom, for example, will offer rWGS as part of the care for all seriously ill children from 2019.30 However, few guidelines yet exist for use of rapid genomic sequencing. In particular, it is unclear what proportion of critically ill infants should receive rapid genomic sequencing, at what stage during an ICU workup genomic sequencing should be ordered, what turnaround time is acceptable, whether singletons or trios should be tested, and whether rWGS or rWES has superior analytic or diagnostic performance. The second Newborn Sequencing in Genomic Medicine and Public Health (NSIGHT2) clinical trial was designed to provide initial answers to these questions.31
Subjects and Methods
Subjects and Study Design
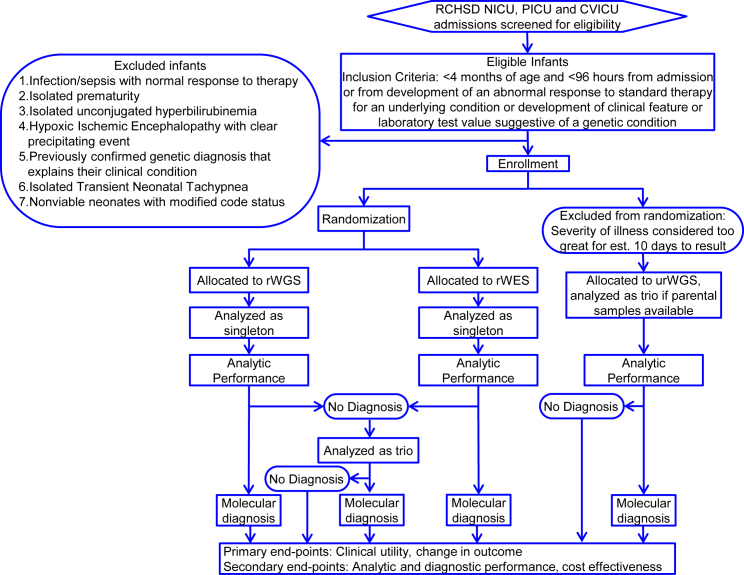
NSIGHT2 was a prospective, randomized, controlled, blinded trial (RCT) in acutely ill infants, primarily from the NICU, PICU, and CVICU at Rady Children’s Hospital, San Diego (RCHSD) to compare the effectiveness and outcomes between rWGS and rWES, with analysis as singleton probands and familial trios (ClinicalTrials.gov NCT03211039, Figure 1). NSIGHT2 was approved by the institutional review board at RCHSD, was designated non-significant risk by the Food and Drug Administration in an Investigational Device Exemption pre-submission, and was performed in accordance with the Declaration of Helsinki. Informed consent was obtained from at least one parent or guardian. The inclusion criteria were age <4 months and time from admission or time from development of a feature suggestive of a genetic condition of <96 h (Figure 1). Infants in whom there was a very low likelihood that a genetic disease diagnosis would change management were excluded (Figure 1). Infants who were gravely ill received urWGS, with the fastest possible time to diagnosis. All other infants whose parents provided informed consent were randomized to receive either rWES or rWGS. Genome interpretation was performed as singleton probands. Infants undiagnosed as singletons were re-analyzed as familial trios.
Figure 1.
Flow Diagram of the NSIGHT2 Randomized, Controlled, Blinded Trial
Singleton analyses included variant validation and segregation analysis in trios, if samples were available, by Sanger sequencing or microarrays. RCHSD, Rady Children’s Hospital San Diego; NICU, level IV neonatal intensive care unit; PICU, regional pediatric intensive care unit; CVICU, regional cardiovascular intensive care unit; rWES, rapid whole-exome sequencing; rWGS, rapid whole-genome sequencing; urWGS, ultra-rapid whole-genome sequencing.
Clinical, Rapid Whole-Genome and -Exome Sequencing, Analysis, and Interpretation
Clinical urWGS, rWGS, and rWES were performed in laboratories accredited by the College of American Pathologists and certified through the Clinical Laboratory Improvement Amendments. Each step included benchmarked quality assessment. Experts selected a few clinical features representative of each child’s illness from the Electronic Health Record (Epic) and mapped them to simple genetic diseases with VAAST.32 Trio EDTA-blood samples were obtained where possible and all samples were sequenced upon receipt. Genomic DNA was isolated with an EZ1 Advanced XL robot and the EZ1 DSP DNA Blood kit (QIAGEN). DNA quality was assessed with the Quant-iT Picogreen dsDNA assay kit (ThermoFisher Scientific) using the Gemini EM Microplate Reader (Molecular Devices). Genomic DNA was fragmented by sonication (Covaris), and bar-coded, paired-end, PCR-free libraries were prepared for rWGS with TruSeq DNA LT kits (Illumina) or Hyper kits (KAPA Biosystems). Sequencing libraries were analyzed with a Library Quantification Kit (KAPA Biosystems) and High Sensitivity NGS Fragment Analysis Kit (Advanced Analytical), respectively. 2 × 101 nucleotide rWGS and urWGS was performed to at least 40-fold coverage with Illumina HiSeq 2500 (rapid run mode), HiSeq 4000, or NovaSeq 6000 (S1 or S2 flow cell) instruments, as described.12
Sample preparation and sequencing for rWES was performed by an external clinical laboratory (GeneDx). Exome enrichment was with the xGen Exome Research Panel v1.0 (Integrated DNA Technologies), and amplification used the Herculase II Fusion polymerase (Agilent).33 FASTQ files for rWES were transferred to Rady Children’s Institute for Genomic Medicine (RCIGM) for analysis and interpretation.
urWGS, rWGS, and rWES sequences were aligned to human genome assembly GRCh37 (hg19), and variants were identified with the Illumina DRAGEN (Dynamic Read Analysis for GENomics) Bio-IT Platform (v.2.5.1, Illumina; Table S2).12 Structural variants were identified with Manta and CNVnator (using DNAnexus), a combination that provided the highest sensitivity and precision.12 Structural variants were filtered to retain those affecting coding regions of known disease-associated genes and with allele frequencies < 2% in the RCIGM database. Nucleotide and structural variants were annotated, analyzed, and interpreted by clinical molecular geneticists using Opal Clinical (Fabric Genomics), according to standard guidelines.34 Interpretation was initially performed with proband sequences alone, to determine the diagnostic yield of singleton sequencing (Figure 1). If no diagnosis was forthcoming, interpretation was performed again, with parental samples if available, in order to determine the net increase in diagnostic yield of duo or trio sequencing (Figure 1). Opal annotated variants with respect to pathogenicity, generated a rank ordered differential diagnosis based on the disease gene algorithm VAAST, a gene burden test, and the algorithm PHEVOR (Phenotype Driven Variant Ontological Re-ranking), which combined the observed HPO phenotype terms from infants, and re-ranked disease genes based on the phenotypic match and the gene score.12 Automatically generated, ranked results were manual interpreted through iterative Opal searches. Initially, variants were filtered to retain those with allele frequencies of < 1% in the Exome Variant Server, 1000 Genomes Samples, and Exome Aggregation Consortium database.12 Variants were further filtered for de novo, recessive, and dominant inheritance patterns. The evidence supporting a diagnosis was then manually evaluated by comparison with the published literature. Analysis, interpretation, and reporting required an average of 6 h of expert effort. If rWGS or rWES established a provisional diagnosis for which a specific treatment was available to prevent morbidity or mortality, this was immediately conveyed to the clinical team. All causative variants were confirmed by Sanger sequencing or chromosomal microarray, as appropriate. Secondary findings were not reported, but medically actionable incidental findings were reported if families consented to receiving this information.
Measurement of Analytic Performance
Analytic performance metrics were calculated using MOON (Diploid).12 Data sources and versions were ClinVar: 2018-04-29; dbNSFP: 3.5; dbSNP: 150; dbscSNV: 1.1; Apollo: 2019-04-29; Ensembl: 37; gnomAD: 2.0.1; HPO: 2019-02-12; DGV: 2016-03-01; dbVar: 2018-06-24; MOON: 3.0.3; KB: 2019-05-29; Mitomap: 2019-01-14; Mitimpact: 2.9.1; Mastermind: 2018-11-26. For ClinVar, we retained variants with at least one Likely Pathogenic or Pathogenic classification that did not have any Benign or Likely Benign classifications. For splicing variants, only splice acceptor and donor variants were counted.
Statistical Analysis
Tests for binary outcomes were conducted with a chi-square test or, if the expected counts were less than 5, Fisher’s exact test. For continuous outcomes, p values were determined with the Wilcoxon signed-rank test due to deviations from normality. McNemar’s test was used to test for differences in diagnostic rates between trio and singleton genomic testing. To account for multiple comparisons, Bonferroni-Hochberg adjusted p values less than 0.05 were considered significant. All analyses were conducted in R v3.3.5.
Results
NSIGHT2 was a prospective RCT to compare the effectiveness (rate of diagnosis, time to diagnosis, clinical utility, perceived family utility, and cost) and outcomes of two methods of rapid genomic sequencing (rWGS or rWES) and two methods of analysis (singleton probands and familial trios) in acutely ill infants (Figure 1). Here we report results of analytic and diagnostic performance, including time to diagnosis.
Enrolled Infants
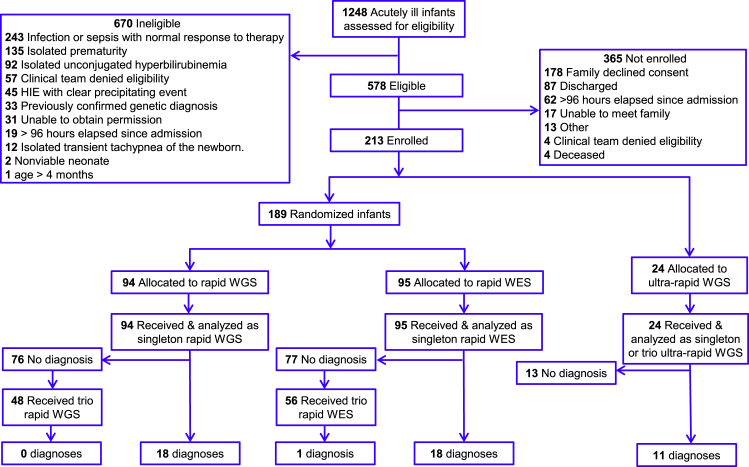
Between 6/29/2017 and 10/9/2018 (467 days), 1,248 infants of age less than 4 months were screened shortly after admission for eligibility (Figure 2). Those screened represented 98% of RCHSD regional NICU, PICU, and CVICU admissions. There were 578 infants (46%) eligible for enrollment (Figure 2). Herein, the inclusion criteria were purposefully broad, because we sought to compare outcomes following use of genomic sequencing early in the evaluation of infants with diseases of unknown etiology (Figure 1). The most common reasons for ineligibility were exclusionary criteria representative of infants in whom it was considered highly unlikely that genomic sequencing would identify a genetic disorder that changed inpatient management (Figure 1). Common exclusion criteria were sepsis with normal response to therapy (243, 19% of those screened), isolated prematurity (135, 11% of those screened), and isolated hyperbilirubinemia (92, 7% of those screened; Figure 2). Only 2% (20) infants were ineligible because of failure to meet inclusion criteria (age greater than 4 months or greater than 96 h had elapsed since admission; Figure 2).
Figure 2.
CONSORT Flow Diagram of the Number of Infants in ICUs Who Were Screened for Eligibility in NSIGHT2, Sequenced, and Received a Genetic Disease Diagnosis that Explained Their Presentation
HIE, hypoxic ischemic encephalopathy.
Some infants have genetic diseases in which specific treatment must be started within the first week of life for optimal outcomes. In previous studies of the utility of rWES and rWGS, genomic sequencing was performed relatively late in the hospital course, which decreased the potential for results to change management. Herein, enrollment was restricted to within 96 h of admission, or within 96 h of development of a new presentation, in order to evaluate outcomes when genomic sequencing was used as a first-tier test (Figure 1). Of the 578 eligible infants, 213 (37%) were enrolled. The most common reasons for failure to enroll were parents refused to provide consent (178, 31% of those eligible), consent not obtained within 96 h of admission (62, 11% of those eligible), and infants discharged prior to consent obtained (87, 15% of eligible infants; Figure 2).
Randomization
Of 213 enrolled infants, 24 (11%) were considered to be too severely ill at enrollment to be randomized. Instead, they were excluded from randomization and received urWGS, with the fastest possible time to diagnosis (Figure 2). The remaining 189 infants were randomized, with 95 receiving rWES and 94 rWGS. No statistically significant differences were found in sex, race, ethnicity, age, birth weight, location, or intensity of medical therapy at time of enrollment between the rWGS and rWES groups (Table 1). Infants who received urWGS differed significantly from those who were randomized by age at symptom onset (median day of life 3.1 days versus 0.5 days, respectively; p = 0.002), receipt of antibiotics at enrollment (88% versus 44%, respectively; p = 0.0001), and short-term mortality (21% versus 2%, respectively; p = 0.0005; Table 1). Furthermore, infants who received urWGS tended to differ from those who were randomized in proportion admitted from home (as opposed to transferred from a birthing hospital; 54% in the urWGS group versus 29% in the randomized group, respectively, p = 0.01), and proportion enrolled from the CVICU (4% versus 30%, respectively, p = 0.01, Table 1).
Table 1.
Demographic and Clinical Characteristics of the 213 NSIGHT2 Probands
| rWES (n = 95) | rWGS (n = 94) | urWGS (n = 24) | rWES versus rWGS p Value | rWES + rWGS versus UrWGS p Value | |
|---|---|---|---|---|---|
| Sexa | |||||
| Female, n (%) | 43 (45%) | 33 (35%) | 8 (33%) | 0.15 | 0.52 |
| Race | |||||
| White, n (%) | 55 (58%) | 58 (62%) | 17 (71%) | 0.59 | 0.30 |
| Asian, n (%) | 2 (2%) | 6 (6%) | 0 (0%) | 0.17 | 0.60 |
| Native Hawaiian or Other Pacific Islander, n (%) | 5 (5%) | 2 (2%) | 0 (0%) | 0.44 | 1.00 |
| American Indian/Alaskan Native, n (%) | 0 (0%) | 0 (0%) | 1 (4%) | 1.00 | 0.11 |
| African American/Black, n (%) | 2 (2%) | 3 (3%) | 1 (4%) | 0.68 | 0.52 |
| Other, n (%) | 27 (28%) | 20 (21%) | 3 (12%) | 0.26 | 0.18 |
| Unknown, n (%) | 4 (4%) | 5 (5%) | 2 (8%) | 0.75 | 0.36 |
| Ethnicity | |||||
| Hispanic, n (%) | 42 (44%) | 36 (38%) | 10 (42%) | 0.41 | 0.97 |
| Unknown/Undetermined, n (%) | 1 (1%) | 2 (2%) | 0 (0%) | 0.62 | 1.00 |
| Age | |||||
| Gestational age, weeks, median (range) | 39 (27–42) | 38 (27–41) | 39 (33–42) | 0.23 | 0.12 |
| Age at symptom onset, day of life, median (range) | 0.6 (0–120) | 0.4 (0–102) | 3.1 (0–63) | 0.64 | 0.002b |
| Age at admission, day of life, median (range) | 2 (0–121) | 2 (0–103) | 6 (0–64) | 0.47 | 0.36 |
| Age at enrollment, day of life, median (range) | 5 (1–121) | 4 (1–105) | 7.5 (2–67) | 0.42 | 0.56 |
| Low birth weight (<2,500 g), n (%) | 17 (18%) | 24 (26%) | 3 (12%) | 0.20 | 0.42 |
| Location at Enrollment | |||||
| NICU, n (%) | 59 (62%) | 63 (67%) | 21 (88%) | 0.48 | 0.02 |
| CVICU, n (%) | 28 (29%) | 28 (30%) | 1 (4%) | 0.96 | 0.01b |
| PICU, n (%) | 7 (7%) | 2 (2%) | 2 (8%) | 0.17 | 0.36 |
| Other, n (%) | 1 (1%) | 1 (1%) | 0 (0%) | 1.00 | 1.00 |
| Admitted from home rather than birthing hospital, n (%) | 31 (33%) | 23 (24%) | 13 (54%) | 0.21 | 0.01b |
| Medical Management at Enrollment | |||||
| Received antibiotics, n (%) | 41 (43%) | 43 (46%) | 21 (88%) | 0.72 | 0.0001b |
| Inotropic support, n (%) | 15 (16%) | 24 (26%) | 6 (25%) | 0.10 | 0.62 |
| Conventional or high-frequency oscillatory ventilation, or ECMO, n (%) | 19 (20%) | 28 (30%) | 8 (33%) | 0.12 | 0.37 |
| Mortality | |||||
| 28-day mortality, n (%) | 3 (3%) | 0 (0%) | 5 (21%) | 0.25 | 0.0005b |
No enrolled infants had unknown or undetermined sex. ECMO, extra-corporeal membrane oxygenation.
Benjamini-Hochberg adjusted p value (not shown) < 0.05.
Analytic Performance of rWGS and rWES
rWGS and urWGS were performed at RCIGM with a quality requirement of at least 35-fold average coverage. Similarly, the sample preparation and sequencing for rWES were performed at an external clinical laboratory (GeneDx) with a requirement at least 20-fold coverage of at least 95% of RefSeq protein-coding nucleotides. In practice rWES had at least 20-fold coverage of 98% of RefSeq protein-coding nucleotides. The resultant analytic performance of rWGS and urWGS were the same, but superior to rWES (Tables 2 and S1). Whole-genome sequencing provided more even coverage than exome sequencing. Thus, the median proportion of nucleotides with greater than 10-fold coverage was 98.0% (range 97.7%–98.5%) with whole-genome sequencing and 94.5% (range 93.8%–95.1%) with exome sequencing (Tables 2 and S1). The disparity in values for rWES reflects coverage for RefSeq protein-coding nucleotides versus coverage for all nucleotides with aligned reads. Whole-genome sequencing identified 121-fold more nucleotide variants than exome sequencing (median 4,669,310 [range 4,445,016–5,570,158] versus 38,901 [range 36,763–46,938], p < 0.0001), 258-fold more nucleotide insertion-deletion variants (indels, median 881,669 [range 756,073–1,044,121] versus 3,401 [range 36,763–4,445,016], p < 0.0001), and 85-fold more rare variants (minor allele frequency < 1%; median 240,628 [range 198,728–415,172] versus 2,703 [range 2,046–9,795], p < 0.0001; Tables 2 and S1). The number of nucleotide variants identified by 35-fold clinical whole-genome sequencing was similar to prior studies.13, 14, 15 Of more relevance to the diagnosis of simple genetic diseases, whole-genome sequencing compared to whole-exome sequencing identified: (1) 12% more coding domain variants (median 26,080 [range 24,305–31,345] versus 23,421 [range 22,193–28,390], p < 0.0001), (2) 37% more variants of types likely to affect protein function (missense, nonsense, altered splice sites, frameshift indels, disrupted start codons, in-frame indels, copy number variants, and variants predicted to create cryptic splice sites; median 1,028 [range 762–3,585] versus 766 [range 555–2,930], p < 0.0001), and (3) twice as many variants annotated as pathogenic or likely pathogenic by ClinVar (median 6.0 [range 2–16] versus 3.0 [range 0–8], p < 0.0001). Relevant to the veracity of these variant calls, 56 nucleotide variants of types likely to affect protein function were orthogonally tested by Sanger sequencing in 40 probands. All were confirmed. They included 38 nucleotide variants in probands who received rWGS or urWGS and 18 nucleotide variants in probands who received rWES. In silico tools are starting to provide useful pathogenicity assessments for non-exonic variants.35, 36 WGS identified 6.5-fold more variants with predicted splice-altering consequences than rWES (median 7 [range 3–18] versus 1 [range 0–6], p < 0.0001).35
Table 2.
Comparison of the Analytic Performance of rWES (n = 95) and Rapid Genome Sequencing (rWGS/urWGS, n = 118)
| Value | rWES or rWGS | % Coding Nt with ≥10× Coverage | % Coding Nt with ≥15× Coverage | % Coding Nt with ≥20× Coverage | Total Variants | Copy Number Variants | Single Nt Variants | Indels | Heterozygous Variants | Homozygous Variants | Coding Variants | Coding Single Nt Variants | Coding Indels | Rare Variants (MAF < 1%) | Variants in OMIM Disease Genes | Missense Variants | Non-sense Variants | Altered Canonical Splice Sites | Frameshift Indels | Disrupted Start Codons | In-frame Indels | P/LP Variant per ClinVar | Non-coding Splice Variants |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | rWES | 94.5 | 94.3 | 94.0 | 38,901 | 7.0 | 35,465 | 3,401 | 22,134 | 13,157 | 23,421 | 22,644 | 783 | 2,703 | 670 | 558 | 14.0 | 15.0 | 46.0 | 3.0 | 122.0 | 3.0 | 1.0 |
| Median | rWGS | 98.0 | 97.6 | 96.3 | 4,669,310 | 9.0 | 3,792,213 | 881,699 | 2,312,709 | 1,420,249 | 26,080 | 25,062 | 1,033 | 240,648 | 48,231 | 687 | 16.0 | 82.0 | 85.0 | 4.0 | 138.0 | 6.0 | 7.0 |
| Fold difference | 1.04 | 1.03 | 1.02 | 121 | 1.3 | 107 | 258 | 108 | 112 | 1.12 | 1.1 | 1.3 | 85 | 69 | 1.27 | 1.29 | 5.41 | 1.82 | 1.26 | 1.14 | 2.05 | 6.5 | |
| Wilcoxon signed-rank p values | <.0001a | 0.31 | <.0001a | 0.001a | <.0001a | ||||||||||||||||||
Only variants passing quality filters were counted. OMIM, Online Mendelian Inheritance in Man; Nt, nucleotide; indel, oligonucleotide insertion deletion variants; P/LP, pathogenic or likely pathogenic; Non-coding Splice Variants, variants predicted to cause cryptic splicing.35
Benjamini-Hochberg adjusted p value < 0.05.
Diagnoses and Incidental Findings
Genetic diseases were diagnosed by identification of pathogenic or likely pathogenic variants in genes known to cause diseases with similar presentations to those observed in the respective infants. Including six incidental findings (pathogenic variants in genes with known disease associations but with features unrelated to those observed in the respective infants), 54 genetic diseases were identified by genomic sequencing in 52 of the 213 (24%) enrolled infants (Tables 3 and S2). There were 49 primary genetic findings (diagnoses) in 48 (23%) of 213 NSIGHT2 infants (Table 3). In 33 (5%) of 670 ineligible infants, genetic diseases were identified prior to enrollment (Figure 2). Four of 24 (17%) infants, who were eligible but not enrolled, were subsequently diagnosed by rWGS (performed outside the NSIGHT2 study at subsequent clinician request). Assuming that the 213 (37%) enrolled infants were representative of 578 eligible infants, the minimum incidence of simple genetic diseases in regional NICU, PICU, and CVICU infants of age less than 4 months was 14% ((52∗578/213 + 33)/1,248). This was a minimum value since it assumed that the 33 of 670 ineligible infants diagnosed with genetic diseases prior to enrollment were the only ineligible infants with genetic diseases, and that there was no bias in genetic disease incidence among eligible parents who elected to enroll or declined.
Table 3.
49 Genetic Diseases Diagnosed by Genomic Sequencing in 48 of 213 NSIGHT2 Infants
| ID | Solo,Duo,Trio | rWES,rWGS,UrWGS | Disease | Affected Gene(s) or Chromosomal Segment | Variant 1 | Variant 2 | Inheritance | De novo Inherited | Complete/Partial Diagnosis | Diagnosis Made with Solo |
|---|---|---|---|---|---|---|---|---|---|---|
| 201 | trio | rWES | Prader-Willi syndromea | 15q11.2-q12 del | chr15:23684685-26108259 | AD | de novo | complete | yes | |
| 205 | trio | UrWGS | Dursun syndrome (G6PC3 Deficiency) | G6CP3 | c.199_218+1delCTCAACCTCATCTTCAAGTGG | c.207dupC (p.Ile70HisfsTer17) | AR | inherited | complete | yes |
| 208 | trio | rWGS | alpha-1-antitrypsin deficiency | SERPINA1 | c.1096G>A (p.Glu366Lys) (homozygous) | AR | inherited | partial | yes | |
| 210 | trio | rWES | CHARGE syndrome | CHD7 | c.8962dupG (p.Asp2988GlyfsTer2) | AD | de novo | complete | yes | |
| 213 | duo | rWGS | heterotaxy, visceral, 5 | NODAL | c.778G>A (p.Gly260Arg) | AD | inherited | complete | yes | |
| 216 | trio | rWES | stroke, hemorrhagic, susceptibility to | COL4A2 | c.3272G>A (p.Gly1091Asp) | AD | de novo | partial | no | |
| 217 | trio | rWGS | DiGeorge syndrome | 22q11.21 del | chr22:18873201-21565900 | AD | de novo | complete | yes | |
| 222 | trio | rWGS | focal dermal hypoplasia | PORCN | c.1356delA (p.Cys453AlafsTer8) | XLD | de novo | complete | yes | |
| 223 | trio | rWGS | Williams-Beuren syndrome | 7q11.23 del | chr7:72521701-74158700 | AD | de novo | complete | yes | |
| 224 | trio | rWGS | Mowat-Wilson syndrome | ZEB2 | c.1297C>T (p.Gln433Ter) | AD | de novo | complete | yes | |
| 227 | solo | rWGS | visceral heterotaxy 5 | NODAL | c.824G>A (p.Arg275His) | AD | n.d. | complete | yes | |
| 233 | trio | rWGS | tuberous sclerosis 1 | TSC1 | c.1498C>T (p.Arg500Ter) | AD | de novo | complete | yes | |
| 239 | duo | rWGS | atrioventricular septal defect 3 | GJA1 | c.1085G>A (p.Arg362Gln) | AD | inherited | complete | yes | |
| 243 | trio | UrWGS | epilepsy, pyridoxine-dependent | ALDH7A1 | c.328C>T (p.Arg110Ter) | c.1279G>C (p.Glu427Gln) | AR | inherited | complete | yes |
| 244 | duo | UrWGS | 14q31.2q32.2 del | 14q31.2q32.2 del | chr14:84783523-96907490 | AD | de novo | complete | yes | |
| 247 | trio | rWES | CHARGE syndrome | CHD7 | c.496C>T (p.Gln166Ter) | AD | de novo | complete | yes | |
| 253 | duo | rWES | optic atrophy plus syndrome | OPA1 | c.556+1G>A | AD | inherited | complete | yes | |
| 259 | trio | rWES | DiGeorge syndrome | 22q11.21 del | chr22:18893883-21568208 | AD | de novo | complete | yes | |
| 263 | trio | UrWGS | early infantile epileptic encephalopathy 7 | KCNQ2 | c.727C>G (p.Leu243Val) | AD | de novo | complete | yes | |
| 276 | trio | rWGS | transient neonatal diabetes mellitus 2 | ABCC8 | c.4591A>C (p.Thr1531Pro) | AD | inherited | complete | yes | |
| 282 | solo | rWES | Turner syndrome | monosomy X | XLD | de novo | complete | yes | ||
| 286 | trio | rWES | Prader-Willi syndromea | 15q11.2-q13.1 del | chr15:22833478-28566610 | AD | de novo | complete | yes | |
| 296 | trio | rWGS | Stickler syndrome, type I | COL2A1 | c.2908_2909dupCC (p.Pro971HisfsTer58) | AD | inherited | complete | yes | |
| 301 | solo | rWGS | Noonan syndrome 1 | PTPN11 | c.1510A>G (p.Met504Val) | AD | de novo | complete | yes | |
| 302 | trio | UrWGS | chromosome 17q12 deletion syndrome | 17q12 del | chr17:34759401-36284600 x1 | AD | inherited | partial | yes | |
| 309 | trio | rWES | 19p12q13.11 dup | 19p12q13.11 dup | chr19:23158251-37100999x3 | AD | de novo | complete | yes | |
| 311 | duo | UrWGS | Barth syndrome | TAZ | c.811C>T (p.Gln271Ter) | XLR | inherited | complete | yes | |
| 313 | trio | rWES | Sotos syndrome 1 | NSD1 | c.5431C>T (p.Arg1811Ter) | AD | de novo | complete | yes | |
| 314 | trio | rWES | Mowat-Wilson syndrome | ZEB2 | c.1387delG (p.Val463PhefsTer24) | AD | de novo | complete | yes | |
| 319 | trio | rWES | Down syndrome | trisomy 21 | 47XY+21 | AD | de novo | complete | yes | |
| 322 | trio | UrWGS | Muenke syndrome | FGFR3 | c.749C>G (p.Pro250Arg) | AD | inherited | complete | yes | |
| 325 | solo | rWES | campomelic dysplasia w. sex reversal | SOX9 | c.196G>T (p.Glu66Ter) | AD | n.d. | complete | yes | |
| 326 | trio | rWGS | Dubin-Johnson syndrome | ABCC2 | c.3399_3400delTT (p.Tyr1134CysfsTer43) | c.3851G>A (p.Trp1284Ter) | AR | inherited | partial | yes |
| 336 | duo | rWES | hypobetalipoproteinemia, 1q21 del | APOB, 1q21.1-q21.2 del | c.2988_2994delCGGGGAC (p.Gly997ProfsTer3) | chr1:146631123-147416271 | AD | n.d. | complete | yes |
| 341 | trio | UrWGS | maple syrup urine disease | BCKDHB | c.212T>G (p.Met71Arg); c.249C>A (p.Asn83Lys) | c.410C>T (p.Ala137Val) | AR | inherited | complete | yes |
| 352 | trio | UrWGS | permanent neonatal diabetes mellitus | INS | c.26C>G (p.Pro9Arg) | AD | de novo | complete | yes | |
| 354 | trio | rWGS | 6q24-q25 deletion syndrome | 6q24.2-q25.1 del | chr6:144951601-150260400 | AD | de novo | complete | yes | |
| 356 | trio | rWES | Emanuel syndrome | der(22)t(11;22)(q23;q11) | chr11:116691508-134257793 | chr22:17038511-20307516 | AD | de novo | complete | yes |
| 361 | trio | rWES | DiGeorge syndrome | 22q11.21 del | chr22:18893883-21562619 | AD | de novo | complete | yes | |
| 364 | duo | rWES | congenital heart defect | NOTCH1 | c.1810delA (p.Ile604SerfsTer27) | AD | inherited | complete | yes | |
| 366 | trio | rWGS | Costello syndrome | HRAS | c.34G>A (p.Gly12Ser) | AD | de novo | complete | yes | |
| 377 | trio | rWGS | lymphedema, hereditary, IA | FLT4 | c.3121C>T (p.Arg1041Trp) | AD | inherited | complete | yes | |
| 400 | trio | UrWGS | Brugada syndrome 1 | SCN5A | c.4534C>T (p.Arg1512Trp) | AD | inherited | complete | yes | |
| 401 | trio | rWGS | renal hypodysplasia/aplasia 3 | GREB1L | c.3194C>T (p.Thr1065Ile) | AD | inherited | complete | yes | |
| 405 | trio | rWES | spinal muscular atrophy | SMN1 | Del | AR | inherited | complete | yes | |
| 408 | duo | rWES | susceptibility to Hirschsprung disease 1 | RET | c.712G>T (p.Glu238Ter) | AD | ND | complete | yes | |
| 412 | trio | UrWGS | benign neonatal seizures 1 | KCNQ2 | c.1051C>G (p.Leu351Val) | AD | inherited | complete | yes | |
| 415 | trio | rWGS | Kabuki syndrome 1 | KMT2D | c.3968dupG (p.Arg1324Ter) | AD | de novo | complete | yes | |
Abbreviations: AD, autosomal dominant; AR, autosomal recessive; XLD, X-linked dominant; XLR, X-linked recessive; ND, not determined.
Confirmed by methylation study.
Forty-five (82%) of 55 genetic diseases explained the infants’ presentations completely, while 4 (7%) partially explained their presentations, and 6 genetic diseases or actionable carrier states (11%) were considered incidental findings (Tables 3, S2, and S3). The rate of incidental findings was 3% (6 of 213). Of 62 pathogenic and likely pathogenic variants, 33 (53%) were single-nucleotide variants, 18 (29%) were copy number or structural variants, and 11 (18%) were small insertion-deletion variants (Tables 3, S2, and S3). The most common inheritance mode was autosomal dominant (46 of 55, 84%), with 11% (6 of 55) being autosomal recessive and 5% (3 of 55) X-linked. Twenty-five (57%) of the 44 diagnoses for which parental genotypes were available occurred de novo (Tables 3, S2, and S3). De novo variants accounted for 23 (66%) of 35 diagnoses that were autosomal dominant and for which inheritance information was known, and for 2 of 2 X-linked dominant disorder diagnoses.
Singleton, Trio, and Inheritance Testing
Parent-child trio samples were available from 147 (69%) of 213 families. Forty-eight (98%) of 49 diagnoses that explained presentations were made by solo genomic sequencing. Importantly, most were confirmed by downstream targeted inheritance testing. Especially for novel variants, the incorporation of inheritance information was critical to the variant classification. Although this led to a delay in reporting, in certain cases it was necessary to classify the variant. In 104 infants with negative solo sequencing, parental samples were available. The incremental diagnostic yield of trio sequencing was 1 (1%) of 104. Thus, the diagnostic rate of trio sequencing was not significantly higher than singleton sequencing (p = 1.0, McNemar’s test). However, trio sequencing did aid in a faster turn-around time for positive and negative cases. Of note, infants receiving urWGS were run as trios if parent samples were available to avoid the delay of confirmatory testing for phasing variants in recessive disorders and determination of inheritance, which can upgrade or downgrade their pathogenicity classification.
Comparison of the Diagnostic Rate of rWGS, rWES, and urWGS
With 189 randomized subjects and current interpretation guidelines, the diagnostic rate of rWGS (18 of 94, 19%) was not significantly different than rWES (19 of 95, 20%; Table 4). With this sample size, our study had 80% power to detect an 18% difference between groups (group 1 diagnostic rate 20%, group 2 diagnostic rate 38%). One diagnosis by rWGS, renal hypodysplasia/aplasia 3 (GREB1L c.3194C>T, p.Thr1065Ile [MIM: 617805]), would have been missed had infant 401 been randomized to rWES since the chromosomal region containing that gene had no sequence coverage in the 240 rWES sequences.
Table 4.
Rate of Molecular Diagnosis and Time to Diagnosis with rWGS, rWES, and urWGS in NICU, PICU, and CVICU Infants
| rWES (n = 95) | rWGS (n = 94) | urWGS (n = 24) | rWES versus rWGS p Value | urWGS versus rWES+rWGS p Value | |
|---|---|---|---|---|---|
| Infants diagnosed, n (%) | 19 (20%) | 18 (19%) | 11 (46%) | 0.88 | 0.004a |
| Time, sample receipt to first positive or negative report (days), median (range) | 11.2 (4.3–38.6) | 11.0 (3.3–49.1) | 4.6 (1.1–14) | 0.65 | <.0001a |
| Time, sample receipt to first positive report (days), median (range) | 11.4 (8.1–38.6) | 12.4 (3.3–41.2) | 2.3 (1.1–14) | 1.00 | 0.0001a |
| Time, sample receipt to interpretation (days), median (range) | 5.3 (2.6–11.9) | 3.2 (1.6–16.4) | 2.2 (0.9–3.3) | <.0001a | <.0001a |
| Time, sample receipt to interpretation minus shipping time (days), median (range) | 4 (2–11) | 3 (1–16) | 2 (1–3) | <.0001a | <.0001a |
| Interpretation time to first positive or negative report (days), median (range) | 5.1 (0.9–33.1) | 7.3 (1.1–44.4) | 2.4 (0.1–12.7) | 0.0008a | <.0001a |
| Positive or negative result within 7 days of accession, n (%) | 4 (4%) | 10 (11%) | 17 (71%) | 0.10 | <.0001a |
| Positive result within 7 days of accession, n (%) | 0 (0%) | 3 (3%) | 8 (33%) | 0.12 | <.0001a |
| Positive result prior to ICU discharge, n (%) | 12 (13%) | 13 (14%) | 8 (33%) | 0.81 | .02a |
Benjamini-Hochberg adjusted p value < 0.05.
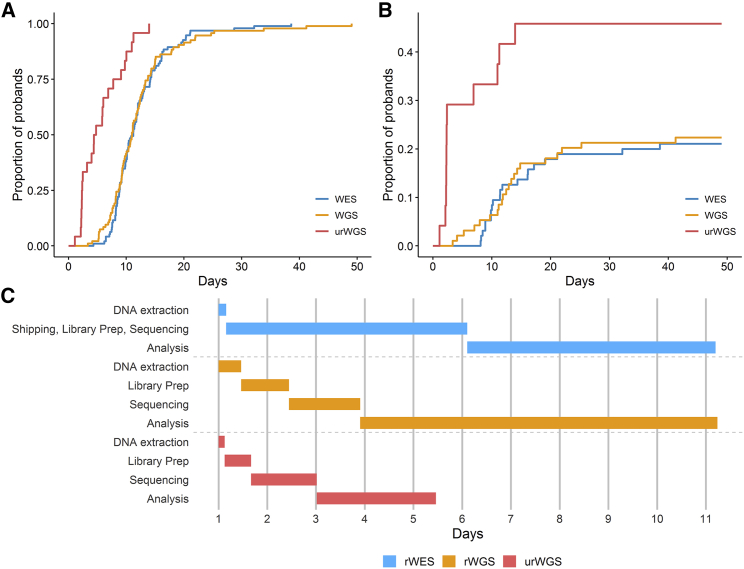
The time to report of results of rWGS and rWES varied widely (Table 4, Figure 3). This variability had two principal sources. First, as noted above, some variants required confirmation in order to make a diagnosis. Furthermore, structural variants were confirmed by send-out tests with a turnaround time of several weeks. Second, 13 of 49 (27%) genetic disease diagnoses informed immediate treatments that had the potential to improve outcomes; these provisional diagnoses were reported prior to confirmation (Figure 2). The median time to report of results was not significantly different between rWGS and rWES (Table 4). However, the median time from sample accessioning to interpretation of rWGS (3.2 days) was shorter than rWES (5.3 days, p < 0.0001, Table 4, Figure S1). This difference remained significant after accounting for shipping time of rWES to an external reference laboratory. Excluding urWGS, the proportion of rWGS results reported within 7 days (11%) was greater than rWES (4%), although this difference was not statistically significant with a sample size of 189 (p = 0.10).
Figure 3.
Comparison of Time to Return of Results by rWES, rWGS, and urWGS in 213 Infants
(A and B) Kaplan-Meier curves of time from sample receipt to first positive or negative report (A) or first positive report (B).
(C) Gantt chart of the median times of the major components in clinical genomic sequencing for diagnosis of genetic diseases. Diagnoses that informed acute changes in management that had the potential to improve outcomes were reported verbally to clinicians provisionally before confirmatory testing of variants. Upon confirmation, all diagnoses were reported in writing.
The diagnostic rate of urWGS (11 of 24, 46%) was considerably higher than rWES and rWGS (37 of 189, 20%, p = 0.004; Table 4, Figure 3). All turnaround times with urWGS were significantly less than rWES and rWGS (Table 4, Figures 3 and S1). In particular, the median time to positive report of urWGS (2.3 days) was faster than rWES and rWGS (11.8 days, p = 0.0001). As measured by return of provisional results, the proportion of urWGS infants diagnosed with genetic diseases that informed immediate treatments that had the potential to improve outcomes (7 of 24, 29%) was greater than rWES and rWGS (6 of 189, 3%, p = 0.0001, Figure 2).
Discussion
Prior to this study, there was considerable evidence that rapid genomic sequencing was a useful diagnostic test for infants in ICUs who were suspected to have genetic diseases (Table 5). However, consensus with regard to optimal methods and scope of use was still lacking. In this report of the NSIGHT2 RCT, we addressed the relative analytic and diagnostic performance of urWGS, rWGS, and rWES in seriously ill infants, evaluated as singleton probands and trios. Longitudinal data related to relative clinical utility, outcomes, and cost effectiveness is being collected and will be reported when complete.
Table 5.
Prior Studies of the Diagnostic Performance of rWES, rWGS, and Rapid Panel Tests in Seriously Ill Infants
| Reference | Date | Study Type | Sequencing Type | NICU and PICU Enrollment Criteria | Study Size | Rate of Diagnosis | Time to Result (days) |
|---|---|---|---|---|---|---|---|
| 15 | 2015 | cohort | rWGS | <4 mo of age; suspected actionable genetic disease | 35 | 57% | 23 |
| 11 | 2017 | cohort | rWES | <100 days of life; suspected genetic disease | 63 | 51% | 13 |
| 37 | 2017 | cohort | rapid panel | infants; suspected genetic disease | 23 | 30% | 12 |
| 14 | 2018 | RCT | rWGS | <4 mo of age; suspected genetic disease | 32 | 41% | 13 |
| 13 | 2018 | cohort | rWGS | infants; suspected genetic disease | 42 | 43% | 23 |
| 21 | 2018 | cohort | rWGS | PICU children with suspected genetic disease | 24 | 42% | 9 |
| 20 | 2018 | cohort | rWES | acutely ill children with suspected genetic diseases | 40 | 53% | 16 |
| 26 | 2019 | cohort | rWGS | 4 months-18 years; PICU; suspected genetic disease | 38 | 48% | 14 |
| 25 | 2019 | cohort | rWGS | NICU and PICU; suspected genetic disease | 195 | 21% | 21 |
Most prior studies of rapid genomic sequencing examined diagnostic utility for simple genetic diseases in selected subsets of infants in regional ICUs, such as those with disorders of unknown etiology that were suspected to be genetic (Table 5). These narrow criteria resulted in less than 20% of regional ICU infants being eligible in those studies. Correspondingly, the rate of genetic disease diagnosis in prior studies ranged from 21% to 57% (Table 5) and was dependent on pre-test likelihood of genetic disease. A distinguishing feature of NSIGHT2 was much broader eligibility in order to inform a less biased estimate of the incidence of genetic disease in regional ICU infants and to assess diagnostic utility when the prior probability of a genetic disease was lower. Herein, we pre-screened 98% of NICU, PICU, and CVICU infant admissions under 4 months of age. Of these, 46% were eligible for enrollment, and all eligible families were approached for enrollment. Based on 2016 records, this represented ∼1.0% of births in San Diego County.38 In NSIGHT2 we also sought to examine the feasibility of deploying genomic sequencing as a first-tier diagnostic test in infants with disorders of unknown etiology upon ICU admission. We found that it was technically feasible to limit rapid genomic sequencing to the first 96 h of admission in regional ICU infants: 37% of eligible infants were enrolled, and short-term follow-up did not disclose any false positive results. However, limiting enrollment to the first 96 h of admission did result in 24 (7%) of 365 infants, who had been eligible but were not enrolled, subsequently receiving rWGS outside the study. Of these, 4 (17%) infants were diagnosed with genetic disorders.
The incidence of genetic diseases among infants enrolled in NSIGHT2 was 24% (including incidental findings), which suggested a substantially higher incidence of simple genetic diseases than previous studies whose enrollment was limited to infants in whom a genetic disease had been suspected. Including 33 ineligible infants who were known to have a genetic disease at pre-screening, this suggested the incidence of genetic diseases in infants in San Diego regional ICUs to be 14%. This estimate represents a minimal incidence since the assumption was that no ineligible infants had undiagnosed genetic diseases and that there were no false negative results. One prior study explored the use of WES as a screening test in newborns in a regional ICU and found the incidence of genetic disease to be 16%.39 Together, data from our study and that of Ceyhan-Birsoy imply that genetic diseases are substantially under-recognized in infants in ICUs. Furthermore, the pre-test probability of a genetic disease in infants in regional ICUs provides evidence of recommend genomic sequencing as a first-tier test.
A second distinguishing feature of NSIGHT2 was that enrollment was restricted to infants admitted within the past 96 h or within 96 h of development of a new clinical feature, which ensured that genomic sequencing was performed as a first-tier diagnostic test. Thus, many infants were enrolled prior to subspecialty consults, including medical genetics. Early enrollment for rapid genomic sequencing was feasible in ICU settings: only 11% of families were excluded due to inability to make enrollment decisions in the allotted time. Most prior studies evaluated the diagnostic and clinical utility of genomic sequencing performed late during NICU and PICU stay. Infants in those studies had received multiple consultations and negative tests, changing the prior probability of a genetic disease. Those studies were also biased toward evaluation of infants with prolonged length of stay. In such cases, a considerable proportion of medical and surgical decisions had already been made empirically, prior to return of genomic sequencing results. Thus the clinical utility of positive and negative results was decreased. An insight from our prior studies was that a determinant of the cost effectiveness of genomic sequencing was time from admission to return of results, and, particularly, prior to medical decision making with long-term consequences for care.13
The third distinguishing feature was that most NSIGHT2 enrollment occurred before broad use of urWGS was technologically possible. Until February 2018, urWGS could be performed only for singleton proband samples, and at considerably higher cost (requiring a full run on the HiSeq 2500 in rapid run mode). This forced us to adopt a study design where only a subset—24 (11%) of 213 enrolled infants and 5% of infant NICU, PICU, and CVICU admissions—were selected for urWGS. These were the most unstable infants in whom the differential diagnosis included rare genetic diseases with specific treatments or interventions whose effectiveness required early institution. Delayed diagnosis in these infants was likely to be associated with permanent morbidity or mortality. They were not randomized between rWGS and rWES or analyzed as singleton first with secondary trio analysis, but instead received urWGS, with the fastest possible time to diagnosis (median 2.3 days). Selection of such infants by NSIGHT2 clinicians was relatively effective. Infants selected for urWGS were diagnosed with genetic diseases more frequently than those who were randomized (11 of 24 [46%] versus 37 of 189 [20%]; p = 0.004). In addition, the proportion of infants who had urWGS leading to diagnoses that informed immediate interventions or treatments that had the potential to improve outcomes was greater than in those who were randomized (7 of 24 [29%] versus 6 of 189 [3%], p = 0.0001).
In comparison to rWES, the analytic performance of rWGS identified: (1) 107-fold more single-nucleotide variants; (2) 91-fold more variants in OMIM disease-associated genes; (3) 7-fold more noncoding variants predicted to cause cryptic splicing, as expected from capture probe distribution; and (4) 259-fold more nucleotide insertion deletion variants, reflecting strong purifying selection of exonic indels relative to exonic single-nucleotide variants (41). Thus, as expected, rWGS sampled non-exonic variation much more comprehensively than rWES. Less expected, however, was the finding that rWGS identified 11% more nucleotide coding variants than rWES, 26% more missense variants, 21% more nonsense variants, 33% more disrupted stop codons, 81% more frameshift indels, and 5.5 times more altered splice sites. WES is known to have methodologic characteristics that decrease exonic variant sensitivity, particularly for heterozygous variants. These include PCR amplification, which tends toward lower coverage in GC-rich regions, preferential capture of reference sequence alleles (particularly for multiple nucleotide variants and indels), and more heterogeneous read coverage over target regions.40, 41 Of greatest relevance for clinical sequencing, rWGS identified twice as many nucleotide variants with ClinVar pathogenic and likely pathogenic assertions as rWES.
Given the greater number of potentially disease-causing variants detected by rWGS, it was surprising that rWES and rWGS had very similar rates of diagnosis (19 of 95 [20%] versus 18 of 94 [19%], respectively). Four previous within-cohort comparisons of WES and WGS reported 4%–7% increased diagnostic yield with WGS.42, 43, 44, 45 Conditions that resulted in diagnosis by WGS but not WES included non-coding variants in neurodevelopmental and skeletal disorders, pseudogenes in polycystic kidney disease, and structural variants.45, 46, 47, 48, 49, 50 Herein, one diagnosis (6%) by WGS, renal hypodysplasia/aplasia 3 (GREB1L c.3194C>T) would have been missed had that infant been randomized to WES, since that gene lacks sequence coverage by WES. Failure to show a difference in the diagnostic yield of rWGS and rWES herein likely resulted from differences in experimental design from previous studies, and the limited statistical power to detect a difference within a moderate sample size. As our ability to interpret the pathogenicity of noncoding nucleotide and structural variants associated with cryptic splicing or gene regulation improves, the difference in proportion diagnosed by rWGS and rWES should increase. Currently, however, interpretation is largely limited to coding variation. Furthermore, the ability to make a genetic diagnosis is significantly limited by published scientific knowledge on pathogenicity of genetic variation. Therefore, it is not unreasonable to expect higher diagnostic rate by rWGS compared to rWES in the near future given the superior analytic performance of rWGS.
As a result of differences in sequencing and analysis methods, urWGS results were reported much faster than in those who were randomized (median time to positive report 2.3 days versus 11.8 days, p = 0.0001). In contrast, time to results of rWGS and rWES did not differ significantly. As expected, time from sample receipt to interpretation was faster by rWGS, since it had much simpler library preparation than rWES. Of note, 13% of infants randomized to rWES received a diagnosis prior to discharge from the ICU, compared with 14% with rWGS (p = 0.81) and 33% with urWGS (p = 0.02). Thus, the timeliness of urWGS was superior to rWGS and rWES. With the introduction of the Illumina NovaSeq 6000, rWGS and urWGS have similar scalability to rWES in a clinical laboratory setting. The principal rationale in favor of rWES has been lower test cost. In our clinical laboratory, at volumes of at least 1,000 per annum, proband rWES costs $1,500 less than rWGS and urWGS. A direct comparison of the diagnostic performance of urWGS and rWES is warranted, with larger sample size than herein, and, ideally, performance of both tests in each proband.
Finally, in 147 (69%) of 213 families with trio samples, we examined whether the diagnostic rate of genomic sequencing of parent-child trios was higher than that of singletons with genotype confirmation in trios. Traditionally, trios were considered superior for genetic disease diagnosis, since they facilitated detection of de novo variants and allowed phasing of compound heterozygous variants during interpretation. Determination of de novo occurrence often upgrades the pathogenicity classification of variants. Herein, however, 98% (50) of 51 diagnoses were made with singleton genomic sequencing and trio genotype confirmation, where possible. Trio sequencing only yielded one new diagnosis (0.7%). Thus, the diagnostic rate of trio sequencing was not significantly higher than singleton sequencing. In contrast, a recent meta-analysis of five cohorts showed trio genomic sequencing to have twice the odds of diagnosis than singleton genomic sequencing (95% CI 1.62–2.56, p <.0001).51 A likely explanation of the disparity in results herein was the difference in experimental design: herein, trios were used to confirm phasing and de novo occurrence of likely causative variants detected by singleton sequencing, but not in the other studies. Of note, de novo variants are frequently novel (absent from allele frequency databases), facilitating their presumptive identification in singletons during interpretation, before confirmation as de novo. Likewise, the occurrence of two pathogenic or likely pathogenic variants in a singleton in a recessive disorder that fits an infant’s presentation is sufficient to merit nomination for confirmatory phasing. While trio genomic sequencing is about twice as costly as singleton testing, the latter adds ∼5 days to the time to result in cases requiring confirmatory testing for variant phasing or determination of de novo occurrence. This was a common occurrence: 32 (63%) of the diagnoses herein were associated with de novo or compound heterozygous variants. For this reason, all infants selected for urWGS received trio genomic sequencing. Recurrent presentations for which urWGS merits consideration include neonatal encephalopathy, organ failure, severe metabolic abnormalities, and infants in whom major surgeries or palliative care are being considered.
In conclusion, NSIGHT2 demonstrated that rapid genomic sequencing can be performed as a first-tier diagnostic test in infants with diseases of unknown etiology at time of admission to regional ICUs. In unstable infants and those in whom a genetic diagnosis was likely to impact immediate management, urWGS had optimal analytic and diagnostic performance, by virtue of shortest time to result. We did not find significant differences in the diagnostic performance of rWES or rWGS, nor of singleton testing with confirmatory testing of trios as indicated or trio testing. A larger study of the diagnostic performance of genomic sequencing methods in ICU infants is warranted since the underpinning technologies and the breadth of knowledge of variant pathogenicity are improving rapidly. While analytic and diagnostic performance are important measures of the utility of clinical tests, these conclusions will be modified by results of the clinical utility and outcomes in the NSIGHT2 cohort.
Data and Material Availability
All data associated with this study are present in the paper or Supplemental Data or are available at the Longitudinal Pediatric Data Resource under a data use agreement and subject to the limitations of the informed consent documents for each subject (Accession Number nbs000003.v1.p, http://www.nbscn.org/longitudinal-pediatric-data-resource.htm).
Consortia
The RCIGM Investigators are as follows: Zaira Bezares, Cinnamon Bloss, Joshua J.A. Braun, Carlos Diaz, Dana Mashburn, Dorjee Tamang, Daniken Orendain, Jenni Friedman, Joe Gleeson, Jaime Barea, George Chiang, Casey Cohenmeyer, Nicole G. Coufal, Marva Evans, Jose Honold, Raymond L. Hovey, Amy Kimball, Brian Lane, Crystal Le, Jennie Le, Sandra Leibel, Laurel Moyer, Patrick Mulrooney, Daeheon Oh, Paulina Ordonez, Albert Oriol, Maria Ortiz-Arechiga, Laura Puckett, Mark Speziale, Denise Suttner, Lucitia Van Der Kraan, Gail Knight, Charles Sauer, Richard Song, Sarah White, Audra Wise, and Catherine Yamada.
Declaration of Interests
C.I.K. and P.S. are employees and shareholders of Diploid Inc., have a management relationship with Diploid Inc., and have a patent related to this work. The remaining authors declare no competing interests.
Acknowledgments
This study was supported by grant U19HD077693 from NICHD and NHGRI to S.F.K., grant UL1TR002550 from NCATS to E.J. Topol, and gifts from the Liguori Family, John Motter and Effie Simanikas, Ernest and Evelyn Rady, and Rady Children’s Hospital.
Published: September 26, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.08.009.
Contributor Information
Stephen F. Kingsmore, Email: skingsmore@rchsd.org.
the RCIGM Investigators:
Zaira Bezares, Cinnamon Bloss, Joshua J.A. Braun, Carlos Diaz, Dana Mashburn, Dorjee Tamang, Daniken Orendain, Jenni Friedman, Joe Gleeson, Jaime Barea, George Chiang, Casey Cohenmeyer, Nicole G. Coufal, Marva Evans, Jose Honold, Raymond L. Hovey, Amy Kimball, Brian Lane, Crystal Le, Jennie Le, Sandra Leibel, Laurel Moyer, Patrick Mulrooney, Daeheon Oh, Paulina Ordonez, Albert Oriol, Maria Ortiz-Arechiga, Laura Puckett, Mark Speziale, Denise Suttner, Lucitia Van Der Kraan, Gail Knight, Charles Sauer, Richard Song, Sarah White, Audra Wise, and Catherine Yamada
Web Resources
ClinicalTrials.gov, https://clinicaltrials.gov
Database of Human Structural Variants (dbVar) Statistics, https://www.ncbi.nlm.nih.gov/dbvar?term=(%22clin%20pathogenic%22%5BFilter%5D)%20AND%20homo%20sapiens%5BOrganism%5D)
Longitudinal Pediatric Data Resource, http://www.nbscn.org/longitudinal-pediatric-data-resource.htm
March of Dimes 2016 Databook, https://www.marchofdimes.org/March-of-Dimes-2016-Databook.pdf
OMIM, https://www.omim.org/
OMIM Entry Statistics, https://www.omim.org/statistics/geneMap
R statistical software, https://www.r-project.org/
Supplemental Data
P values were determined with the Wilcoxon signed-rank test due to deviations from normality. Abbreviations: OMIM: Online Mendelian Inheritance in Man. Indel: oligonucleotide insertion deletion variants. P: Pathogenic variant. LP: Likely pathogenic variant. SpliceAI Passing Filter: Noncoding variants predicted to cause cryptic splicing.38
References
- 1.Murphy S.L., Xu J., Kochanek K.D., Arias E. 2018. Mortality in the United States, 2017, NCHS Data Brief.https://www.ncbi.nlm.nih.gov/pubmed/30500322 [PubMed] [Google Scholar]
- 2.Arth A.C., Tinker S.C., Simeone R.M., Ailes E.C., Cragan J.D., Grosse S.D. Inpatient Hospitalization Costs Associated with Birth Defects Among Persons of All Ages - United States, 2013. MMWR Morb. Mortal. Wkly. Rep. 2017;66:41–46. doi: 10.15585/mmwr.mm6602a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry M.A., Shah P.S., Brouillette R.T., Hellmann J. Predictors of mortality and length of stay for neonates admitted to children’s hospital neonatal intensive care units. J. Perinatol. 2008;28:297–302. doi: 10.1038/sj.jp.7211904. [DOI] [PubMed] [Google Scholar]
- 4.Daoud H., Luco S.M., Li R., Bareke E., Beaulieu C., Jarinova O., Carson N., Nikkel S.M., Graham G.E., Richer J. Next-generation sequencing for diagnosis of rare diseases in the neonatal intensive care unit. CMAJ. 2016;188:E254–E260. doi: 10.1503/cmaj.150823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malam F., Hartley T., Gillespie M.K., Armour C.M., Bariciak E., Graham G.E., Nikkel S.M., Richer J., Sawyer S.L., Boycott K.M., Dyment D.A. Benchmarking outcomes in the Neonatal Intensive Care Unit: Cytogenetic and molecular diagnostic rates in a retrospective cohort. Am. J. Med. Genet. A. 2017;173:1839–1847. doi: 10.1002/ajmg.a.38250. [DOI] [PubMed] [Google Scholar]
- 6.Weiner J., Sharma J., Lantos J., Kilbride H. How infants die in the neonatal intensive care unit: trends from 1999 through 2008. Arch. Pediatr. Adolesc. Med. 2011;165:630–634. doi: 10.1001/archpediatrics.2011.102. [DOI] [PubMed] [Google Scholar]
- 7.Petrikin J.E., Willig L.K., Smith L.D., Kingsmore S.F. Rapid whole genome sequencing and precision neonatology. Semin. Perinatol. 2015;39:623–631. doi: 10.1053/j.semperi.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith L.D., Willig L.K., Kingsmore S.F. Whole-Exome Sequencing and Whole-Genome Sequencing in Critically Ill Neonates Suspected to Have Single-Gene Disorders. Cold Spring Harb. Perspect. Med. 2015;6:a023168. doi: 10.1101/cshperspect.a023168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saunders C.J., Miller N.A., Soden S.E., Dinwiddie D.L., Noll A., Alnadi N.A., Andraws N., Patterson M.L., Krivohlavek L.A., Fellis J. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci. Transl. Med. 2012;4:154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller N.A., Farrow E.G., Gibson M., Willig L.K., Twist G., Yoo B., Marrs T., Corder S., Krivohlavek L., Walter A. A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome Med. 2015;7:100. doi: 10.1186/s13073-015-0221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng L., Pammi M., Saronwala A., Magoulas P., Ghazi A.R., Vetrini F., Zhang J., He W., Dharmadhikari A.V., Qu C. Use of Exome Sequencing for Infants in Intensive Care Units: Ascertainment of Severe Single-Gene Disorders and Effect on Medical Management. JAMA Pediatr. 2017;171:e173438. doi: 10.1001/jamapediatrics.2017.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark M.M., Hildreth A., Batalov S., Ding Y., Chowdhury S., Watkins K., Ellsworth K., Camp B., Kint C.I., Yacoubian C. Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Sci. Transl. Med. 2019;11:11. doi: 10.1126/scitranslmed.aat6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farnaes L., Hildreth A., Sweeney N.M., Clark M.M., Chowdhury S., Nahas S., Cakici J.A., Benson W., Kaplan R.H., Kronick R. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom. Med. 2018;3:10. doi: 10.1038/s41525-018-0049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrikin J.E., Cakici J.A., Clark M.M., Willig L.K., Sweeney N.M., Farrow E.G., Saunders C.J., Thiffault I., Miller N.A., Zellmer L. The NSIGHT1-randomized controlled trial: rapid whole-genome sequencing for accelerated etiologic diagnosis in critically ill infants. NPJ Genom. Med. 2018;3:6. doi: 10.1038/s41525-018-0045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willig L.K., Petrikin J.E., Smith L.D., Saunders C.J., Thiffault I., Miller N.A., Soden S.E., Cakici J.A., Herd S.M., Twist G. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir. Med. 2015;3:377–387. doi: 10.1016/S2213-2600(15)00139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farnaes L., Nahas S.A., Chowdhury S., Nelson J., Batalov S., Dimmock D.M., Kingsmore S.F., RCIGM Investigators Rapid whole-genome sequencing identifies a novel GABRA1 variant associated with West syndrome. Cold Spring Harb. Mol. Case Stud. 2017;3:a001776. doi: 10.1101/mcs.a001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hildreth A., Wigby K., Chowdhury S., Nahas S., Barea J., Ordonez P., Batalov S., Dimmock D., Kingsmore S., RCIGM Investigators Rapid whole-genome sequencing identifies a novel homozygous NPC1 variant associated with Niemann-Pick type C1 disease in a 7-week-old male with cholestasis. Cold Spring Harb. Mol. Case Stud. 2017;3:a001966. doi: 10.1101/mcs.a001966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanford E., Watkins K., Nahas S., Gottschalk M., Coufal N.G., Farnaes L., Dimmock D., Kingsmore S.F., RCIGM Investigators Rapid whole-genome sequencing identifies a novel AIRE variant associated with autoimmune polyendocrine syndrome type 1. Cold Spring Harb. Mol. Case Stud. 2018;4:a002485. doi: 10.1101/mcs.a002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen D.Y., Chowdhury S., Farnaes L., Friedman J.R., Honold J., Dimmock D.P., Gold J.J. Rapid Diagnosis of KCNQ2-Associated Early Infantile Epileptic Encephalopathy Improved Outcome. Pediatr. Neurol. 2018;86:69–70. doi: 10.1016/j.pediatrneurol.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stark Z., Lunke S., Brett G.R., Tan N.B., Stapleton R., Kumble S., Yeung A., Phelan D.G., Chong B., Fanjul-Fernandez M., Melbourne Genomics Health Alliance Meeting the challenges of implementing rapid genomic testing in acute pediatric care. Genet. Med. 2018;20:1554–1563. doi: 10.1038/gim.2018.37. [DOI] [PubMed] [Google Scholar]
- 21.Mestek-Boukhibar L., Clement E., Jones W.D., Drury S., Ocaka L., Gagunashvili A., Le Quesne Stabej P., Bacchelli C., Jani N., Rahman S. Rapid Paediatric Sequencing (RaPS): comprehensive real-life workflow for rapid diagnosis of critically ill children. J. Med. Genet. 2018;55:721–728. doi: 10.1136/jmedgenet-2018-105396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanford E., Farnaes L., Batalov S., Bainbridge M., Laubach S., Worthen H.M., Tokita M., Kingsmore S.F., Bradley J. Concomitant diagnosis of immune deficiency and Pseudomonas sepsis in a 19 month old with ecthyma gangrenosum by host whole-genome sequencing. Cold Spring Harb. Mol. Case Stud. 2018;4:a003244. doi: 10.1101/mcs.a003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soden S.E., Saunders C.J., Willig L.K., Farrow E.G., Smith L.D., Petrikin J.E., LePichon J.B., Miller N.A., Thiffault I., Dinwiddie D.L. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci. Transl. Med. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briggs B., James K.N., Chowdhury S., Thornburg C., Farnaes L., Dimmock D., Kingsmore S.F., RCIGM Investigators Novel Factor XIII variant identified through whole-genome sequencing in a child with intracranial hemorrhage. Cold Spring Harb. Mol. Case Stud. 2018;4:a003525. doi: 10.1101/mcs.a003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.French C.E., Delon I., Dolling H., Sanchis-Juan A., Shamardina O., Mégy K., Abbs S., Austin T., Bowdin S., Branco R.G., NIHR BioResource—Rare Disease. Next Generation Children Project Whole genome sequencing reveals that genetic conditions are frequent in intensively ill children. Intensive Care Med. 2019;45:627–636. doi: 10.1007/s00134-019-05552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanford E.F., Clark M.M., Farnaes L., Williams M.R., Perry J.C., Ingulli E.G., Sweeney N.M., Doshi A., Gold J.J., Briggs B., RCIGM Investigators Rapid Whole Genome Sequencing Has Clinical Utility in Children in the PICU. Pediatr. Crit. Care Med. 2019;(Jun):19. doi: 10.1097/PCC.0000000000002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stark Z., Dolman L., Manolio T.A., Ozenberger B., Hill S.L., Caulfied M.J., Levy Y., Glazer D., Wilson J., Lawler M. Integrating Genomics into Healthcare: A Global Responsibility. Am. J. Hum. Genet. 2019;104:13–20. doi: 10.1016/j.ajhg.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman J.M., Bombard Y., Cornel M.C., Fernandez C.V., Junker A.K., Plon S.E., Stark Z., Knoppers B.M., Paediatric Task Team of the Global Alliance for Genomics and Health Regulatory and Ethics Work Stream Genome-wide sequencing in acutely ill infants: genomic medicine’s critical application? Genet. Med. 2019;21:498–504. doi: 10.1038/s41436-018-0055-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borghesi A., Mencarelli M.A., Memo L., Ferrero G.B., Bartuli A., Genuardi M., Stronati M., Villani A., Renieri A., Corsello G., their respective Scientific Societies Intersociety policy statement on the use of whole-exome sequencing in the critically ill newborn infant. Ital. J. Pediatr. 2017;43:100. doi: 10.1186/s13052-017-0418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.U.K. Department of Health and Social Care . 2018. Matt Hancock announces ambition to map 5 million genomes.https://www.gov.uk/government/news/matt-hancock-announces-ambition-to-map-5-million-genomes [Google Scholar]
- 31.Berg J.S., Agrawal P.B., Bailey D.B., Jr., Beggs A.H., Brenner S.E., Brower A.M., Cakici J.A., Ceyhan-Birsoy O., Chan K., Chen F. Newborn Sequencing in Genomic Medicine and Public Health. Pediatrics. 2017;139:e20162252. doi: 10.1542/peds.2016-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flygare S., Hernandez E.J., Phan L., Moore B., Li M., Fejes A., Hu H., Eilbeck K., Huff C., Jorde L. The VAAST Variant Prioritizer (VVP): ultrafast, easy to use whole genome variant prioritization tool. BMC Bioinformatics. 2018;19:57. doi: 10.1186/s12859-018-2056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 34.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 36.Lee M., Roos P., Sharma N., Atalar M., Evans T.A., Pellicore M.J., Davis E., Lam A.N., Stanley S.E., Khalil S.E. Systematic Computational Identification of Variants That Activate Exonic and Intronic Cryptic Splice Sites. Am. J. Hum. Genet. 2017;100:751–765. doi: 10.1016/j.ajhg.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Diemen C.C., Kerstjens-Frederikse W.S., Bergman K.A., de Koning T.J., Sikkema-Raddatz B., van der Velde J.K., Abbott K.M., Herkert J.C., Löhner K., Rump P. Rapid Targeted Genomics in Critically Ill Newborns. Pediatrics. 2017;140:e20162854. doi: 10.1542/peds.2016-2854. [DOI] [PubMed] [Google Scholar]
- 38.San Diego County Crude Birth Rate, https://www.sandiegocounty.gov/content/dam/sdc/hhsa/programs/phs/documents/MchSt-BirthCrude.pdf.
- 39.Ceyhan-Birsoy O., Murry J.B., Machini K., Lebo M.S., Yu T.W., Fayer S., Genetti C.A., Schwartz T.S., Agrawal P.B., Parad R.B., BabySeq Project Team Interpretation of Genomic Sequencing Results in Healthy and Ill Newborns: Results from the BabySeq Project. Am. J. Hum. Genet. 2019;104:76–93. doi: 10.1016/j.ajhg.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meynert A.M., Ansari M., FitzPatrick D.R., Taylor M.S. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinformatics. 2014;15:247. doi: 10.1186/1471-2105-15-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lionel A.C., Costain G., Monfared N., Walker S., Reuter M.S., Hosseini S.M., Thiruvahindrapuram B., Merico D., Jobling R., Nalpathamkalam T. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet. Med. 2018;20:435–443. doi: 10.1038/gim.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor J.C., Martin H.C., Lise S., Broxholme J., Cazier J.B., Rimmer A., Kanapin A., Lunter G., Fiddy S., Allan C. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat. Genet. 2015;47:717–726. doi: 10.1038/ng.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alfares A., Aloraini T., Subaie L.A., Alissa A., Qudsi A.A., Alahmad A., Mutairi F.A., Alswaid A., Alothaim A., Eyaid W. Whole-genome sequencing offers additional but limited clinical utility compared with reanalysis of whole-exome sequencing. Genet. Med. 2018;20:1328–1333. doi: 10.1038/gim.2018.41. [DOI] [PubMed] [Google Scholar]
- 45.Shashi V., Schoch K., Spillmann R., Cope H., Tan Q.K., Walley N., Pena L., McConkie-Rosell A., Jiang Y.H., Stong N., Undiagnosed Diseases Network A comprehensive iterative approach is highly effective in diagnosing individuals who are exome negative. Genet. Med. 2019;21:161–172. doi: 10.1038/s41436-018-0044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., Wu X., Du L., Zheng J., Deng S., Bi X., Chen Q., Xie H., Férec C., Cooper D.N. Identification of compound heterozygous variants in the noncoding RNU4ATAC gene in a Chinese family with two successive foetuses with severe microcephaly. Hum. Genomics. 2018;12:3. doi: 10.1186/s40246-018-0135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilfert A.B., Sulovari A., Turner T.N., Coe B.P., Eichler E.E. Recurrent de novo mutations in neurodevelopmental disorders: properties and clinical implications. Genome Med. 2017;9:101. doi: 10.1186/s13073-017-0498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yip R.K.H., Chan D., Cheah K.S.E. Mechanistic insights into skeletal development gained from genetic disorders. Curr. Top. Dev. Biol. 2019;133:343–385. doi: 10.1016/bs.ctdb.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Khan M.J., Nazli R., Ahmed J., Basit S. Whole Genome Sequencing instead of Whole Exome Sequencing is required to identify the Genetic Causes of Polycystic Ovary Syndrome in Pakistani families. Pak. J. Med. Sci. 2018;34:540–545. doi: 10.12669/pjms.343.14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ali H., Al-Mulla F., Hussain N., Naim M., Asbeutah A.M., AlSahow A., Abu-Farha M., Abubaker J., Al Madhoun A., Ahmad S., Harris P.C. PKD1 Duplicated regions limit clinical Utility of Whole Exome Sequencing for Genetic Diagnosis of Autosomal Dominant Polycystic Kidney Disease. Sci. Rep. 2019;9:4141. doi: 10.1038/s41598-019-40761-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clark M.M., Stark Z., Farnaes L., Tan T.Y., White S.M., Dimmock D., Kingsmore S.F. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom. Med. 2018;3:16. doi: 10.1038/s41525-018-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P values were determined with the Wilcoxon signed-rank test due to deviations from normality. Abbreviations: OMIM: Online Mendelian Inheritance in Man. Indel: oligonucleotide insertion deletion variants. P: Pathogenic variant. LP: Likely pathogenic variant. SpliceAI Passing Filter: Noncoding variants predicted to cause cryptic splicing.38