Abstract
We investigate the variability in the dynamics of the disordered N-terminal domain of amyloid-β fibrils (Aβ), comprising residues 1–16 of Aβ1–40, due to post-translational modifications and mutations in the β-bend regions known to modulate aggregation properties. Using 2H static solid-state NMR approaches, we compare the dynamics in the wild-type Aβ fibrils in the threefold symmetric polymorph with the fibrils from three post-translational modification sequences: isoaspartate-D7, the phosphorylation of S8, and an N-terminal truncation ΔE3. Additional comparisons are made with the mutants in the β-bend region (residues 21–23) corresponding to the familial Osaka E22Δ deletion and D23N Iowa mutation. We also include the aggregates induced by Zn2+ ions. The dynamics are probed at the F4 and G9 positions. The main motional model involves two free states undergoing diffusion and conformational exchanges with the bound state in which the diffusion is quenched because of transient interactions involving fibril core and other intrastrand contacts. The fraction of the bound state increases in a sigmoidal fashion with a decrease in temperature. There is clear variability in the dynamics: the phosphorylation of S8 variant is the most rigid at the G9 site in line with structural studies, the ΔE3 fibrils are more flexible at the G9 site in line with the morphological fragmentation pattern, the Zn-induced aggregates are the most mobile, and the two β-bend mutants have the strongest changes at the F4 site toward higher rigidity. Overall, the changes underlie the potential role of conformational ensembles in setting the stage for aggregation-prone states.
Significance
In this work, we demonstrate variations of motions in the disordered region of amyloid-β fibrils due to modifications induced either by naturally occurring mutations or post-translational modifications. The results indicate a clear variability in the flexibility of the disordered domain and its potential role in modulating aggregation-prone states.
Introduction
Amyloid-β peptide (Aβ) aggregation remains to be viewed as one of the causative factors in Alzheimer’s disease (AD) as a part of the amyloid hypothesis (1, 2, 3, 4). It is believed that Aβ accumulation is a trigger that initiates a pathological cascade implicating τ protein, synuclein, and other aggregation-prone proteins (2,3). The structural polymorphism of amyloid fibrils is a challenging and potentially pathologically important factor in the molecular basis of AD (5, 6, 7, 8, 9). This polymorphism occurs at multiple levels (10), leading to many possible fibrillar structures from the same Aβ protein as well as the existence of multiple molecular variants of Aβ, including 1–39, 1–40, and 1–42, and the variants of these with post-translational modifications (PTMs) and mutations. The majority of PTMs occur in the flexible N-terminal region of the fibrils, encompassing residues 1–16, and are thought to trigger or accelerate the fibrillation of wild-type Aβ peptides (2,11,12). However, the pathological roles of modified Aβ in AD have not been determined in detail. It has been suggested that PTMs may be responsible for sporadic cases of AD, which encompass 90% of all patients (2,11). On the contrary, many familial-type mutations that cause the early onset of the disease occur around the β-bend region of Aβ, comprising residues 21–23, and are associated with altered folds, higher aggregation propensities, and higher toxicities (13, 14, 15, 16). Metal ions are also known to alter aggregation propensities. In particular, animal models show that Zn2+ coordination can play a crucial role in the formation of plaques in vivo (17, 18, 19). At the concentrations of what is found at the synapses, Zn2+ specifically binds to Aβ and promotes aggregation (20,21). The resulting multitude of the structural ensembles, combined with the differential seeding abilities of the variants, can have profound implications on the existence of the most aggressive forms as well as on the effectiveness of the initiation of the pathological cascades of the aggregation of other related proteins.
One of the ways in which to characterize the ensembles is probing the flexibility of the structures (10). We compare the dynamics in wild-type Aβ1–40 in one of its more toxic polymorphs possessing threefold symmetry (22) with two PTMs occurring around the “hot-spot” six to eight region (10) of the N-terminal domain, an N-terminal truncation PTM, and two mutants corresponding to familial mutations in the β-bend 21–23 region of the fibrils. In particular, we investigate PTMs corresponding to the phosphorylation of S8 (pS8) and aspartate to isoaspartate at position D7 (isoD7); the truncation variant corresponds to the cleavage of the peptide bond between positions 2 and 3 (ΔE3) as well as the Osaka E22Δ deletion mutant and D23N Iowa mutant of the β-bend region. Additionally, we include in the comparison the results for the aggregates formed by wild-type Aβ1–40 in the presence of Zn2+ ions (Zn). Below, we present the relevant backgrounds of these variants.
The E22Δ Aβ1–40 deletion mutation has been found to be more neurotoxic in rat primary neuron cultures than wild-type Aβ (23). Previous works have characterized the twofold symmetric structure displaying the in-register parallel β-sheet as well as determined the mass per length (MPL) measurements (24). The cross section of the fibril can be approximated by a rectangle defined by an additional intermolecular salt bridge between the Glu3 and Lys28 side chains, which seems to be absent in wild-type models. A unique feature of the fibrils is that they form extremely fast and show very low levels of thioflavin T fluorescence with no lag phase (25). The E22Δ variant has also been shown to efficiently cross-seed the wild-type protein (26).
One of the essential features of the D23N Aβ1–40 Iowa mutant is its ability to form an antiparallel β-sheet structure (27, 28, 29, 30). The polymorph of D23N with the antiparallel β-sheet structures are relatively short and curved fibril-like intermediates. They are metastable and eventually convert into mature D23N fibrils, which have parallel β-sheet structures similar to wild-type Aβ fibrils (28,30,31). The structural unit is a monomer in this case. The hydrophobic core is somewhat less compact than the wild-type.
The pS8 modification has recently been shown to have a strong cross-seeding ability for wild-type Aβ (32), and its structure as well as the dynamics of the core region have been determined by solid-state NMR (33). S8 phosphorylation has been shown to play an important role in late-onset sporadic AD (34,35) and is especially associated with the symptomatic pathology (36). It may also display increased nucleation-dependent fibrillation and enhance Aβ-mediated toxicity (11,37). The pS8 fibrils show the twofold symmetric structure with the in-register parallel β-sheet and striated-ribbon morphology similar to the twofold wild-type Aβ1–40 when grown under similar conditions (33). However, the fibrils are ∼2.5 nm wider. The highest structural distinction were observed in the N-terminal region. In particular, the variant showed strong intrastrand interactions between the N-terminus and the rest of the amyloid core. Additionally, the hydrophobic core packing was more pronounced than the wild-type, and the side-chain dynamics were more restricted in the core.
The isoD7 PTM has been found to increase aggregation propensities and zinc-dependent oligomerization (12,38,39). In parenchymal plaque core preparations, isoaspartate is the predominant form at position number 7 (12,40). In vitro, this type of isomerization has also been suggested to lead to an enhanced propensity to form β-sheets as well as an enhanced insolubility and resistance to enzymatic degradation (12,41, 42, 43). The intriguing study by Fonseca et al. suggested that the amount of isoD7 in plaques can serve as an indication of plaque age (44). Fukuda et al. (42) demonstrated that Aβ1–42 does not form fibrils in the presence of isoD7 modification. In this study, we work with the isoD7 sequence cross-seeded with wild-type Aβ1–40 to promote fibrillation. For this variant, as the structure is not available, we supplement an NMR investigation with the MPL characterization of these fibrils based on tilted beam transmission electron microscopy (TEM) measurements (45).
It has been suggested that the shortening of the N-terminus may lead to a higher aggregation propensity (12,46,47). One example is the 3-glutamate truncation (ΔE3) seen in plaques originating from late AD cases as well as in mouse-based models of AD (48). This truncation also serves as a starting point for the generation of the pyroglutamate-3 aggressive modification. The enhanced aggregation propensity in these PTMs may be at least partially attributed to the reduced polarity and enhanced hydrophobicity from the deletion of charged residues at N-termini (47). Morphologies and fibrillation kinetics have been found to be similar between the ΔE3 and pyroglutamate-3 variants and in general faster than the fibrillation of other N-terminal PTM subtypes (49). Scheidt et al. (50) utilized solid-state NMR to compare the structures of the pyroglutamate-3 and the wild-type fibrils and concluded that the cores are rather similar structurally, although the N-terminus shows an alteration in the dynamics based on the backbone order parameters probed at the Cα sites. This study did not tackle the ΔE3 precursor itself, however.
This work focuses on investigating the dynamics of the flexible N-terminal region of the fibrils, which is known to be important for the regulation of aggregation control (51, 52, 53, 54, 55, 56, 57). The general flexibility of noncore regions in Aβ has been demonstrated by multiple techniques, such as NMR (58, 59, 60, 61, 62, 63, 64, 65, 66), electron paramagnetic resonance (67), hydrogen-deuterium exchange (68, 69, 70), x-ray crystallography (71), and fluorescence spectroscopy (72). Fawzi et al. utilized solution NMR saturation transfer approaches to probe the binding of monomeric Aβ to the surface of protofibrils (73,74). The site-specific characterization of dynamics in aggregated forms remains challenging because of the difficulties of obtaining site-specific resolution. Many structures (including wild-type structures) (60) have been unable to include the first nine residues of Aβ because of the mobility of these sites.
We recently investigated the dynamics of the N-terminal subdomain in wild-type fibrils by probing the motions of hydrophobic side chains using 2H solid-state NMR methods combined with residue-specific deuteration (75,76). The N-terminal domain participates in a number of motional modes, including the overall diffusion-like motion at two different timescales and the conformational exchange process that could be attributed to the transient interactions of the N-terminal domain with the structured hydrophobic core. At 37°C, we observed a progressive freezing of the dynamics along the sequence, characterized by an increased fraction of the bound state and reduction in the overall diffusive motion of the domain.
The goal of this work is the detailed comparison of the N-terminal domain dynamics among the variants described above. The dynamics are probed at the F4 site belonging to the more flexible N-terminal end and the G9 site at which the dynamics are significantly reduced in wild-type fibrils (Fig. 1; (76)). The differential dynamics in all these variants underscores conformational sampling, which likely contributes to differentiating the aggregation propensities and cross-seeding aggressiveness.
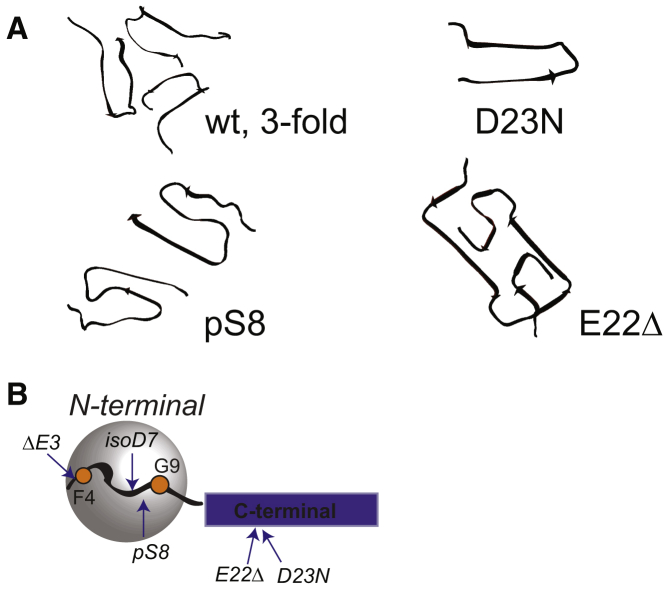
Figure 1.
(A) Quaternary structure for the variants studied in this work for which the structural coordinates are available: wild-type threefold symmetric structure (Protein Data Bank [PDB]: 2LMP) (22); pS8 PTM with the twofold symmetric structure (PDB: 6OC9) (33); monomeric structure of the Iowa D23N mutant with the antiparallel β-sheet structure (PDB: 2LNQ) (30); and Osaka E22Δ mutation with the rectangular cross section (PDB: 2MVX) (24). (B) Shown is a schematic representation of the domains of Aβ1–40, with the flexible N-terminal domain (residues 1–16, gray sphere) and C-terminal domain spanning the core (blue rectangle). Modified sequences investigated in this work are marked with arrows; ΔE3, isoD7, and pS8 are located in the N-terminal domain, and E22Δ and D23N are located in the β-bend region of the C-terminal domain. The sites of the deuterium isotope labels on the side chains of F4 and G9 are shown as orange dots. The labeling patterns are ring-D5 for F4 and αCD2 for G9. To see this figure in color, go online.
Materials and Methods
Peptide synthesis
The Aβ1−40 peptides were prepared by using solid-state peptide synthesis (performed by Thermo Fisher Scientific, Rockford, IL). Fluorenylmethyloxycarbonyl-phenylalanine-ring-d5 and fluorenylmethyloxycarbonyl-glycine-Cα-d2 were purchased from Cambridge Isotopes Laboratories (Andover, MA). Further details are listed in the Supporting Materials and Methods. The resulting peptides had isotopic labels in only one chosen residue, either F4 or G9.
Fibrils preparation
The pS8 and ΔE3 fibrils were grown following the generation seeding protocol developed previously (33,77). IsoD7 cross-seeded with wild-type Aβ1−40; the seeds consisted of the wild-type Aβ1–40 fibrils in the threefold symmetric polymorph and were used in 1:10 molar ratio of the wild-type Aβ1–40 seeds to the isoD7. E22Δ fibrils were prepared at 0.3 mg/mL concentration, and TEM images were taken immediately upon dissolving in the buffer and at 2–3 h and 24–30 h after dissolution. Details of these procedures are listed in the Supporting Materials and Methods. Preparation of the D23N fibrils with antiparallel β-sheet structure utilized a two-step seeding/filtration cycle that takes advantage of the differences in fibril formation rate between the parallel and antiparallel structures (30). Zn2+-induced aggregates were prepared as described in the earlier work (76). After pelleting and lyophilizing of the fibrils, the hydrated state with a water content of 200% by weight was achieved by pipetting deuterium-depleted H2O. The samples were packed in 5-mm NMR tubes (cut to 21-mm length) using Teflon tape to center the sample volume in the coil of the NMR probe.
NMR spectroscopy
Line shape experiments were performed with a quadrupole echo pulse sequence (78). The R1ρ experiments were performed at 9.4 T and 37°C using the methodology described in previous work (79) with the pulse sequence of Fig. S1. The relaxation decay curves M(t) corresponding to the integration of the central narrow component (up to the half-height intensity) were fitted to a single exponential function with an offset: . 2H QCPMG time domain measurements (Fig. S1) (80) were performed at 14.1 T field strength and 37°C. Integrated echo intensities were fitted to a single exponential function with no offset. Further details are listed in Supporting Materials and Methods.
Fitting of freezing curves
The data for pbound were fitted to the function as follows:
| (1) |
in which Tm is the midpoint of the freezing curve, σ is the characteristic width of the transition region, and a and b are the higher and lower temperature baselines, respectively.
Results
Morphological patterns
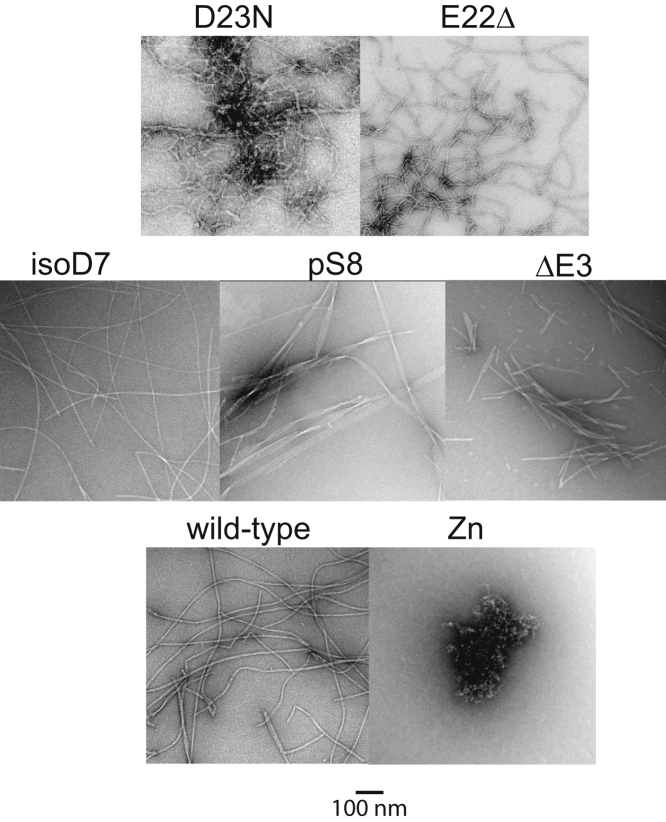
The fibrils of the Aβ1–40 variants (Fig. 1) exhibited different morphological patterns characterized by the negatively stained TEM technique (Fig. 2, additional images in Fig. S2), consistent with the findings in the literature and our previous work (25,30,32,33,81, 82, 83). The twisted threefold symmetric morphology of the wild-type fibrils (Fig. 2, previous analysis of the wild-type fibrils (76,83)) contrasts with the straighter and thicker fibril filaments for the pS8 fibrils, relatively short straight fibrils for the ΔE3, curvy and short D23N fibrils, long and curvy E22Δ fibrils, and amorphous Zn2+-induced aggregates. Two additional observations are worth noting; for the E22Δ mutant, the fibrils reached the final morphologies in 2–3 h (Fig. S3), in line with previous studies (25). In addition, the fragmentation pattern observed for the ΔE3 morphology was similar to that of the pyroglutamate-3 fibrils (Fig. S4; Wulff et al. (49)).
Figure 2.
Typical examples of the negatively stained TEM images of fibrils from different variants investigated in this study.
For the isoD7 variant cross-seeded with wild-type threefold Aβ, the morphology was largely conserved. To further quantify the dimensions of the fibrils resulting from cross-seeding isoD7 with the wild-type fibrils in the threefold polymorph, we performed a detailed MPL analysis. Specifically, we employed tilted beam TEM (dark-field imaging), which is effective for the statistical characterization of the MPL of fibrils (45,84,85). Both the twofold and the threefold polymorphs of native Aβ1–40 (60,81) have previously been analyzed using this technique (45). The results (Fig. S5) indicated that the predominant polymorph is closer in its MPL to the threefold wild-type variant (around 90% of the fibrils), with a small proportion of the twofold characteristics. Further studies are needed to assess whether this trend is general; in this study, we suffice with characterization of our bulk samples used in the NMR analysis.
Overview of the general model of N-terminal flexibility and approaches
For each of the fibril types, two samples were prepared for the NMR analysis: one containing a deuterium label at the F4 position, ring-D5, and another at the G9 position, αCD2.
We recently demonstrated that 2H rotating frame relaxation (R1ρ) and quadrupolar Carr-Purcell-Meiboom-Gill (QCPMG) transverse relaxation measurements provide a comprehensive view of conformational exchange processes (75). Although both these techniques are sensitive to motions on the biologically relevant μs-ms timescales, they provide complementary information on the motional modes.
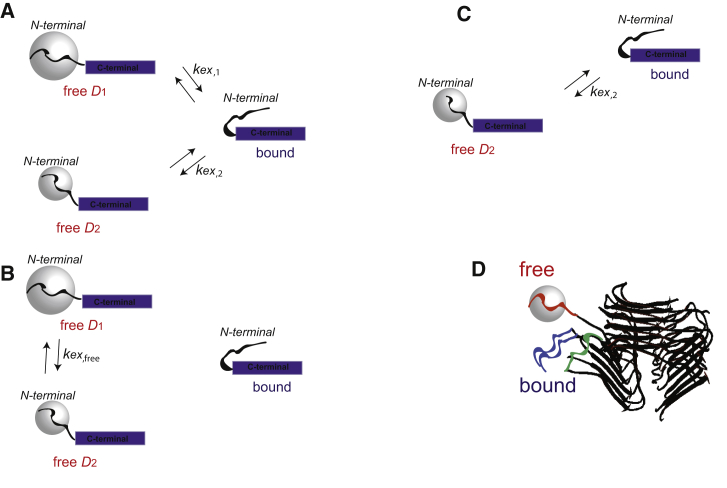
In this section, we review the motional models employed to fit all the experimental data. The motional model in Fig. 3 A, which was developed based on the data for the wild-type protein (75), includes three states of the N-terminal domain: two “free” mobile states in which the N-terminal undergoes large-angle diffusion motion and one bound state in which the diffusive motion in quenched, most likely because of transient interactions with the structured fibril core or transient intrastrand interactions with N-terminal domains of neighboring chains (Fig. 3 D). In the free state, restricted rotational diffusion is very pronounced, and we assume an isotropic scenario for the local motional axis (i.e., phenyl axis for the case of F4 and twofold rotation axis for the case of G9 CD2 group). This is not to be confused with global isotropic diffusion of macromolecules in solution—the fibrils are only 200% by weight hydrated with most of the water contributing to the hydration shell.
Figure 3.
Modeling schemes for the motions of the disordered N-terminal domain (residues 1–16) of the Aβ1–40 fibrils. (A) Three-state motional model in which the N-terminal domain (curved line) transiently interacts with the structured C-terminal domain (blue rectangle) (75). In the two free states, the N-terminal domain is assumed to undergo isotropic diffusion with the diffusion coefficients D1 and D2, , represented by the gray spheres, whereas in the bound state, the interactions quench this mode. The timescales of the interactions are given by the two chemical exchange rate constants kex,1 and kex,2, respectively. Most sites were fit with this full model. (B) Shown is a modification of model A for which there is no conformational exchange between the free and bound states on the experimental timescales but instead exchange between the two free states with the rate constant kex,free, which is applicable to the Zn2+ aggregates at the G9 site. (C) Shown is the submodel of (A) in which there is no fast free diffusion state, only a slow free diffusion state, which is applicable to the D23N sample at the G9 site. (D) Shown is a schematic representation of free (orange wiggly line) and bound (blue and green wiggly lines) states of N-terminal subdomain with the threefold wild-type fibrils structure. The threefold symmetric fibril core is taken from PDB: 2LMP (22) and is shown as a black ribbon diagram. The bound state can arise either because of transient interactions with the core (green lines) or transient tight stacking with the neighboring N-terminal strand (blue lines). In the free state (orange wiggly line), the domain can undergo relatively large-scale fluctuations, which are not possible in any of the bound states. To see this figure in color, go online.
The diffusion coefficients in the two free states differ by about two orders of magnitude difference between the two states, . Each of the free states undergoes a conformational exchange process with the bound state, with the rate constants of kex,1 and kex,2, respectively. For the G9 sites, in two of the variants, this model cannot fit the data. For the Zn-induced aggregates, no conformational exchange that involves the bound state is detected; rather, the exchange between the two free states is invoked (Fig. 3 B). For the D23N fibrils, only a single free state has the “slow” diffusion coefficient of D2 (Fig. 3 C).
The model of Fig. 3 A has been developed as the simplest scheme based on the extensive data on the dynamics of the wild-type Aβ fibrils. It does not incorporate any structural constraints other than a physical intuition on what would be, in general, consistent with the known structures of the fibrils. In particular, we do not assume any specific structural constraints for the transient interactions between the free and bound states. Additionally, the approximation of the isotropic diffusion and two different conformational states of the N-terminal domain is possibly reflecting a much more complex situation with anisotropic motions and an ensemble of multiple states, in analogy to the existence of tethered states found for monomers on the surface of protofibrils (73,74). Within the available dynamics data, invoking these more complex models would be a clear overfitting of the data. For consistency, we keep the model the same for all of the Aβ variants, unless a clear need arises to introduce modifications based on the data. We also note that the line shape data alone could be modeled based on an assumption of two static nonexchanging fractions with rigid and motionally narrowed tensors. However, our previous analysis of the longitudinal relaxation data for the wild-type fibrils in conjunction with R1ρ measurements precludes the use of such a model.
We show that depending on the quadrupolar interaction magnitude and timescale of motion, QCPMG and R1ρ experiments are sensitive to different conformational exchange processes. For the F4 residues, the effective quadrupolar tensor is narrow, with an effective quadrupolar coupling constant of Cq = 36.6 kHz after averaging over the rotameric motions of the side chain. On the contrary, for the G9, the effective Cq is 77.6 kHz after averaging over the two-site jumps of the CD2 group. As a result, the QCPMG experiment primarily probes the conformational exchange between the free state with a fast diffusion state and the bound state (D1, rate constant kex,1) for the F4 sites and the free slow diffusion state and the bound state (D2, rate constant kex,2) for the G9 sites. The situation is reversed for the R1ρ experiment; it is most sensitive to the conformational exchanges between the fast diffusion free state and bound state for the G9 sites as well as between the slow diffusion state and bound state for the F4 sites. The relative populations of the two free states are obtained by fitting the data to the full model in Fig. 3 A.
The fraction of the bound state pbound was taken from the line shape analysis. pbound as a function of temperature follows a sigmoidal curve indicative of a relatively abrupt freezing of the diffusion motions. This approach is not based on any assumptions regarding possible structural changes induced by lowering the temperature (i.e., pbound only reports on the cumulative increase of intra- and intermolecular interactions that lead to line broadening). The midpoint freezing temperature Tm (see Eq. 1) increases along the N-terminal domain chain for the wild-type protein. Although, for the residues close to the N-terminal end (A2 to H6 sites), it appears to be dictated by the freezing of the hydration layer (∼267 K), Tm increases along the sequence and is at 284 K for the G9 (76). pbound at the physiological temperature increases from 8 to 10% in the A2–H6 region, to 35% for the G9, and to 85% at the V12 site. Thus, to probe the dynamics of all the variants, we selected labels at the F4 and G9 locations to capture the details of the dynamics in the more flexible N-terminal domain region and the region in which the dynamics can be more restricted because of more pronounced interactions with the core. R1ρ measurements are conducted for four values of spin-locking field strength and QCPMG measurements for at least seven values of interpulse spacing. Between these relaxation data and pbound determination from the line shape analysis, there are enough data points to determine all model parameters.
2H NMR line shape analysis yields the fraction of the bound state
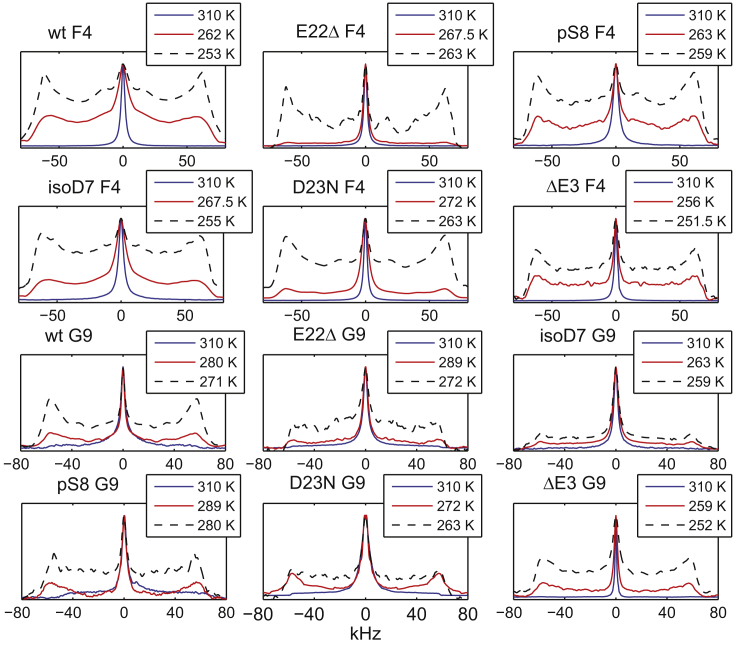
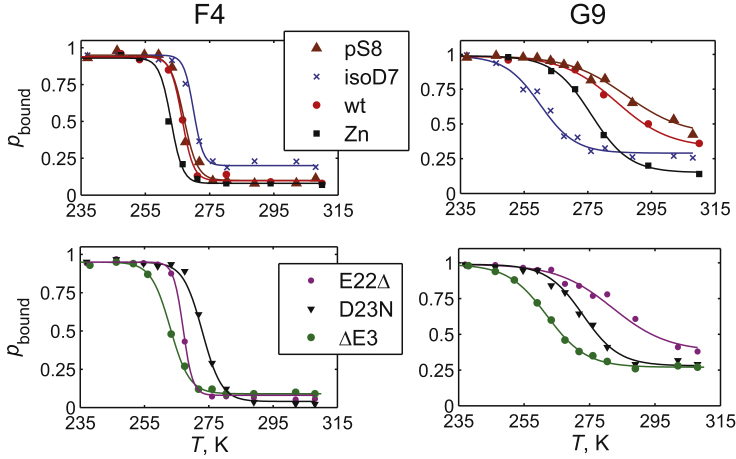
The 2H line shape is a simple one-dimensional experiment performed with a quadrupolar echo pulse sequence (78) that provides a wealth of information on the dynamics close to the timescale of the effective quadrupolar coupling constant. At the physiological temperature, for all variants, the line shapes significantly narrow and they approach solution-like line shapes (Fig. 4), indicating the presence of large-scale motions on the relevant timescale. With a decrease in temperature, the wide component of the powder pattern becomes apparent. The narrow component is attributed to the free states undergoing motions approximated by the isotropic diffusion. The wide component is attributed to the “bound” state, which lacks diffusive motions, presumably because of interactions with the rigid fibril core. The pbound for the F4 residue is quantified by decomposing the line shape into its Lorenzian and non-Lorenzian components (76), the latter of which is attributed to that of the bound state (see the examples in Fig. S6). Similar to the treatment of the wild-type fibrils (76), the analysis for the G9 sites is somewhat more complex, with pbound spanning the subset of the non-Lorenzian contribution corresponding to the rigid pattern. Interestingly, for the pS8 G9 site at high temperatures, it appears that the bound state has a different chemical shift than the free states (Fig. S6). The pbound for all residues (Fig. 5) follows the characteristic sigmoidal behavior observed for the wild-type protein, and this can be fitted to Eq. 1, which yields the midpoint of the freezing curves Tm and width of the transition σ, summarized in Fig. 6 along with the results for the fraction of the bound state pbound at the physiological temperature.
Figure 4.
Representative 2H static solid-state NMR line shape data in the hydrated fibril samples of Aβ1–40 at 310 K and two intermediate temperatures. The isotope labeling patterns are F4-ring-D5 and G9-CαD2. The data for the wild-type fibrils are taken from an earlier work (76). To see this figure in color, go online.
Figure 5.
pbound derived from the line shape decomposition as a function of temperature. The solid lines represent the fits according to Eq. 1. The data for the wild-type fibrils and Zn2+-induced aggregates are taken from an earlier work (76). To see this figure in color, go online.
Figure 6.
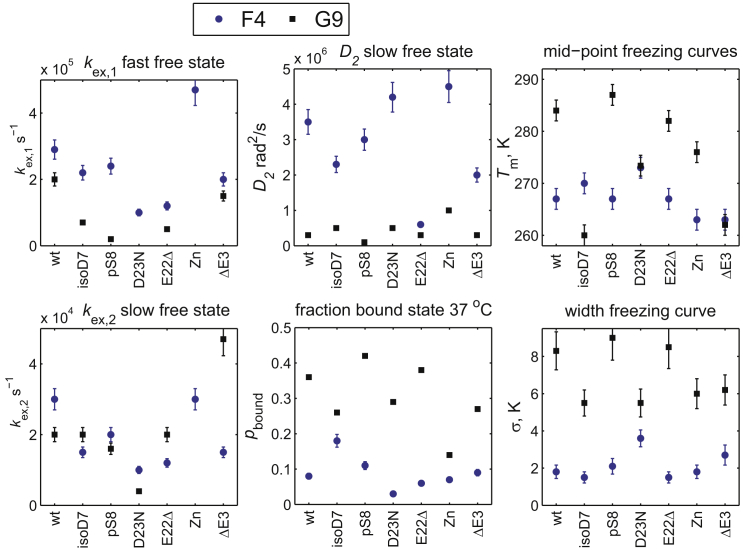
Summary of the model parameters according to the full model in Fig. 3A and the fits of the freezing curves. The rate constants are kex,1, kex,2, D2, pbound, Tm, and σ. D1 is in the fast limit and is at least 0.7–1 × 108 rad2/s. The relative percentages of the populations of the free to slow diffusion state are as follows: 70/30 for the F4 sites of wild-type and Zn; 60/40 for the F4 sites of ΔE3, isoD7, E22Δ, and pS8; 20/80 for the F4 site of D23N; and 5/95 to 10/90 for all the G9 sites, except for D23N for which it is zero (see the model in Fig. 3C) and Zn for which it is around 50/50. The Zn G9 site is fitted with the model in Fig. 3B with kex,free = 250 s−1. The errors were obtained by the inverse covariance matrix method calculated with Monte-Carlo sampling of the model parameters. Error bars smaller than the sizes of the symbols are not shown. To see this figure in color, go online.
The higher value of Tm signifies that the onset of the freezing of the diffusion motions starts at a higher temperature upon sample cooling. Most variants display a gap in Tm value between the F4 and G9 sites of 5–20°. For the ΔE3 fibrils, the value of Tm is 262–263 K for both sites; for D23N, it is 273 K for both sites (Fig. 6). The largest gap of 20° is for the pS8 fibrils. Interestingly, the width of the transition is maintained at a higher value for the G9 sites (average value of 7°) in comparison with F4 for all samples (with an average value of 2.1°). The higher values of σ may reflect stronger interactions with the core with a distribution of distances between the probed site and contacts within the core, leading to a wider transition. Further, the value of pbound at 37°C is consistently higher (0.14–0.42) than that of F4 (0.06–0.18) for all samples (Fig. 6). There is a clear variability in the fitted values of Tm between the different Aβ variants. For example, the pS8 variant has the highest Tm and highest pbound for the G9 site. This result is in line with a recent structural study (33) pointing to a more structured N-terminal domain. The Tm values at the G9 sites are much lower for the ΔE3, isoD7, and D23N variants. It is also of note that the line shapes at the physiological temperature are wider for the D23N variant for both sites, pointing to the possibility of a slower conformational exchange between the free and bound states, as detailed later in the text.
2H QCPMG and R1ρ relaxation measurements provide the conformational exchange rate constants and diffusion coefficients of the free states
To probe the timescales of the conformational exchange and obtain the most accurate values of the diffusion coefficients, it is necessary to perform more complicated measurements known to be sensitive to conformational exchange processes at the μs to ms timescale, such as the QCPMG and R1ρ measurements (86, 87, 88, 89) briefly described in Overview of the general model of N-terminal flexibility and approaches above. Our earlier work describes the technical details of these measurements and the fitting procedure based on the models using the full Liouvillian treatment of relaxation (75,79). Here, we focus on the results, which were obtained at the physiological temperature.
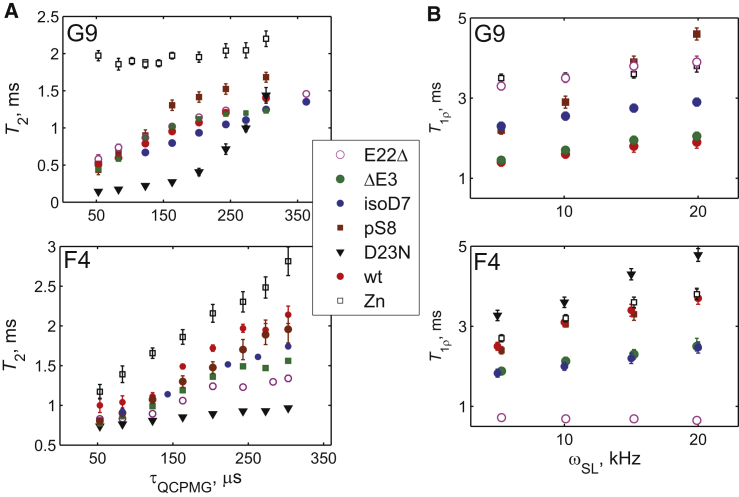
The 2H QCPMG experiment under static conditions (see Supporting Materials and Methods; Fig. S1 A) is performed by measuring the transverse relaxation rates as a function of the spacing of the refocusing pulses, τQCPMG, using a time domain approach. Motions on the timescale of τQCPMG interfere with the refocusing of magnetization, and the extent of this interference depends on the interpulse spacing. Interestingly, the dispersion profiles are the reverse of those traditionally seen in solution CPMG experiments, likely because of the anisotropic nature of the quadrupolar interactions (75,88). For the 2H R1ρ experiment under static conditions (Fig. S1 B) (79), refocusing is accomplished by the variable spin-locking field, ωSL.
Fig. 7 summarizes the experimental relaxation times, and Fig. S7 provides examples of the raw data. The variability between the samples is immediately apparent for both experiments. For the D23N sample at the G9 site, the R1ρ rate was too fast to be measured accurately.
Figure 7.
(A) Experimental transverse relaxation time T2 as a function of τQCPMG in the hydrated fibril samples of Aβ1–40 derived from the time domain QCPMG experiment, collected at 37°C and 14.1 T. (B) Experimental 2H T1ρ = 1/R1ρ relaxation times at 37°C and 9.4 T are shown. The errors in the rates were obtained by the inverse covariance matrix method. Error bars smaller than the sizes of the symbols are not shown. To see this figure in color, go online.
As noted, defining the magnitude of the quadrupolar interaction is important because this governs which timescales can be obtained from the two experiments. For the G9 site, we used the tensor with a quadrupolar coupling constant Cq of 77.6 kHz and the asymmetry parameter η = 1, as previously determined (76). The tensor corresponds to the effective value averaged over fast two-site jumps of the CD2 group, which followed from the longitudinal relaxation results for the wild-type samples. For F4, we started with the tensor corresponding to the averaging over the fast two-site π-flips of the ring, consistent with the longitudinal relaxation data. However, this tensor leads to rate constants for the F4 site in the wild-type fibrils that are orders of magnitude smaller than those found for the A2 and H6 sites, inferring the presence of additional motions that affect the QCPMG and R1ρ rates and lead to a lower effective Cq value. We thus invoke a two-site rotameric exchange involving the χ1 and χ2 dihedral angles. Specifically, we assume the existence of the rotamers found by Drobny and co-workers for one of the phenylalanine residues of the mineral recognition domain of the biomineralization protein salivary statherin adsorbed onto its native hydroxyapatite (90); conformer one has χ1 = 186.2° and χ2 = 88.6°, whereas conformer two has χ1 = 274.7° and χ2 = 54.8°. The ratio of the populations is taken as 1:1. In the fast limit, this leads to an effective tensor of Cq = 36.6 kHz and η of 0.94. This tensor provides diffusion coefficients and rate constants for the wild-type fibrils perfectly in line with those of the A2 and H6 residues, allowing us to conclude that the additional rotameric motion is a good approximation of the additional mode affecting the relaxation rates.
All the F4 sites followed the model in Fig. 3 A. For the G9 sites, there are two exceptions; as noted, D23N in the G9 site had a very fast R1ρ decay that could not be measured. Based on the analysis of the model for the wild-type fibrils (75), this signifies the almost complete absence of the fast diffusion state, and thus, this site is fitted with a single free state (Fig. 3 C). For the G9 site in the Zn2+-induced aggregates, the model in Fig. 3 A cannot be used; however, the model in which the exchange is dominated by the two free states (Fig. 3 B) is adequate. The QCPMG-T2 data are qualitatively different in this variant, with higher T2 values and, most importantly, negligible dispersion.
The variations in the experimental rates are reflected in the fitted values of D2, kex,1, and kex,2. Fig. 6 summarizes the main parameters of the model, and Fig. S8 provide examples of the fits of the experimental relaxation rates. The diffusion constant for the fast diffusion state D1 is in the fast limit, and thus, we can only state its lower limit, which falls to 0.7–1 × 108 rad2/s. The conformational exchange rate constant kex,1 for the wild-type for this state is 3 × 105 s−1 for the A2, F4, and H6 sites and 2 × 105 s−1 for the G9 sites. The variants’ range for kex,1 is 1.2–4.7 × 105 s−1 for F4 and 0.7–5 × 105 s−1 for G9. The slow diffusion state values of D2 are 3.5 × 106 rad2/s and 3.0∙× 105 rad2/s for the wild-type F4 and G9 sites, respectively. The range of the variants is 0.6–4.5 × 106 rad2/s for the F4 sites and 3–10 × 105 rad2/s for the G9 sites. The conformational exchange rate constant kex,2 is 3 × 104 s−1 for the A2, F4, and H6 sites in the wild-type protein and 2 × 104 s−1 for G9. The ranges of the variants are 1–3 × 104 s−1 and 0.4–4.7 × 104 s−1 for the F4 and G9 sites, respectively. For the G9 site of the Zn2+-induced aggregates, the exchange rate constant between the two free states (see model in Fig. 3 B) is 250 s−1. The relative percentage of the fast to slow diffusion free states at the F4 site is the largest for the wild-type and Zn variants (70/30%) and decreases somewhat (60/40%) for the other variants, except for D23N in which it is drastically reduced to 20/80%. The fast state “freezes” to 5–10% for all samples for the G9 site and to zero for the D23N site, whereas it remains at around 50/50% for the Zn G9 site.
Discussion
The N-terminal domain remains flexible for all the considered variants, and the models of motions are generally similar; however, the extent of this flexibility varies among sequences. The three-state model involving two free states undergoing isotropic diffusion and one bound state (Fig. 3 A) is applicable to most sites, with the exceptions noted for the D23N fibrils and Zn2+-induced aggregates at the G9 position. Relatively significant variations exist in the parameters of the motions such as the conformational exchange rate constants and diffusion coefficients of the slower diffusion free states. All the protein sequences also display sigmoidal behavior in the fraction of the bound state upon lowering the temperature as well as variability in the midpoints of the freezing curves.
The patterns of mobility modulations in all the variants are complex; however, certain essential features emerge from the analysis, as summarized in Table 1. The pS8 fibrils, which were determined to have accelerated seeded fibrillation kinetics and enhanced toxicities as well as a more rigid core and a more structured N-terminus than the wild-type protein using structural techniques (33), appear to have the most rigid dynamics. Because of a larger extent of intrastrand contacts in the structure between the N-terminal domain and the core for the fibrils in pS8, one might also expect a larger value of σ for the G9 site, but this is not the case based on the experimental data. The ΔE3 fibrils tend toward higher mobility at the G9 site in line with the morphological fragmentation. A morphologically similar fragmentation is observed for the fibrils in the presence of the cyclization to the pyroglutamate-3 (49) (see also Fig. S4), suggesting that similar changes in the N-terminal dynamics are likely to be present for this PTM as well. There is no clear pattern for the isoD7 fibrils cross-seeded with wild-type Aβ, which has a mixture of changed dynamical features. The most drastic change is seen for the value of Tm at the G9 site, which is ∼15–20° lower than for most of the other types of fibrils. This underscores the need for high resolution structural data for the isoD7 fibrils. Based on the structural, kinetics, morphological, and N-terminal dynamics information available for the ΔE3, pyroglutamate-3, pS8, and isoD7 variants (11,32,33,42,49,50,91), all these PTMs are likely to have different cross-seeding abilities, which remains to be elucidated in follow-up work.
Table 1.
Summary of the Most Essential Changes (with at Least Three SDs) Compared with the Wild-Type
| Variant | Parameters Significantly Different from the Wild-Type Fibrils | Overall Conclusion |
|---|---|---|
| pS8 | kex,1-G9, D2-G9, Tm-G9 | Most rigid of all fibrils at G9, in line with structural information |
| ΔE3 | Tendency toward higher mobility at G9, in line with morphology fragmentation | |
| isoD7 cross-seeded with wild-type | pbound-F4, kex,1-G9, | Mixture of higher and lower mobility features; no clear patterns |
| D23N | Tm-F4; kex,1-F4, σ-F4, kex,2-F4, , no fast diffusion state for G9, kex,2-G9 | Most differences of all fibrils at F4 (more rigid) |
| E22Δ | D2-F4, kex,2-F4, kex,1-F4, kex,1-G9 | Significant differences at F4 toward rigidity |
| Zn2+ aggregates | Overall enhanced mobility but more so at G9 site |
The change toward the more rigid value is underlined with the solid lines; the change toward more enhanced dynamics is underlined with the dashed lines.
For the β-bend region mutation, the most interesting observation is that the modulation of the dynamics compared with the wild-type protein appears to occur closer to the N-terminal end. Indeed, many of the changes toward the restriction of the dynamics can be seen at the F4 site rather than the G9 site. The salt bridge involving E3 may play a role in this restriction for the case of E22Δ. However, no such salt bridge was seen for the antiparallel β-sheet structure of the D23N fibrils. In general, these mutations lead to a less compact fibrils core (30), and thus, the restriction of the dynamics cannot be explained solely by the structural features in this case. Moreover, the pbound value at the F4 site of the D23N fibrils is considerably lower than those of the other residues (Fig. 6). However, this does not indicate an enhancement of the dynamics compared with the other fibrils as the line shapes are significantly wider because of the slow exchange dynamics. The Zn2+-induced aggregates have a clear pattern of overall enhanced mobility in the N-terminus with more pronounced changes at the G9 sites, which is in line with the expectation that the hydrophobic core is less defined in these types of amorphous aggregates, leading to weaker interactions with the N-terminal domain.
Overall, these changes indicate that the dynamics information is complementary to the structural data, and often, it is not straightforward to predict in which direction the dynamics will change upon structural modifications. The results imply that the presence of intricate conformational ensembles can play an important role in the onset of aggregation-prone states and in the differentiation of cross-seeding aggressiveness, thus contributing to the overall polymorphism paradigm. Similar conformational exchange processes may be present in the toxic oligomeric forms as conformational exchange has been detected for the monomer on the surface of the protofibrils by solution NMR measurements (73,74). Our study underlines the need to consider both structural and dynamics information when characterizing the disordered regions of the various toxic variants of Aβ and other aggregation-prone proteins along AD pathways.
Author Contributions
L.V. conceived and coordinated the project, designed and performed NMR experiments, and wrote the article with input from all the authors. D.F.A. prepared some of the fibrils samples and performed TEM analysis with contribution from B.K. and performed some of the NMR measurements. Z.-w.H. prepared pS8 fibrils. W.Q. prepared D23N fibrils and performed MPL TEM measurements and analysis. D.O. performed modeling with input from L.V. R.F. contributed to the development of NMR methodology and data collection. All authors discussed the results.
Acknowledgments
This work was supported by a National Institutes of Health grant 1R15-GM111681 and National Science Foundation grant 1726947. Some of the experiments were performed at the National High Magnetic Field Laboratory, which is supported by National Science Foundation Cooperative Agreement NSF/DMR-1644779, the State of Florida, and the United States Department of Energy.
Editor: Keir Neuman.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.09.004.
Supporting Citations
References (92, 93, 94, 95, 96, 97, 98, 99, 100, 101) appear in the Supporting Material.
Supporting Material
References
- 1.Hardy J., Selkoe D.J. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Barykin E.P., Mitkevich V.A., Makarov A.A. Amyloid β modification: a key to the sporadic Alzheimer’s disease? Front. Genet. 2017;8:58. doi: 10.3389/fgene.2017.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musiek E.S., Holtzman D.M. Three dimensions of the amyloid hypothesis: time, space and ‘wingmen’. Nat. Neurosci. 2015;18:800–806. doi: 10.1038/nn.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardy J.A., Higgins G.A. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 5.Paravastu A.K., Leapman R.D., Tycko R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tycko R. Amyloid polymorphism: structural basis and neurobiological relevance. Neuron. 2015;86:632–645. doi: 10.1016/j.neuron.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiang W., Yau W.M., Tycko R. Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature. 2017;541:217–221. doi: 10.1038/nature20814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J.X., Qiang W., Tycko R. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen J., Mahler J., Jucker M. Amyloid polymorphisms constitute distinct clouds of conformational variants in different etiological subtypes of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2017;114:13018–13023. doi: 10.1073/pnas.1713215114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubin E., van Nuland N.A.J., Pauwels K. Transient dynamics of Aβ contribute to toxicity in Alzheimer’s disease. Cell. Mol. Life Sci. 2014;71:3507–3521. doi: 10.1007/s00018-014-1634-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roher A.E., Kokjohn T.A., Beach T.G. APP/Aβ structural diversity and Alzheimer’s disease pathogenesis. Neurochem. Int. 2017;110:1–13. doi: 10.1016/j.neuint.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kummer M.P., Heneka M.T. Truncated and modified amyloid-beta species. Alzheim. Res. Ther. 2014;6:28. doi: 10.1186/alzrt258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gessel M.M., Bernstein S., Bowers M.T. Familial Alzheimer’s disease mutations differentially alter amyloid β-protein oligomerization. ACS Chem. Neurosci. 2012;3:909–918. doi: 10.1021/cn300050d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatami A., Monjazeb S., Glabe C.G. Familial Alzheimer’s disease mutations within the amyloid precursor protein alter the aggregation and conformation of the amyloid-β peptide. J. Biol. Chem. 2017;292:3172–3185. doi: 10.1074/jbc.M116.755264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang H., Arce F.T., Nussinov R. Familial Alzheimer’s disease Osaka mutant (ΔE22) β-barrels suggest an explanation for the different Aβ1-40/42 preferred conformational states observed by experiment. J. Phys. Chem. B. 2013;117:11518–11529. doi: 10.1021/jp405389n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumketner A., Bernstein S.L., Shea J.E. Structure of the 21-30 fragment of amyloid beta-protein. Protein Sci. 2006;15:1239–1247. doi: 10.1110/ps.062076806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bush A.I. The metal theory of Alzheimer’s disease. J. Alzheimers Dis. 2013;33(Suppl 1):S277–S281. doi: 10.3233/JAD-2012-129011. [DOI] [PubMed] [Google Scholar]
- 18.Hamley I.W. The amyloid beta peptide: a chemist’s perspective. Role in Alzheimer’s and fibrillization. Chem. Rev. 2012;112:5147–5192. doi: 10.1021/cr3000994. [DOI] [PubMed] [Google Scholar]
- 19.Ayton S., Lei P., Bush A.I. Biometals and their therapeutic implications in Alzheimer’s disease. Neurotherapeutics. 2015;12:109–120. doi: 10.1007/s13311-014-0312-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suh S.W., Jensen K.B., Frederickson C.J. Histochemically-reactive zinc in amyloid plaques, angiopathy, and degenerating neurons of Alzheimer’s diseased brains. Brain Res. 2000;852:274–278. doi: 10.1016/s0006-8993(99)02096-x. [DOI] [PubMed] [Google Scholar]
- 21.Lovell M.A., Robertson J.D., Markesbery W.R. Copper, iron and zinc in Alzheimer’s disease senile plaques. J. Neurol. Sci. 1998;158:47–52. doi: 10.1016/s0022-510x(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 22.Paravastu A.K., Qahwash I., Tycko R. Seeded growth of beta-amyloid fibrils from Alzheimer’s brain-derived fibrils produces a distinct fibril structure. Proc. Natl. Acad. Sci. USA. 2009;106:7443–7448. doi: 10.1073/pnas.0812033106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ovchinnikova O.Y., Finder V.H., Glockshuber R. The Osaka FAD mutation E22Δ leads to the formation of a previously unknown type of amyloid β fibrils and modulates Aβ neurotoxicity. J. Mol. Biol. 2011;408:780–791. doi: 10.1016/j.jmb.2011.02.049. [DOI] [PubMed] [Google Scholar]
- 24.Schütz A.K., Vagt T., Meier B.H. Atomic-resolution three-dimensional structure of amyloid β fibrils bearing the Osaka mutation. Angew. Chem. Int. Ed. Engl. 2015;54:331–335. doi: 10.1002/anie.201408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cloe A.L., Orgel J.P., Meredith S.C. The Japanese mutant Aβ (ΔE22-Aβ(1-39)) forms fibrils instantaneously, with low-thioflavin T fluorescence: seeding of wild-type Aβ(1-40) into atypical fibrils by ΔE22-Aβ(1-39) Biochemistry. 2011;50:2026–2039. doi: 10.1021/bi1016217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomiyama T., Nagata T., Mori H. A new amyloid beta variant favoring oligomerization in Alzheimer’s-type dementia. Ann. Neurol. 2008;63:377–387. doi: 10.1002/ana.21321. [DOI] [PubMed] [Google Scholar]
- 27.Grabowski T.J., Cho H.S., Greenberg S.M. Novel amyloid precursor protein mutation in an Iowa family with dementia and severe cerebral amyloid angiopathy. Ann. Neurol. 2001;49:697–705. doi: 10.1002/ana.1009. [DOI] [PubMed] [Google Scholar]
- 28.Qiang W., Yau W.M., Tycko R. Structural evolution of Iowa mutant β-amyloid fibrils from polymorphic to homogeneous states under repeated seeded growth. J. Am. Chem. Soc. 2011;133:4018–4029. doi: 10.1021/ja109679q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tycko R., Sciarretta K.L., Meredith S.C. Evidence for novel beta-sheet structures in Iowa mutant beta-amyloid fibrils. Biochemistry. 2009;48:6072–6084. doi: 10.1021/bi9002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiang W., Yau W.M., Tycko R. Antiparallel β-sheet architecture in Iowa-mutant β-amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2012;109:4443–4448. doi: 10.1073/pnas.1111305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sgourakis N.G., Yau W.M., Qiang W. Modeling an in-register, parallel “Iowa” aβ fibril structure using solid-state NMR data from labeled samples with rosetta. Structure. 2015;23:216–227. doi: 10.1016/j.str.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 32.Hu Z.W., Ma M.R., Li Y.M. Phosphorylation at Ser8 as an intrinsic regulatory switch to regulate the morphologies and structures of Alzheimer’s 40-residue β-Amyloid (Aβ40) fibrils. J. Biol. Chem. 2017;292:2611–2623. doi: 10.1074/jbc.M116.757179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Z.W., Vugmeyster L., Qiang W. Molecular structure of an N-terminal phosphorylated β-amyloid fibril. Proc. Natl. Acad. Sci. USA. 2019;116:11253–11258. doi: 10.1073/pnas.1818530116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S., Walter J. Phosphorylation of amyloid beta (Aβ) peptides - a trigger for formation of toxic aggregates in Alzheimer’s disease. Aging (Albany N.Y.) 2011;3:803–812. doi: 10.18632/aging.100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S., Wirths O., Walter J. Early intraneuronal accumulation and increased aggregation of phosphorylated Abeta in a mouse model of Alzheimer’s disease. Acta Neuropathol. 2013;125:699–709. doi: 10.1007/s00401-013-1107-8. [DOI] [PubMed] [Google Scholar]
- 36.Rijal Upadhaya A., Kosterin I., Thal D.R. Biochemical stages of amyloid-β peptide aggregation and accumulation in the human brain and their association with symptomatic and pathologically preclinical Alzheimer’s disease. Brain. 2014;137:887–903. doi: 10.1093/brain/awt362. [DOI] [PubMed] [Google Scholar]
- 37.Kumar S., Rezaei-Ghaleh N., Walter J. Extracellular phosphorylation of the amyloid β-peptide promotes formation of toxic aggregates during the pathogenesis of Alzheimer’s disease. EMBO J. 2011;30:2255–2265. doi: 10.1038/emboj.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsvetkov P.O., Popov I.A., Kozin S.A. Isomerization of the Asp7 residue results in zinc-induced oligomerization of Alzheimer’s disease amyloid beta(1-16) peptide. ChemBioChem. 2008;9:1564–1567. doi: 10.1002/cbic.200700784. [DOI] [PubMed] [Google Scholar]
- 39.Kozin S.A., Cheglakov I.B., Makarov A.A. Peripherally applied synthetic peptide isoAsp7-Aβ(1-42) triggers cerebral β-amyloidosis. Neurotox. Res. 2013;24:370–376. doi: 10.1007/s12640-013-9399-y. [DOI] [PubMed] [Google Scholar]
- 40.Roher A.E., Lowenson J.D., Ball M.J. Structural alterations in the peptide backbone of beta-amyloid core protein may account for its deposition and stability in Alzheimer’s disease. J. Biol. Chem. 1993;268:3072–3083. [PubMed] [Google Scholar]
- 41.Kuo Y.M., Webster S., Roher A.E. Irreversible dimerization/tetramerization and post-translational modifications inhibit proteolytic degradation of A beta peptides of Alzheimer’s disease. Biochim. Biophys. Acta. 1998;1406:291–298. doi: 10.1016/s0925-4439(98)00014-3. [DOI] [PubMed] [Google Scholar]
- 42.Fukuda H., Shimizu T., Shirasawa T. Synthesis, aggregation, and neurotoxicity of the Alzheimer’s Abeta1-42 amyloid peptide and its isoaspartyl isomers. Bioorg. Med. Chem. Lett. 1999;9:953–956. doi: 10.1016/s0960-894x(99)00121-3. [DOI] [PubMed] [Google Scholar]
- 43.Fabian H., Szendrei G.I., Otvös L., Jr. Synthetic post-translationally modified human A beta peptide exhibits a markedly increased tendency to form beta-pleated sheets in vitro. Eur. J. Biochem. 1994;221:959–964. doi: 10.1111/j.1432-1033.1994.tb18811.x. [DOI] [PubMed] [Google Scholar]
- 44.Fonseca M.I., Head E., Tenner A.J. The presence of isoaspartic acid in beta-amyloid plaques indicates plaque age. Exp. Neurol. 1999;157:277–288. doi: 10.1006/exnr.1999.7058. [DOI] [PubMed] [Google Scholar]
- 45.Chen B., Thurber K.R., Tycko R. Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc. Natl. Acad. Sci. USA. 2009;106:14339–14344. doi: 10.1073/pnas.0907821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pike C.J., Overman M.J., Cotman C.W. Amino-terminal deletions enhance aggregation of beta-amyloid peptides in vitro. J. Biol. Chem. 1995;270:23895–23898. doi: 10.1074/jbc.270.41.23895. [DOI] [PubMed] [Google Scholar]
- 47.Dunys J., Valverde A., Checler F. Are N- and C-terminally truncated Aβ species key pathological triggers in Alzheimer’s disease? J. Biol. Chem. 2018;293:15419–15428. doi: 10.1074/jbc.R118.003999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Güntert A., Döbeli H., Bohrmann B. High sensitivity analysis of amyloid-beta peptide composition in amyloid deposits from human and PS2APP mouse brain. Neuroscience. 2006;143:461–475. doi: 10.1016/j.neuroscience.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 49.Wulff M., Baumann M., Fändrich M. Enhanced fibril fragmentation of N-terminally truncated and pyroglutamyl-modified Aβ peptides. Angew. Chem. Int. Ed. Engl. 2016;55:5081–5084. doi: 10.1002/anie.201511099. [DOI] [PubMed] [Google Scholar]
- 50.Scheidt H.A., Adler J., Huster D. Fibrils of truncated pyroglutamyl-modified Aβ peptide exhibit a similar structure as wildtype mature Aβ fibrils. Sci. Rep. 2016;6:33531. doi: 10.1038/srep33531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazzitelli S., Filipello F., Matteoli M. Amyloid-β 1-24 C-terminal truncated fragment promotes amyloid-β 1-42 aggregate formation in the healthy brain. Acta Neuropathol. Commun. 2016;4:110. doi: 10.1186/s40478-016-0381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brännström K., Öhman A., Olofsson A. The N-terminal region of amyloid β controls the aggregation rate and fibril stability at low pH through a gain of function mechanism. J. Am. Chem. Soc. 2014;136:10956–10964. doi: 10.1021/ja503535m. [DOI] [PubMed] [Google Scholar]
- 53.Morris C., Cupples S., Du D. N-terminal charged residues of Amyloid-β peptide modulate amyloidogenesis and interaction with lipid membrane. Chemistry. 2018;24:9494–9498. doi: 10.1002/chem.201801805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nussbaum J.M., Schilling S., Bloom G.S. Prion-like behaviour and tau-dependent cytotoxicity of pyroglutamylated amyloid-β. Nature. 2012;485:651–655. doi: 10.1038/nature11060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kummer M.P., Hermes M., Heneka M.T. Nitration of tyrosine 10 critically enhances amyloid β aggregation and plaque formation. Neuron. 2011;71:833–844. doi: 10.1016/j.neuron.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 56.Rezaei-Ghaleh N., Amininasab M., Zweckstetter M. Phosphorylation modifies the molecular stability of β-amyloid deposits. Nat. Commun. 2016;7:11359. doi: 10.1038/ncomms11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu L., Nussinov R., Ma B. Allosteric stabilization of the amyloid-β peptide hairpin by the fluctuating N-terminal. Chem. Commun. (Camb.) 2016;52:1733–1736. doi: 10.1039/c5cc08107f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheidt H.A., Morgado I., Huster D. Dynamics of amyloid β fibrils revealed by solid-state NMR. J. Biol. Chem. 2012;287:2017–2021. doi: 10.1074/jbc.M111.308619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lührs T., Ritter C., Riek R. 3D structure of Alzheimer’s amyloid-beta(1-42) fibrils. Proc. Natl. Acad. Sci. USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petkova A.T., Yau W.M., Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bertini I., Gonnelli L., Nesi A. A new structural model of Aβ40 fibrils. J. Am. Chem. Soc. 2011;133:16013–16022. doi: 10.1021/ja2035859. [DOI] [PubMed] [Google Scholar]
- 62.Olofsson A., Sauer-Eriksson A.E., Ohman A. The solvent protection of alzheimer amyloid-beta-(1-42) fibrils as determined by solution NMR spectroscopy. J. Biol. Chem. 2006;281:477–483. doi: 10.1074/jbc.M508962200. [DOI] [PubMed] [Google Scholar]
- 63.Whittemore N.A., Mishra R., Serpersu E.H. Hydrogen-deuterium (H/D) exchange mapping of Abeta 1-40 amyloid fibril secondary structure using nuclear magnetic resonance spectroscopy. Biochemistry. 2005;44:4434–4441. doi: 10.1021/bi048292u. [DOI] [PubMed] [Google Scholar]
- 64.Prade E., Barucker C., Reif B. Sulindac sulfide induces the formation of large oligomeric aggregates of the Alzheimer’s disease Amyloid-β peptide which exhibit reduced neurotoxicity. Biochemistry. 2016;55:1839–1849. doi: 10.1021/acs.biochem.5b01272. [DOI] [PubMed] [Google Scholar]
- 65.Linser R., Sarkar R., Reif B. Dynamics in the solid-state: perspectives for the investigation of amyloid aggregates, membrane proteins and soluble protein complexes. J. Biomol. NMR. 2014;59:1–14. doi: 10.1007/s10858-014-9822-6. [DOI] [PubMed] [Google Scholar]
- 66.Wang T., Jo H., Hong M. Water distribution, dynamics, and interactions with Alzheimer’s β-amyloid fibrils investigated by solid-state NMR. J. Am. Chem. Soc. 2017;139:6242–6252. doi: 10.1021/jacs.7b02089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Török M., Milton S., Langen R. Structural and dynamic features of Alzheimer’s Abeta peptide in amyloid fibrils studied by site-directed spin labeling. J. Biol. Chem. 2002;277:40810–40815. doi: 10.1074/jbc.M205659200. [DOI] [PubMed] [Google Scholar]
- 68.Kheterpal I., Zhou S., Wetzel R. Abeta amyloid fibrils possess a core structure highly resistant to hydrogen exchange. Proc. Natl. Acad. Sci. USA. 2000;97:13597–13601. doi: 10.1073/pnas.250288897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang S.S., Tobler S.A., Fernandez E.J. Hydrogen exchange-mass spectrometry analysis of beta-amyloid peptide structure. Biochemistry. 2003;42:9507–9514. doi: 10.1021/bi0342766. [DOI] [PubMed] [Google Scholar]
- 70.Kheterpal I., Chen M., Wetzel R. Structural differences in Abeta amyloid protofibrils and fibrils mapped by hydrogen exchange--mass spectrometry with on-line proteolytic fragmentation. J. Mol. Biol. 2006;361:785–795. doi: 10.1016/j.jmb.2006.06.066. [DOI] [PubMed] [Google Scholar]
- 71.Sawaya M.R., Sambashivan S., Eisenberg D. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 72.Liu H., Morris C., Du D. Residue-specific dynamics and local environmental changes in Aβ40 oligomer and fibril formation. Angew. Chem. Int. Ed. Engl. 2018;57:8017–8021. doi: 10.1002/anie.201802490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fawzi N.L., Libich D.S., Clore G.M. Characterizing methyl-bearing side chain contacts and dynamics mediating amyloid β protofibril interactions using 13C(methyl)-DEST and lifetime line broadening. Angew. Chem. Int. Ed. Engl. 2014;53:10345–10349. doi: 10.1002/anie.201405180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fawzi N.L., Ying J., Clore G.M. Atomic-resolution dynamics on the surface of amyloid-β protofibrils probed by solution NMR. Nature. 2011;480:268–272. doi: 10.1038/nature10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vugmeyster L., Au D.F., Fu R. Deuteron solid-state NMR relaxation measurements reveal two distinct conformational exchange processes in the disordered N-terminal domain of amyloid-β fibrils. Chemphyschem. 2019;20:1680–1689. doi: 10.1002/cphc.201900363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Au D.F., Ostrovsky D., Vugmeyster L. Solid-state NMR reveals a comprehensive view of the dynamics of the flexible, disordered N-terminal domain of amyloid-β fibrils. J. Biol. Chem. 2019;294:5840–5853. doi: 10.1074/jbc.RA118.006559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiao Y., Ma B., Ishii Y. Aβ(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat. Struct. Mol. Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vold R.L., Vold R.R. Deuterium relaxation in molecular solids. In: Warren W., editor. Advances in Magnetic and Optical Resonance. Acadenic Press; 1991. pp. 85–171. [Google Scholar]
- 79.Vugmeyster L., Ostrovsky D. Deuterium rotating frame NMR relaxation measurements in the solid state under static conditions for quantification of dynamics. Chemphyschem. 2019;20:333–342. doi: 10.1002/cphc.201800454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Larsen F.H., Jakobsen H.J., Nielsen N.C. High-field QCPMG-MAS NMR of half-integer quadrupolar nuclei with large quadrupole couplings. Mol. Phys. 1998;95:1185–1195. doi: 10.1006/jmre.1997.1341. [DOI] [PubMed] [Google Scholar]
- 81.Petkova A.T., Leapman R.D., Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 82.Noy D., Solomonov I., Sagi I. Zinc-amyloid beta interactions on a millisecond time-scale stabilize non-fibrillar Alzheimer-related species. J. Am. Chem. Soc. 2008;130:1376–1383. doi: 10.1021/ja076282l. [DOI] [PubMed] [Google Scholar]
- 83.Vugmeyster L., Clark M.A., Hoatson G.L. Flexibility and solvation of amyloid-β hydrophobic core. J. Biol. Chem. 2016;291:18484–18495. doi: 10.1074/jbc.M116.740530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iadanza M.G., Jackson M.P., Ranson N.A. MpUL-multi: software for calculation of amyloid fibril mass per unit length from TB-TEM images. Sci. Rep. 2016;6:21078. doi: 10.1038/srep21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kurouski D. Supramolecular organization of amyloid fibrils. In: Fernandez-Escamilla A.-M., editor. Exploring New Findings on Amyloidosis. InTech; 2016. [Google Scholar]
- 86.Palmer A.G., III Chemical exchange in biomacromolecules: past, present, and future. J. Magn. Reson. 2014;241:3–17. doi: 10.1016/j.jmr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schanda P., Ernst M. Studying dynamics by magic-angle spinning solid-state NMR spectroscopy: principles and applications to biomolecules. Prog. Nucl. Magn. Reson. Spectrosc. 2016;96:1–46. doi: 10.1016/j.pnmrs.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tollinger M., Sivertsen A.C., Schanda P. Site-resolved measurement of microsecond-to-millisecond conformational-exchange processes in proteins by solid-state NMR spectroscopy. J. Am. Chem. Soc. 2012;134:14800–14807. doi: 10.1021/ja303591y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krushelnitsky A., Gauto D., Saalwächter K. Microsecond motions probed by near-rotary-resonance R1ρ15N MAS NMR experiments: the model case of protein overall-rocking in crystals. J. Biomol. NMR. 2018;71:53–67. doi: 10.1007/s10858-018-0191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li K., Emani P.S., Drobny G.P. A study of phenylalanine side-chain dynamics in surface-adsorbed peptides using solid-state deuterium NMR and rotamer library statistics. J. Am. Chem. Soc. 2014;136:11402–11411. doi: 10.1021/ja504677d. [DOI] [PubMed] [Google Scholar]
- 91.Adler J., Scheidt H.A., Huster D. Local interactions influence the fibrillation kinetics, structure and dynamics of Aβ(1-40) but leave the general fibril structure unchanged. Phys. Chem. Chem. Phys. 2014;16:7461–7471. doi: 10.1039/c3cp54501f. [DOI] [PubMed] [Google Scholar]
- 92.Harris J.R., Horne J.W. Negative staining. In: Harris J.R., editor. Electron Microscopy in Biology. IRL Press; 1991. pp. 203–228. [Google Scholar]
- 93.Dubochet J., Groom M., Mueller-Neuteboom S. The mounting of macromolecules for electron microscopy with particular reference to surface phenomena and the treatment of support films by glow discharge. In: Barrer R., Cosslett V.E., editors. Advances in Optical and Electron Microscopy. Academic Press; 1982. pp. 107–135. [Google Scholar]
- 94.Gor’kov P.L., Chekmenev E.Y., Brey W.W. Using low-E resonators to reduce RF heating in biological samples for static solid-state NMR up to 900 MHz. J. Magn. Reson. 2007;185:77–93. doi: 10.1016/j.jmr.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 95.Persson P.-O., Strang G. A simple mesh generator in MATLAB. SIAM Rev. 2004;46:329–345. [Google Scholar]
- 96.Namba K., Stubbs G. Structure of tobacco mosaic virus at 3.6 A resolution: implications for assembly. Science. 1986;231:1401–1406. doi: 10.1126/science.3952490. [DOI] [PubMed] [Google Scholar]
- 97.Parr W.C., Schucany W.R. Minimum distance and robust estimation. J. Am. Stat. Assoc. 1980;75:616–624. [Google Scholar]
- 98.Vold R.L., Hoatson G.L. Effects of jump dynamics on solid state nuclear magnetic resonance line shapes and spin relaxation times. J. Magn. Reson. 2009;198:57–72. doi: 10.1016/j.jmr.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 99.Vega A.J. MAS NMR spin locking of half-integer quadrupolar nuclei. J. Magn. Reson. 1992;96:50–68. [Google Scholar]
- 100.van der Maarel J.R.C. The relaxation dynamics of spin 1=1 nuclei with a static quadrupolar coupling and a radio-frequency field. J. Chem. Phys. 1993;99:5646–5653. [Google Scholar]
- 101.Abragam A. Clarendon Press; Oxford, UK: 1961. Principles of Nuclear Magnetism. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.