Abstract
Disorders of somatic mosaicism (DoSM) are a diverse group of syndromic and non-syndromic conditions caused by mosaic variants in genes that regulate cell survival and proliferation. Despite overlap in gene space and technical requirements, few clinical labs specialize in DoSM compared to oncology. We adapted a high-sensitivity next-generation sequencing cancer assay for DoSM in 2014. Some 343 individuals have been tested over the past 5 years, 58% of which had pathogenic and likely pathogenic (P/LP) findings, for a total of 206 P/LP variants in 22 genes. Parameters associated with the high diagnostic yield were: (1) deep sequencing (∼2,000× coverage), (2) a broad gene set, and (3) testing affected tissues. Fresh and formalin-fixed paraffin embedded tissues performed equivalently for identification of P/LP variants (62% and 71% of individuals, respectively). Comparing cultured fibroblasts to skin biopsies suggested that culturing might boost the allelic fraction of variants that confer a growth advantage, specifically gain-of-function variants in PIK3CA. Buccal swabs showed high diagnostic sensitivity in case subjects where disease phenotypes manifested in the head or brain. Peripheral blood was useful as an unaffected comparator tissue to determine somatic versus constitutional origin but had poor diagnostic sensitivity. Descriptions of all tested individuals, specimens, and P/LP variants included in this cohort are available to further the study of the DoSM population.
Keywords: somatic overgrowth, vascular malformation, mosaicism, overgrowth syndrome, megalencephaly, brain malformation, macrocephaly, nevus syndrome
Introduction
Disorders of somatic mosaicism (DoSM) are caused by mutations that arise post-zygotically and are present in only part of the body (Figure 1). These mutations alter the function and regulation of key genes in cell survival and proliferation pathways, including the PI3K/AKT/mTOR and RAS/RAF pathways, and show significant overlap with oncogenic mutations identified in cancer. Examples include: gain-of-function PIK3CA (MIM: 171834) variants in breast cancer (MIM: 114480) and PIK3CA-related overgrowth spectrum (PROS), gain-of-function AKT1 (MIM: 164730) variants in non-small cell lung cancer (MIM: 211980) and Proteus syndrome (MIM: 176920), and gain-of-function NRAS (MIM: 164790) mutations in acute myeloid leukemia (MIM: 601626) and congenital melanocytic nevus syndrome (MIM: 137550).1, 2 The manifestations of DoSM are highly variable, depending on when the mutation occurred during embryogenesis, the location of affected cells and tissues within the developing body, allelic diversity, and the function and expression of the altered gene(s).3 Phenotypes can appear before or after birth and may include soft tissue and/or bony overgrowth, digital anomalies, glaucoma, vascular and pigmentary alterations, and brain abnormalities, among others.1, 4
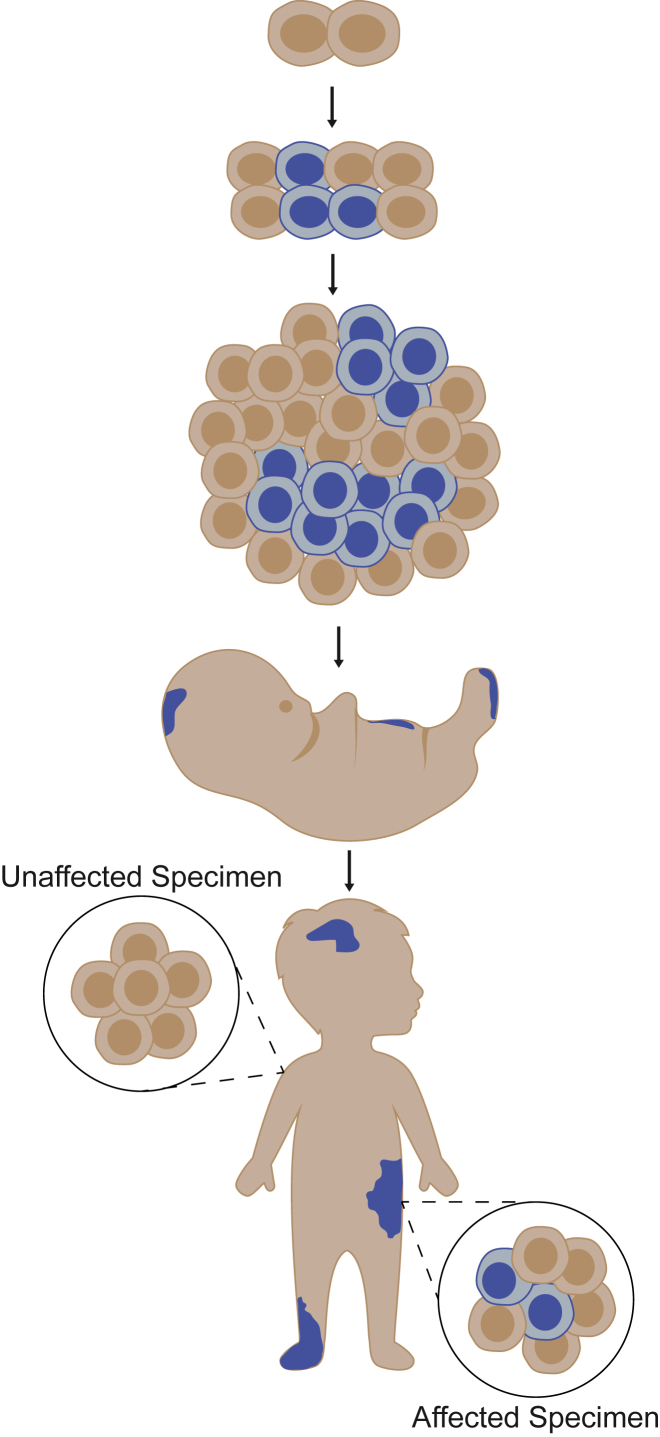
Figure 1.
Disorders of Somatic Mosaicism
Somatic variants can arise during or after the 2-cell stage of embryogenesis. They are restricted to the specific lineage of cells derived from the original mutant (blue) and are absent from other cells and tissues (tan). Proliferative disorders arise when somatic variants impact the function of key genes involved in the regulation of cell growth, division, and senescence. Their phenotypic manifestations are highly variable, depending on the timing of the mutation during or after embryonic development, the location of the variant within the developing body, allelic diversity, and the function and expression of the altered gene(s). Phenotypes can appear before or after birth and may include soft tissue overgrowth, skin lesions, bone malformations, and brain abnormalities, among others.
There are no FDA-approved drugs to treat DoSM; however, we and others have demonstrated that several DoSM are caused by mutations in highly conserved genes, many of which are implicated in cancer (oncogenes). This has transformed our fundamental understanding of the pathobiology of these lesions and revealed potential pharmacologic targets for these developmental but progressive disorders. For example, venous and lymphatic malformations driven by activation of the PI3K/AKT/mTOR pathway were improved by treatment with oral mTOR inhibitors,5, 6, 7 and Venot et al. demonstrated dramatic reversal of disease employing compassionate use of an oral PIK3CA protein inhibitor to treat people with complex vascular anomalies and/or PROS.8 Unfortunately, clinical diagnosis of DoSM is often challenging since phenotypes can be non-specific and presentations vary depending on the affected tissue(s). The use of various historic and specialty-specific acronyms to describe these phenotypes further contributes to confusion in diagnosis and misperceptions regarding prognosis and hinders progress in therapeutic intervention. Thus, molecular diagnoses are critical for management of affected individuals and counseling of affected families.
Testing for somatic variation in the setting of cancer presents unique challenges in comparison to testing for constitutional variants that are present in every nucleated cell. Just as the variants in a tumor are restricted to the transformed cells (usually with oligoclonal distribution), the variants implicated in DoSM are restricted to the discrete lineage of cells descended from the founder cell. As such, reliable detection of somatic variants requires methods with high analytical sensitivity, designed to identify variants that may be present at a very low allelic frequency.9 Sensitivity requirements, coupled with the need to evaluate numerous genetic loci (extending beyond mutational hotspots), makes next-generation sequencing (NGS) an ideal method for DoSM testing.
Here we present the data derived from our experience in NGS testing of 358 unique specimens from 343 individuals for DoSM over the course of 5 years. Although discrete aspects from 16 case subjects (4.7%) have been reported previously,9, 10, 11 this report is a comprehensive review of the aggregate data across the entire cohort. Our findings stress the importance of several factors unique to testing in the somatic setting: multiplexing with broad exonic sequence coverage of multiple gene targets, high unique coverage depth, and directed selection of specimens from disease-affected tissue. Detailed descriptions of the individuals that were tested and their genetic findings are offered as a resource to advance genetic testing for DoSM.
Material and Methods
Biological Materials
Specimens from 343 individuals with suspected DoSM were submitted to Genomics and Pathology Services at Washington University in St. Louis for clinical NGS-based testing between October 2013 and December 2018 (Tables S1 and S2). Accepted specimen types were peripheral blood, buccal swabs, cultured fibroblasts, fresh tissue, and formalin-fixed, paraffin-embedded (FFPE) tissue. Fresh tissue specimens included small punch biopsies, soft tissue masses, brain biopsies, and amputated digits that were transported either frozen or in culture media. Peripheral blood was often submitted for comparison to affected tissue (see Variant Confirmation in Peripheral Blood by Sanger Sequencing below). Specimens were classified as affected or unaffected based on the descriptions provided on test requisitions and other available clinical notes. Buccal swabs were considered affected if the individual exhibited disease phenotypes in the head.
DNA Isolation, Library Preparation, and Sequencing
DNA was isolated from fresh and FFPE tissue using the QIAamp DNeasy blood and tissue kit (QIAGEN) and from peripheral blood, buccal swabs, and cultured fibroblasts using the QIAamp DNA blood mini kit (QIAGEN). Isolated DNA was sheared by sonication to an average size of 160–230 bp. Fragmentation was measured using an Agilent Bioanalyzer 2100, and concentration was measured using a Qubit fluormetric quantitation system. Fragmentation was followed by end repair, A-tailing, and sequencing adaptor ligation using standard, commercial library preparation kits (Table S3). Adaptor-ligated DNA was amplified by selective, limited-cycle PCR. Prepared libraries were hybridized to custom capture probes to enrich for the targeted genes as specified in Table S4. Enriched libraries were PCR amplified, pooled, and sequenced on Illumina instrumentation according to the manufacturer’s recommended protocol to generate paired-end 101 or 151 bp reads. Detailed parameters across multiple assay versions may be found in Table S3.
Bioinformatic Analyses
Details of the bioinformatic pipelines used with each version of the assay described here are listed in Table S3. Briefly, Illumina base call (bcl) files were converted to fastq format and demultiplexed using custom scripts. Reads were aligned to the human reference assembly (UCSC hg19) using Novoalign. Sequencing metrics, including total reads generated, reads mapped to the reference assembly, reads mapped to targeted exons/genes, unique versus duplicate reads, and coverage depth, were calculated for each case subject to ensure that data quality was sufficient to support further analysis (Table S5).
Single-nucleotide variants (SNVs) with variant allele frequency (VAF) greater than 3% and small insertions and deletions (indels, <21 bp) were identified from alignment files using open source software (Table S3). The consequences of any detected variants were determined by Variant Effect Predictor.12 Recurrently mutated positions with clinical relevance were further evaluated to identify pathogenic SNVs between 1% and 3% VAF. Aligned bam files were interrogated using samtools mpileup (v.0.1.1913, 14) at 803 codons across 35 genes in regions associated with recurrent mutations (Table S6). The mpileup output was parsed and filtered by a custom script, and alignment files were manually inspected in Integrative Genomics Viewer15 to confirm the presence of the identified variants.
Variant Interpretation
Variant interpretation was limited to one or more of the specified gene sets described in Table 1. Annotated variants were subjected to expert classification using the ACMG/AMP standards and guidelines for constitutional variant interpretation,16 taking into consideration literature review, association with FDA-approved therapies, professional guidelines, and representation within publically available databases of human genomic variation (e.g., ClinVar, dbSNP, ExAC, and gnomAD). Variant attributes were additionally considered when assessing for pathogenicity including variant location (hotspot or domain), VAF, occurrence and frequency in cancer, type of alteration, sampled tissue type, and clinical phenotype. All detected pathogenic variants, likely pathogenic variants, and variants of uncertain clinical significance are listed in Table S7. Pathogenic variants and likely pathogenic variants were listed as “P/LP” for the purpose of this report. For visual representation, P/LP variants were plotted along the length of coding genes using MutationMapper, which is available from cBioPortal.17, 18
Table 1.
Individuals and Specimens Tested Using Each Gene Set Offered from October 2013 to December 2018
| Gene Set | Date of Offering | Reported Genesa | P/LP Findings/Individuals Testedb | FFPE Specimens | Fresh Tissue Specimens | Fibroblast Cultures | Buccal Swabs | Peripheral Blood Specimens | Total Specimens |
|---|---|---|---|---|---|---|---|---|---|
| PIK3CA-Related overgrowth | Oct 2013 to present | PIK3CA | 20/27 | 3 | 8 | 4 | 13 | 2 | 30 |
| McCune Albright syndrome | Oct 2013 to present | GNAS | 7/33 | 10 | 4 | 1 | 0 | 19 | 34 |
| Segmental overgrowth | Oct 2013 to May 2015 | AKT1, AKT2, AKT3, GNAS, MTOR, PIK3CA, PTEN | 7/10 | 6 | 1 | 1 | 2 | 0 | 10 |
| Somatic overgrowth V3.0 | May 2015 to Aug 2015 | AKT1, AKT2, AKT3, GNAQ, MTOR, PIK3CA, PIK3R2, PTEN | 2/5 | 3 | 1 | 1 | 0 | 0 | 5 |
| Somatic overgrowth V3.1 | Aug 2015 to Feb 2017 | AKT1, AKT2, AKT3, GNAQ, MTOR, PIK3CA, PIK3R2, PTEN, RASA1, TSC1, TSC2 | 42/79 | 9 | 34 | 12 | 25 | 0 | 80 |
| Somatic overgrowth V3.2 | Feb 2017 to present | AKT1, AKT2, AKT3, GNA11, GNAQ, IDH1, IDH2, MAP2K1, MAP2K3, MTOR, PIK3CA, PIK3R2, PTEN, RASA1, SMO, TEK, TSC1, TSC2 | 91/155 | 25 | 81 | 13 | 37 | 2 | 158 |
| Nevus syndrome | Feb 2017 to present | FGFR3, GNA11, GNAQ, HRAS, KRAS, MAP3K3, NRAS, PIK3CA, TEK | 17/18 | 5 | 11 | 1 | 2 | 0 | 19 |
| Curry-Jones syndrome | Feb 2017 to present | SMO | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Maffucci syndrome | Feb 2017 to present | IHD1, IDH2 | 1/1 | 1 | 0 | 0 | 0 | 0 | 1 |
| Rasopathies | Feb 2017 to present | BRAF, CBL, HRAS, KRAS, MAP2K2, MAP2K2, NF1, NRAS, PTPN11, RAF1, RIT1, SHOC2, SOS1, SPRED1 | 2/3 | 2 | 1 | 0 | 0 | 0 | 3 |
| Custom gene set | Oct 2013 to present | variable | 11/18 | 6 | 8 | 1 | 0 | 3 | 18 |
| Total | 349 | 70 | 149 | 34 | 79 | 26 | 358 |
Online Mendelian Inheritance in Man (OMIM) identifiers provided for each gene in Table S4.
Note that multiple specimens were submitted from some individuals and some specimens were tested on multiple gene sets.
Variant Confirmation in Peripheral Blood by Sanger Sequencing
Peripheral blood was obtained as a comparative unaffected specimen alongside the primary affected tissue for 139 of the 199 individuals with P/LP findings (Table S7). In the event that a pathogenic, likely pathogenic, or variant of uncertain clinical significance (VUS) was identified, peripheral blood was submitted to a clinical reference laboratory for confirmation of the variant of interest by Sanger sequencing. Detection of the variant in peripheral blood indicates either a multi-tissue distribution of a mosaic alteration or that the variant has a constitutional origin whereas the absence of the variant in peripheral blood is taken as an indication of mosaicim.
Regulatory Approval
The institutional review board of Washington University School of Medicine deemed this study as not fitting the definition of human subjects research; thus, no IRB protocol was required.
Results
Testing Model
Testing for DoSM was initiated at Washington University in 2013. DoSM gene panels were modified three times between 2013 and 2017, expanding the number of orderable genes from 7 to 35; discrete single-gene testing was also available (e.g., PIK3CA for suspected PROS, GNAS [MIM: 139320] for suspected McCune-Albright syndrome [MIM: 174800]). The latest revision in February 2017 saw a reorganization of the testing program to offer smaller gene sets specifically tailored to individual syndromes and disorders (Table 1). These range from a single-gene analysis for Curry-Jones syndrome (MIM: 601707) to broader panels for disorders known to exhibit locus heterogeneity (e.g., Rasopathies Gene Set, Nevus Gene Set, Somatic Overgrowth Gene Set).
In 17 instances, the ordering physician requested a custom gene set comprised of one or more of the genes that were offered on the available version of the advertised panels. Custom gene sets can be particularly useful in case subjects with complex phenotypes and/or multiple potential etiologies.
Cohort Demographics
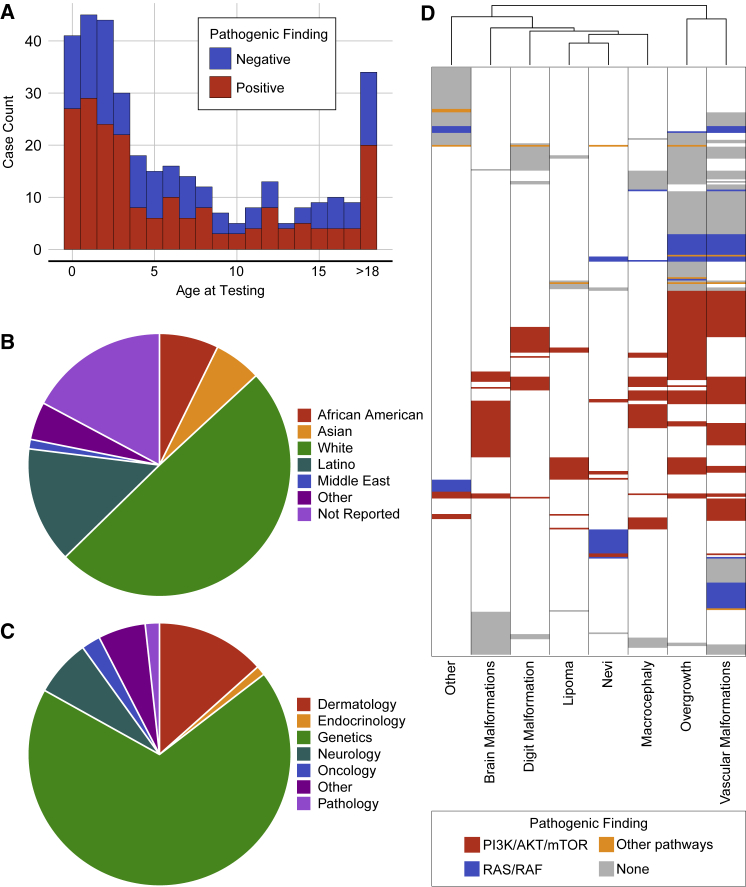
A total of 358 unique specimens from 343 individuals were submitted for NGS testing between October 2013 and December 2018 (Tables 1, S1, and S2). Individuals ranged in age from birth to 62 years (Figure 2A). The average age at the time of testing was 7.77 ± 9.41 years, consistent with the congenital or early-onset of DoSM phenotypes.6 Information about racial ancestry was provided for 284 people; 60% self-identified as white (Figure 2B). However, this is more likely a reflection of the laboratory’s client demographic rather than the true racial distribution of DoSM. The overall sex ratio among the cohort was balanced at 173 females and 170 males; however, skewing was noted for specific conditions (for example: 11 males and 26 females with suspected McCune Albright syndrome). Sixty-eight percent of the individuals were referred for testing by physicians that specialize in genetics; other referring sub-specialties included dermatology, endocrinology, neurology, and pathology among others (Figure 2C).
Figure 2.
Cohort Demographics
(A) A total of 358 unique specimens from 343 people were submitted for testing between October 2013 and December 2018. Individuals ranged in age from birth to 62 years with an average age of 7.77 ± 9.41 years. P/LP findings were not concentrated in any particular age group.
(B) Racial ancestry was listed on the test requisition for most people: 25 African American, 20 Asian, 170 white, 49 Latino, 4 Middle Easterner, 16 of other backgrounds; 59 people did not report their racial background.
(C) Individuals were referred for testing by physicians that represent a total of 18 sub-specialties, including: genetics, dermatology, neurology, oncology, and pathology.
(D) Phenotypes reported for each person were binned into eight discrete categories. People were clustered based on the eight phenotypic categories and the biochemical pathway associated with any P/LP finding. Variants in the PI3K/AKT/mTOR pathway (red) are associated with a wide variety of symptoms, while variants in the RAS/RAF pathway (blue) were often associated with overgrowth, vascular malformations, brain malformations, and nevi.
Test requisitions from 81 people listed a diagnosis based on clinical findings (Table 2); however, most did not have a specific diagnosis at the time of testing. With careful analysis of test requisitions and other available clinical records, people were grouped into eight discrete phenotypic categories (Figure 2D, Table S1). The most prevalent features noted in this cohort were asymmetric overgrowth and vascular malformations (169 and 189 people, respectively). Seventy-three individuals (21.3%) had a single feature indicated on the requisition (e.g., port wine stain, lipoma, or an isolated malformation or overgrowth), while the rest (78.7%) had multiple phenotypes described. Clustering analyses revealed that segmental overgrowth and vascular malformations were likely to co-occur, possibly driven by overlap in the causative genes (Figure 2D). We also observed a degree of overlap between brain malformations and macrocephaly (70 individuals with brain malformations, 57 with macrocephaly, and 24 with both).
Table 2.
Specific Clinical Diagnoses of Individuals Tested for Disorders of Somatic Mosaicism
| Prior Clinical Diagnosis (OMIM Identifier) | AKT1 | AKT3 | GNA11 | GNAQ | GNAS | HRAS | KRAS | IDH1 | PIK3CA | PIK3R1 | PIK3R2 | RASA1 | TSC2 | Negative Cases | Total Cases |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CLOVES (612918) | – | – | – | – | – | – | 1 | – | 9∗ | 1 | – | – | – | 2 | 13 |
| KTS (149000) | – | – | – | – | – | – | 1 | 1 | 2∗ | 1 | – | – | – | 5 | 9 |
| Maffucci syndrome (614569) | – | – | – | – | – | – | – | 2∗ | – | – | – | – | – | 1 | 3 |
| MCAP (602501) | – | 1 | – | – | – | – | – | – | 16∗ | – | – | – | – | 4 | 21 |
| McCune Albright syndrome (174800) | – | – | – | – | 4∗ | – | – | – | – | – | – | – | – | 7 | 11 |
| MPPH (603387, 615937) | – | 0∗ | – | – | – | – | – | – | – | – | 0∗ | – | – | 2 | 2 |
| PROS (n/a) | – | – | 1 | – | – | – | – | – | 0∗ | – | – | – | – | 2 | 3 |
| Proteus syndrome (176920) | 0∗ | – | – | – | – | – | – | – | 3 | – | – | – | 1 | 2 | 6 |
| Parkes Weber syndrome (n/a) | – | – | – | – | – | – | – | – | – | – | – | 0∗ | – | 1 | 1 |
| Nevus sebaceous syndrome (163200) | – | – | – | – | – | 1∗ | 7∗ | – | – | – | – | – | – | 1 | 9 |
| Sturge Weber syndrome (185300) | – | – | – | 2∗ | – | – | – | – | – | – | – | – | – | 1 | 3 |
Abbreviations: OMIM, Online Mendelian Ineritance in Man; CLOVES, congenital lipomatous overgrowth, vascular malformations, epidermal nevi and spinal/skeletal abnormalities; MCAP, megalencephaly-capillary malformation syndrome; MPPH, megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome; PROS, PIK3CA-related overgrowth syndrome. Asterisk (∗) indicates canonical gene(s) associated with the listed syndrome.
Clinically Relevant Findings
NGS was performed on DNA extracted from the submitted specimens, and variants were reported from the gene set selected by the ordering physician (Table 1). Considering all test results, P/LP alterations were identified in 199 of the 343 individuals (58%) in this cohort (Figure 3, Table S7). P/LP variants were found in 22 genes and included 184 missense variants, 5 in-frame indels, 9 frameshifts, 3 nonsense variants, and 5 splice variants (Table 3).
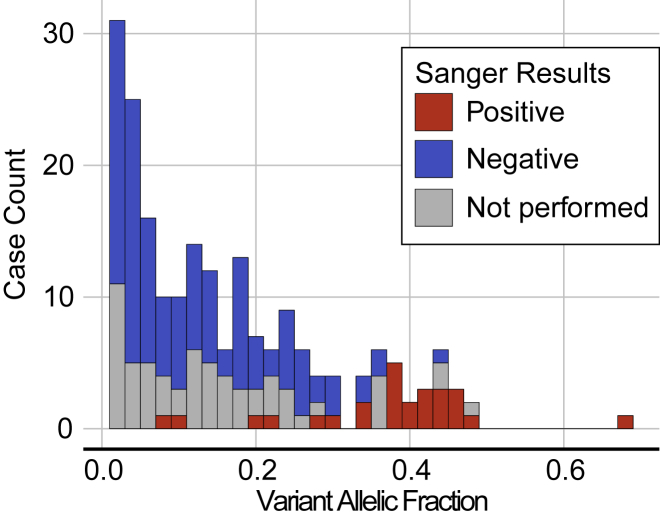
Figure 3.
Allelic Frequency of P/LP Variants
P/LP variants were found with VAF ranging from 1% to 68% with an average VAF of 16.2% ± 13.5%. One hundred sixty of the 206 P/LP variants were found at VAF < 25% and 87 were found with VAF < 10%. Blood specimens were submitted as a comparative sample determine the distribution of the P/LP variant in the body for 139 of the 199 individuals with P/LP findings; 26 of the secondary blood specimens tested positive for the P/LP variant by Sanger sequencing (red) while 113 tested negative (blue). P/LP findings in the peripheral blood were enriched in case subjects where the variant was found at a high VAF in the original specimen used in NGS testing.
Table 3.
P/LP Variants Identified in the Context of Disorders of Somatic Mosaicism
| Gene (OMIM Identifier) | RAS/RAF | Missense Variants | Nonsense Variants | Splice Variants | In-Frame Indels | Frameshifts | Total P/LP Variants |
|---|---|---|---|---|---|---|---|
| PIK3CA (171834) | PI3K/AKT/mTOR | 108 | 0 | 0 | 2 | 2 | 112 |
| PIK3R2 (603157) | PI3K/AKT/mTOR | 6 | 0 | 0 | 0 | 0 | 6 |
| PTEN (601728) | PI3K/AKT/mTOR | 2 | 1 | 2 | 0 | 0 | 5 |
| AKT3 (611223) | PI3K/AKT/mTOR | 4 | 0 | 0 | 0 | 0 | 4 |
| MTOR (601231) | PI3K/AKT/mTOR | 4 | 0 | 0 | 0 | 0 | 4 |
| PIK3R1 (601231) | PI3K/AKT/mTOR | 2 | 0 | 0 | 2 | 0 | 4 |
| AKT1 (164730) | PI3K/AKT/mTOR | 3 | 0 | 0 | 0 | 0 | 3 |
| TSC2 (191092) | PI3K/AKT/mTOR | 1 | 0 | 0 | 0 | 0 | 1 |
| GNA11 (139313) | RAS/RAF | 12 | 0 | 0 | 0 | 0 | 12 |
| KRAS (190070) | RAS/RAF | 10 | 0 | 0 | 1 | 0 | 11 |
| GNAS (139320) | RAS/RAF | 7 | 0 | 0 | 0 | 1 | 8 |
| TEK (600221) | RAS/RAF | 6 | 0 | 0 | 0 | 2 | 8 |
| GNAQ (600998) | RAS/RAF | 7 | 0 | 0 | 0 | 0 | 7 |
| RASA1 (139150) | RAS/RAF | 0 | 1 | 2 | 0 | 2 | 5 |
| HRAS (190020) | RAS/RAF | 3 | 0 | 0 | 0 | 0 | 3 |
| NF1 (613113) | RAS/RAF | 0 | 1 | 1 | 0 | 1 | 3 |
| NRAS (164790) | RAS/RAF | 2 | 0 | 0 | 0 | 0 | 2 |
| BRAF (164757) | RAS/RAF | 1 | 0 | 0 | 0 | 0 | 1 |
| FGFR1 (136350) | RAS/RAF | 1 | 0 | 0 | 0 | 0 | 1 |
| PTPN11 (176876) | RAS/RAF | 1 | 0 | 0 | 0 | 0 | 1 |
| IDH1 (147700) | Krebs cycle | 4 | 0 | 0 | 0 | 0 | 4 |
| IDH2 (147650) | Krebs cycle | 1 | 0 | 0 | 0 | 0 | 1 |
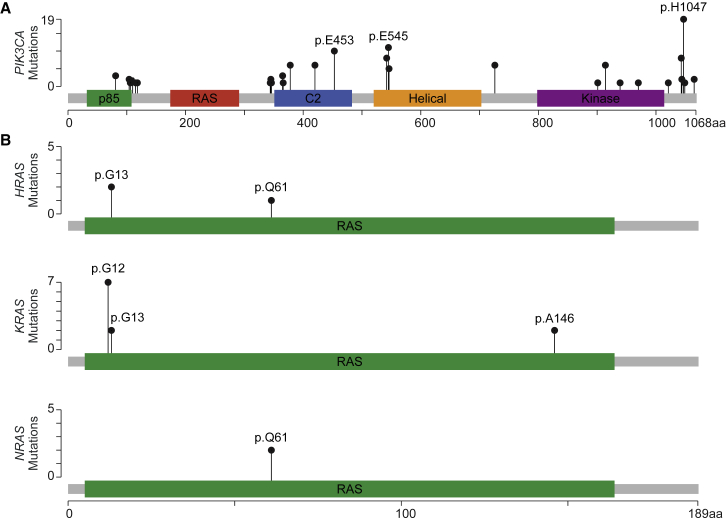
One hundred thirty-nine of the 206 P/LP variants were found in genes of the PI3K/AKT/mTOR pathway; 112 of these were in PIK3CA (Figure 4A, Tables 2 and S7). Nineteen PIK3CA variants occurred at the p.His1047 position and ten affected p.Glu545, both well-known hotspots for mutation in cancer.19 Consistent with previous reports,6 well-characterized, activating variants were found along the full length of PIK3CA (Figure 4A). People harboring these variants can now be considered as candidates for PIK3CA and/or mTOR inhibitor therapy.1, 7, 8
Figure 4.
P/LP Variants in PIK3CA and RAS Genes
(A) P/LP variants were identified along the length of PIK3CA (MIM: 171834) including four of the five outlined protein domains.
(B) Mutational hotspots p.Glu453, p.Glu545, and p.His1047 are common in both DoSM and cancer. In contrast, most of the variants identified in HRAS (MIM: 190020), KRAS (MIM: 190070), and NRAS (MIM: 164790) were found at the p.G12 position, which is commonly mutated in non-small cell lung cancer.
Sixty-seven P/LP variants were found outside the PIK3/ATK/mTOR pathway, primarily in G-protein-coupled receptors and in genes of the RAS/RAF pathway (Table S7). Variants in the RAS/RAF pathway have broad consequences, including overgrowth, brain malformations, vascular malformations, and skin pigment abnormalities.4, 20, 21, 22 Twelve individuals in our cohort had activating GNA11 (MIM: 139313) p.Arg183Cys variants and seven had activating GNAQ (MIM: 600998) p.Arg183Gln variants; all of these had some form of vascular malformation and many had tissue overgrowth as well.22 Eleven individuals had P/LP variants in KRAS (MIM: 190070, Figure 4B), seven were present at the canonical p.Gly12 position. Ten of these presented with nevi, as expected.23, 24 Interestingly, two individuals with KRAS p.Gly12 variants had vascular malformations and overgrowth features that are typically associated with findings in PIK3CA (Table 2). G-protein inhibitor and RAS/RAF pathway inhibitor therapies may prove useful in these people.11
The 81 individuals with clinical diagnoses listed on test requisitions are described in Table 2. Many of the syndromes represented are caused by specific variants in known genes, for example AKT1 variants in Proteus syndrome, PIK3CA variants in Klippel-Trenaunay syndrome (MIM: 149000), and GNAS variants in McCune Albright syndrome. In some instances, the expected variant was detected in the specimen, but unexpected variants were seen in many cases. Specimens referred for the nevus gene set had a high diagnostic rate in general. All P/LP findings associated with nevus sebaceous syndrome (MIM: 163200) were in KRAS and HRAS (MIM: 190020), as expected. In contrast, six individuals were tested for suspected Proteus syndrome, but no AKT1 variants were identified; three individuals had P/LP variants in PIK3CA and one had a P/LP variant in TSC2 (MIM: 191092). While treatment options for AKT1-driven Proteus syndrome are limited, targeted therapies are available for people with variants in PIK3CA and TSC2.
Variant Allelic Frequency for DoSM-Associated Variants
Most of the P/LP variants identified in this dataset were present at a VAF < 50%, indicative of specimens with varying proportions of affected and unaffected cells (Figure 3). One hundred sixty of the 206 P/LP variants were found at VAF < 25% and 87 were found with VAF < 10%. The NGS assay employed in this study is validated for a routine VAF cutoff of 3%; however, a lower cutoff of 1% is possible with manual inspection of the aligned reads at recurrently mutated, hotspot positions (e.g., PIK3CA p.His1047 and p.Glu545, see Table S6). Indeed, this additional step facilitated the identification of 31 variants in 9 genes with VAF between 1% and 3%. When these low VAF variants were reported, the clinical report specifically noted that the finding was near the limit of detection for the assay and should be interpreted in that context; additional specimens were requested for confirmatory testing where needed. If the only P/LP variant found in a case subject was present at <1% VAF, the case subject was reported as negative.
Tissue Distribution Suspected DoSM Variants
Peripheral blood specimens were submitted as a comparative sample to clarify the distribution of the variant for 139 of the 199 individuals with P/LP findings; 26 of the blood specimens tested positive for the P/LP variant by Sanger sequencing (Figure 3, Table S7). Positive findings in the peripheral blood are indicative of multi-tissue and/or constitutional involvement and may necessitate genetic counseling regarding the potential for vertical transmission due to gonadal mosaicism in the proband. The multi-tissue positives were concentrated in case subjects where the P/LP variant was originally identified via NGS at a high (near-heterozygous) VAF in the affected specimen (Figure 3). They were also enriched for indications related to macrocephaly and brain malformations (20 of 26 individuals), conditions that have been described as germline disorders in the literature.25 While Sanger sequencing does not provide a discrete VAF, low peak intensities indicative of a mosaic variant were specifically noted by the reference laboratory in eight case subjects (Table S7). For example, one GNAS variant (p.Arg201His) with a low peak intensity from the peripheral blood specimen had a VAF < 10% in the affected specimen as well. This confirms multi-tissue distribution but suggests that constitutional origin is unlikely (i.e., multi-tissue mosaic).
Both gain- and loss-of-function P/LP variants were identified in the peripheral blood comparator specimens (19 and 7 case subjects, respectively). However, highly penetrant hotspot mutations were infrequently observed in blood, likely due to a deleterious or lethal effect on the developing embryo.25, 26, 27, 28 For example, nine PIK3CA variants were identified in peripheral blood. Only one of these was found at the p.His1047 position, a p.His1047Tyr variant that imparts a weaker activation for the intact phosphatidyl inositol-3 kinase (PI3K) compared to the canonical p.His1047Arg variant.29 Interestingly, low peak intensity was noted in the Sanger chromatogram for the p.His1047Tyr variant in this case subject, consistent with multi-tissue mosaicism.
Several individuals harbored variants associated with germline disorders. For example, all five of the P/LP PTEN (MEM: 601728) variants found in this cohort were detected in peripheral blood,30, 31 as were four of the six PIK3R2 (MIM: 603157) variants.27, 32 Conversely, TEK (MEM: 600221) variants are often implicated in germline syndromes,33 but all four P/LP TEK variants identified in this cohort were absent from peripheral blood. This included one case subject with two separate, confirmed somatic TEK variants (P337_S1 in Table S7).
Impact of Specimen Selection
Five different specimen types were submitted for testing (Figure 5): fresh tissue (n = 149), FFPE tissue (n = 70), cultured fibroblasts (n = 34), buccal swabs (n = 79), and peripheral blood (n = 26). Previous studies have stressed the importance of testing disease-affected tissues.9, 25, 34 Upon review of test requisitions and other medical records, 301 of the 358 submitted specimens were deemed to be representative of disease-affected tissues. We identified P/LP variants in 65% of these affected specimens compared to just 4 of 57 unaffected tissue specimens.
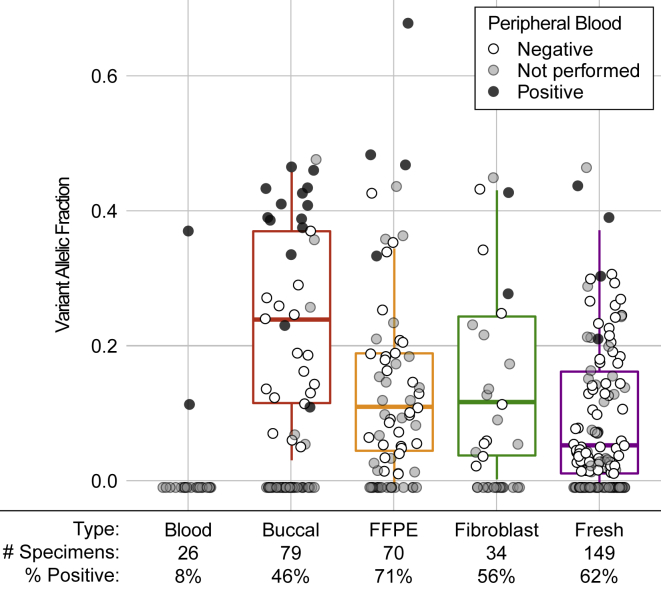
Figure 5.
P/LP Variants Identified from Five Specimen Types
Five specimen types are accepted for testing: peripheral blood (n = 26), buccal swab (n = 79), formalin fixed paraffin embedded (FFPE, n = 70), cultured fibroblasts (n = 34), and fresh, unfixed tissues (n = 149). Case subjects where P/LP variant was identified are plotted under the 0% VAF line, and the 25th, 50th, and 75th percentiles of VAFs of P/LP variants are displayed by boxplots. Shading of dots reflects results of Sanger sequencing of peripheral blood: black dots represent P/LP findings in blood (multi-tissue distribution), white dots represent case subjects from which the P/LP variant was found only in the affected sample (confirmed somatic), and gray dots represent case subjects for which no peripheral blood was submitted. The percent of specimens with P/LP findings are listed for each specimen type.
The highest rates of P/LP findings were reported from fresh and FFPE tissues (62.4% and 71.4%, respectively), likely attributable to the fact that 96% of them were considered disease affected (Table S2). Despite similar sequencing depths, the average VAF of the P/LP variants detected in FFPE tissues was somewhat higher than that of the fresh tissue (Figure 5). This could be due to the fact that FFPE specimens were submitted in severe case subjects that required surgical intervention, and may be expected to contain a higher percentage of abnormal cells. Considering only specimens where the corresponding blood sample tested negative for the P/LP variant (i.e., somatic variants), the average VAFs of variants from fresh and FFPE tissues were comparable (10.1% and 12.8%, respectively). This means that we observed no reduced sensitivity for identifying disease-associated variants in FFPE specimens compared to fresh, un-fixed tissues.
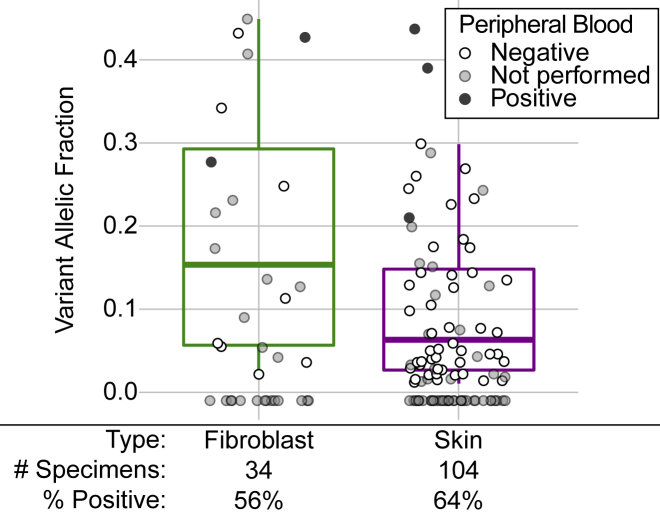
Thirty-four cultured fibroblast specimens were included in this cohort. Culturing fibroblasts provides a way to expand a limited fresh tissue biopsy to support multiple tests or specimen archiving and may increase the proportion of cells carrying variants that confer a growth advantage. Since fibroblasts are often cultured from skin, we compared the 34 fibroblast cultures to 104 fresh skin biopsies (Figure 6); in no case were fibroblasts and skin specimens obtained from the same individual. The overall rate of P/LP findings was comparable between the two specimen types: 55.9% and 63.5% for fibroblast and skin, respectively. However, the average VAF of the P/LP variants found in cultured fibroblasts was higher than those found in skin (18.6% compared to 10.1%, p = 0.005). This finding is also observed when considering only individuals whose peripheral blood specimens tested negative for the P/LP variant: average VAF of 8.8% for 48 variants from skin specimens and average VAF of 16.3% for 8 variants from fibroblast cultures. Furthermore, we noted an overrepresentation in PIK3CA variants among the cultured fibroblasts, representing 15 of the 19 P/LP variants detected in fibroblast (78.9%) as compared to 31 of 66 P/LP variants detected in skin (47.0%). Of the 15 PIK3CA variants found in cultured fibroblast specimens, 2 were at the p.Glu453 position (VAFs of 21.3% and 24.8%) and 3 were at the p.His1047 position (VAFs of 4.2%, 5.5%, and 34.2%). The other P/LP PIK3CA mutations identified in cultured fibroblasts, while known activators, were not located at well-characterized “hotspot” positions.
Figure 6.
P/LP Variants Identified from Cultured Fibroblasts Compared to Skin Biopsies
Fibroblasts are typically cultured from skin biopsies, so the P/LP variants found in cultured fibroblasts (n = 34) were compared to those found in fresh tissue skin biopsies (n = 104). Cases where no P/LP variant was identified are plotted under the 0% VAF line. The 25th, 50th, and 75th percentiles of VAFs of P/LP variants are displayed by boxplots. Shading of dots reflects results of Sanger sequencing of peripheral blood: black dots represent P/LP findings in blood (multi-tissue distribution), white dots represent case subjects from which the P/LP variant was found only in the affected sample (confirmed somatic), and gray dots represent case subjects for which no peripheral blood was submitted. The percent of specimens with P/LP findings are listed for each specimen type.
Forty-six percent of the buccal swab specimens tested positive for P/LP variants (Figure 5). Buccal swabs were often submitted in the context of brain malformations, macrocephaly, or other DoSM with involvement of the head (n = 58, counted as affected specimens in this study). When considering only those individuals with phenotypes involving the head, the rate of P/LP findings in buccal swabs increases to 57%, which is similar to the detection rate from invasive tissue biopsies. In contrast, the P/LP finding rate dropped to 14% for buccal specimens from individuals with phenotypes that do not affect the head. The average VAF of the P/LP variants from buccal swabs was higher compared to other specimen types, likely due to the fact that 14 of the specimens were associated with findings in the peripheral blood (i.e., multi-tissue or constitutional distribution). Considering only specimens for which the peripheral blood comparator tested negative, the average VAF of the P/LP variants detected in buccal swabs was 17.8%.
Peripheral blood showed the lowest diagnostic yield as a primary test specimen, with only two positives from 26 submitted specimens. Nineteen of the 26 peripheral blood specimens were submitted to test for McCune Albright syndrome. This syndrome principally affects bone and endocrine tissues, neither of which is routinely biopsied for genetic testing. Only one peripheral blood specimen tested positive for an activating GNAS variant, the driver of McCune Albright syndrome. The specimens submitted for suspected McCune Albright syndrome are described in Table 4; the highest diagnostic yield was seen in bone and soft tissues (Table 4).
Table 4.
Specimens Submitted to Test for McCune Albright Syndrome
| Specimen Type | Specimens | Specimens with P/LP Findings |
|---|---|---|
| Peripheral blood | 19 | 1 |
| FFPE bone | 4 | 2 |
| Fresh bone | 1 | 1 |
| FFPE thyroid | 1 | 1 |
| FFPE ovary | 4 | 1 |
| FFPE skin | 1 | 1 |
| Fresh skin | 3 | 1 |
| Cultured fibroblast | 1 | 0 |
Variants of Uncertain Clinical Significance
Variants of uncertain clinical significance (VUS) were rarely reported in this cohort (12 of 343 tested individuals). Two VUS were found in specimens that also harbored a P/LP variant. A specimen from a person with macrocephaly (P050, Table S7) had a pathogenic AKT3 variant (MIM: 611223, VAF = 39%) alongside a VUS in PIK3R2 (VAF = 47%; absent from gnomAD v2.1.1), both of which were found in the peripheral blood comparator specimen. A specimen from another person with suspected Proteus syndrome (P287, Table S7) had a pathogenic variant in PIK3CA (VAF = 27%) and a VUS in TEK (VAF = 7%), neither of which was found in the peripheral blood (i.e., confirmed somatic). Venous malformations, a key manifestation of TEK variants,35, 36 were not noted on the test requisition. The specimen from a third person (P263, Table S7) with plexiform neurofibroma had no P/LP variants, only two VUS: one in FGFR1 (MIM: 136350, VAF = 47%; absent from gnomAD v2.1.1, absent from cBioPortal v2.2.1) and another in GNAS (VAF = 47%; present at low population MAF in gnomAD v2.2.1); peripheral blood was not available for testing in this instance.
Nine specimens tested positive for a single VUS, most of which were novel at the time of this study. Three of four people with VUS in MTOR (MIM: 601231) had brain malformations, a key manifestation of pathogenic variants in this gene.11 One somatic VUS in MAP3K3 (MIM: 603539, VAF = 6%) was found in a person with capillary malformations (MIM: 163000), again consistent with the expected phenotype.37 In contrast, two multi-tissue-positive VUS in SMO (MIM: 601500) were detected in people who did not have phenotypes consistent with Curry-Jones syndrome.38 All of the variants identified in this cohort, including VUS, are further described in Table S7.
Discussion
Despite the genetic and technical overlap between DoSM and cancer testing and the increasing appreciation of the importance of somatic variation, clinical NGS for DoSM is relatively uncommon. Here we described the findings obtained from studying a cohort of 343 individuals with DoSM collected over the course of five years. Expert knowledge of DoSM, on-site genetic counseling, and a robust testing process has led to specific molecular diagnoses for 58% of the individuals tested, which is essential for accurate diagnosis, optimal screening for associated malformations, and ongoing surveillance. By identifying the molecular basis of these disorders, we have set the stage for the development and implementation of novel targeted therapies, which has the potential to shift treatment paradigms from surgical debulking of malformed tissue to personalized pharmacologic intervention. Furthermore, the data collected during the testing process have afforded valuable insights into specific testing requirements for this unique clinical population, which will further benefit affected inidividuals in the future with the aim of increasing the diagnostic yield of the assay.
As in the setting of cancer, detecting the variants that drive DoSM requires a very high sensitivity assay. Forty-two percent of the P/LP variants detected in this cohort were found at a VAF < 10%, with some as low as 1%. Reliable detection of low VAF variants requires deep sequencing,39 so we routinely sequence case subjects in excess of 1,500× coverage of high-quality, unique reads (Table S5) and implement specific measures to detect well-characterized, recurrent variants down to 1% VAF through manual review of alignment files (Table S6). Variant detection differs between somatic and constitutional assays, with increased sensitivity in our somatic hybrid-capture assay afforded due to markedly increased depth in concert with a highly tuned detection algorithm. Indeed, we cite four case subjects (see P150, P177, P209, and P253 in Table S7) that tested positive for P/LP variants on our assay after receiving negative results from constitutional assays performed by other laboratories. While we cannot rule out the possibility that the differences in test results were due to specimen selection, the four variants were found at VAF ≤ 35%.
We also stress the need to sequence across the entire exonic sequence of DoSM-related genes rather than single exons or mutation hotspots. Loss-of-function variants in pathway regulators (e.g., PTEN, NF1 [MIM: 613113], RASA1 [MIM: 13950], etc.) are found with a wide distribution spanning multiple protein domains. Activating, gain-of-function variants are often associated with specific positions or functional domains; however, our findings demonstrate that variants present outside these hotspots have demonstrated disease association (see the distribution of PIK3CA variants in Figure 4A).
While the majority of our P/LP findings were in the PI3K/AKT/mTOR pathway, specifically in PIK3CA, the inclusion of a broad array of genes outside this key pathway was essential to the diagnosis of many people. The breadth of the gene set is particularly important given the degree of phenotypic overlap between variants in different genes/pathways. Even when the individual’s symptoms pointed to a specific syndrome, the genetic findings often revealed an unexpected etiology (Table 3). To date, we have reported P/LP variants from 22 genes (Table 3). As gene-disease associations continue to expand, a coordinated re-analysis effort may further increase the diagnostic yield and P/LP variant spectrum in this cohort. This is particularly relevant as the breadth of the gene set analyzed was based on the clinician order and thus varied among our population. It is conceivable that new gene-DoSM associations will necessitate a mechanism for periodic reanalysis, particularly for case subjects that tested negative for P/LP variants despite striking clinical presentations. An increased awareness of the clinical relevance for both the known DoSM-related genes and their associated phenotypes, but also additional phenotypes that may have an underlying component of somatic mosaicism, may promote the expansion of this diverse DoSM group.
Consistent with previous studies, we demonstrated the importance of testing affected tissues for the detection of DoSM variants. While the rate of P/LP findings was relatively high for specimens considered to be “disease affected,” very few P/LP variants were detected in tissues that were not. The highest rate of findings was derived from fresh and FFPE tissues. Of note, FFPE tissues are not commonly accepted specimens for DoSM testing across reference laboratories; however, the proven utility of FFPE tissues in diagnostic NGS opens the possibility of testing archived surgical materials when available rather than performing additional surgeries/biopsies or submitting unaffected samples to avoid repeat procedures.
Although affected tissues are preferred, surgical sampling of diseased tissue is not always practical or possible. We specifically investigated two non-invasive specimen types, blood and buccal swabs, in an effort to determine the conditions for which they may serve as a viable alternative. The rate of P/LP findings in peripheral blood was very low for variants with a true somatic origin. Blood is often submitted in cases of suspected McCune Albright syndrome as an alternative to bone or endocrine tissue biopsies. A study of a European McCune Albright-affected cohort showed that variants were detectable from blood in 21 of 99 individuals tested,40 we identified only one activating GNAS variant from 19 peripheral blood specimens. There are many pigmentary syndromes with features similar to McCune Albright, so the low yield of P/LP GNAS variants may be due to diagnostic confusion in the clinical setting. However, we have not identified any specific conditions for which peripheral blood would serve as an acceptable primary specimen.
In contrast to peripheral blood, buccal swabs (comprised primarily of epithelial cells and leukocytes41) are often perceived as an unaffected tissue type, with studies indicating that P/LP variants are rarely detected from buccal swabs and that the VAFs of the detected variants tend to be low.34 In our experience, however, the rate of findings was similar to more invasive specimen types in case subjects with disease findings involving the head, even in those without obvious macrocephaly and brain malformations (e.g., macroglossia, hemihypertrophy of the face [MIM: 133900], nevi on the head, etc.).
Recently, it was proposed that culturing fibroblasts might increase the detection rate of P/LP variants related to DoSM.42 Theoretically, cells with variants that promote proliferation would have a growth advantage ex vivo, thus increasing the proportion of mutated cells in the culture, and, consequently, the VAF of the variant in NGS data. In light of this finding and in the absence of a prospective paired tissue versus fibroblast analysis, we compared the P/LP variants identified in cultured fibroblasts to those identified in fresh skin biopsies, from which fibroblasts are often cultured. The increased VAF of the P/LP variants detected in fibroblasts compared to skin indirectly supports the hypothesis that ex vivo culture is useful for enrichment of cells harboring variation that confers a proliferative advantage. The apparent overrepresentation of PIK3CA variants in cultured fibroblasts and the conspicuous absence of other P/LP variants that were prevalent in skin specimens warrants consideration. While it could be due to bias in the tested population, it could also be due to the penetrance or independence of the impact of activating PIK3CA variants on cell growth compared to other variants in primary fibroblast cultures. If so, the practice of preparing fibroblast cultures from fresh tissue biopsies—which adds cost and time to the testing process—may be practical only for cases of suspected PROS. We are continuing to culture fibroblasts for case subjects with sufficient fresh tissue in order to more definitively answer this question.
Novel variants, often classified as VUS in clinical testing, may accumulate in the genomes of transformed cells due to unchecked cell division and the breakdown of DNA repair mechanisms. In contrast, VUS were strikingly rare in our DoSM-affected cohort. In individuals with no other P/LP findings, VUS are subject to a higher degree of suspicion and often necessitate further investigation, particularly if the impacted gene has been associated with the person’s specific symptoms. Efforts are currently underway to address the need for functional characterization of VUS in the setting of DoSM (and other diseases) and to collect detailed information on the use of these test results in clinical treatment and management.
Conclusions
Here we present the lessons learned from 5 years of NGS-based testing for DoSM. Three hundred forty-three individuals from diverse backgrounds with diverse clinical presentations were referred for testing. Our findings highlight the importance of deep coverage, high-sensitivity NGS testing, full gene sequencing, the use of broad and frequently updated gene panels, and careful specimen selection (in consultation with clinicians, genetic counselors, and laboratory personnel) in maximizing diagnostic yield. Given these conditions, we detected and characterized a total of 206 P/LP variants in 22 genes and provided specific genetic diagnoses for 58% of the inidivuals tested. A full description of this cohort (Table S1), the tested specimens (Table S2), and the identified variants (including VUS, Table S7) are offered as a resource to the DoSM community to further the diagnosis and care of similarly affected individuals in the future.
Declaration of Interests
B.A.D. serves as a consultant and member of the medical advisory board for Venthera. M.C.S. and J.W.H. consult for Pierian Diagnostics.
Acknowledgments
The authors would like to acknowledge the contributions of the Genomics and Pathology Services team involved in specimen processing, NGS, and variant interpretation. We also thank the referring physicians and individuals referred for testing for their continued support. This work was funded by the Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO.
Published: October 3, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.09.002.
Web Resources
OMIM, https://www.omim.org/
Supplemental Data
References
- 1.Keppler-Noreuil K.M., Parker V.E., Darling T.N., Martinez-Agosto J.A. Somatic overgrowth disorders of the PI3K/AKT/mTOR pathway & therapeutic strategies. Am. J. Med. Genet. C. Semin. Med. Genet. 2016;172:402–421. doi: 10.1002/ajmg.c.31531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kinsler V.A., Thomas A.C., Ishida M., Bulstrode N.W., Loughlin S., Hing S., Chalker J., McKenzie K., Abu-Amero S., Slater O. Multiple congenital melanocytic nevi and neurocutaneous melanosis are caused by postzygotic mutations in codon 61 of NRAS. J. Invest. Dermatol. 2013;133:2229–2236. doi: 10.1038/jid.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keppler-Noreuil K.M., Sapp J.C., Lindhurst M.J., Parker V.E., Blumhorst C., Darling T., Tosi L.L., Huson S.M., Whitehouse R.W., Jakkula E. Clinical delineation and natural history of the PIK3CA-related overgrowth spectrum. Am. J. Med. Genet. A. 2014;164A:1713–1733. doi: 10.1002/ajmg.a.36552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rauen K.A. The RASopathies. Annu. Rev. Genomics Hum. Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.di Blasio L., Puliafito A., Gagliardi P.A., Comunanza V., Somale D., Chiaverina G., Bussolino F., Primo L. PI3K/mTOR inhibition promotes the regression of experimental vascular malformations driven by PIK3CA-activating mutations. Cell Death Dis. 2018;9:45. doi: 10.1038/s41419-017-0064-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keppler-Noreuil K.M., Rios J.J., Parker V.E., Semple R.K., Lindhurst M.J., Sapp J.C., Alomari A., Ezaki M., Dobyns W., Biesecker L.G. PIK3CA-related overgrowth spectrum (PROS): diagnostic and testing eligibility criteria, differential diagnosis, and evaluation. Am. J. Med. Genet. A. 2015;167A:287–295. doi: 10.1002/ajmg.a.36836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boscolo E., Limaye N., Huang L., Kang K.T., Soblet J., Uebelhoer M., Mendola A., Natynki M., Seront E., Dupont S. Rapamycin improves TIE2-mutated venous malformation in murine model and human subjects. J. Clin. Invest. 2015;125:3491–3504. doi: 10.1172/JCI76004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venot Q., Blanc T., Rabia S.H., Berteloot L., Ladraa S., Duong J.P., Blanc E., Johnson S.C., Hoguin C., Boccara O. Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature. 2018;558:540–546. doi: 10.1038/s41586-018-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hucthagowder V., Shenoy A., Corliss M., Vigh-Conrad K.A., Storer C., Grange D.K., Cottrell C.E. Utility of clinical high-depth next generation sequencing for somatic variant detection in the PIK3CA-related overgrowth spectrum. Clin. Genet. 2017;91:79–85. doi: 10.1111/cge.12819. [DOI] [PubMed] [Google Scholar]
- 10.Nemeth K., Szabo S., Cottrell C.E., McNulty S.M., Segura A., Sokumbi O., Browning M., Siegel D.H. Mosaic pathogenic HRAS variant in a patient with nevus spilus with agminated Spitz nevi and parametrial-uterine rhabdomyosarcoma. Br. J. Dermatol. 2018;178:804–806. doi: 10.1111/bjd.16155. [DOI] [PubMed] [Google Scholar]
- 11.Siegel D.H., Cottrell C.E., Streicher J.L., Schilter K.F., Basel D.G., Baselga E., Burrows P.E., Ciliberto H.M., Vigh-Conrad K.A., Eichenfield L.F. Analyzing the Genetic Spectrum of Vascular Anomalies with Overgrowth via Cancer Genomics. J. Invest. Dermatol. 2018;138:957–967. doi: 10.1016/j.jid.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 12.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arafeh R., Samuels Y. PIK3CA in cancer: The past 30 years. Semin. Cancer Biol. 2019 doi: 10.1016/j.semcancer.2019.02.002. S1044-579X(18)30152-4. [DOI] [PubMed] [Google Scholar]
- 20.Hong T., Yan Y., Li J., Radovanovic I., Ma X., Shao Y.W., Yu J., Ma Y., Zhang P., Ling F. High prevalence of KRAS/BRAF somatic mutations in brain and spinal cord arteriovenous malformations. Brain. 2019;142:23–34. doi: 10.1093/brain/awy307. [DOI] [PubMed] [Google Scholar]
- 21.Al-Olabi L., Polubothu S., Dowsett K., Andrews K.A., Stadnik P., Joseph A.P., Knox R., Pittman A., Clark G., Baird W. Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy. J. Clin. Invest. 2018;128:1496–1508. doi: 10.1172/JCI98589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas A.C., Zeng Z., Rivière J.B., O’Shaughnessy R., Al-Olabi L., St-Onge J., Atherton D.J., Aubert H., Bagazgoitia L., Barbarot S. Mosaic Activating Mutations in GNA11 and GNAQ Are Associated with Phakomatosis Pigmentovascularis and Extensive Dermal Melanocytosis. J. Invest. Dermatol. 2016;136:770–778. doi: 10.1016/j.jid.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourdeaut F., Hérault A., Gentien D., Pierron G., Ballet S., Reynaud S., Paris R., Schleiermacher G., Baumann C., Philippe-Chomette P. Mosaicism for oncogenic G12D KRAS mutation associated with epidermal nevus, polycystic kidneys and rhabdomyosarcoma. J. Med. Genet. 2010;47:859–862. doi: 10.1136/jmg.2009.075374. [DOI] [PubMed] [Google Scholar]
- 24.Groesser L., Herschberger E., Ruetten A., Ruivenkamp C., Lopriore E., Zutt M., Langmann T., Singer S., Klingseisen L., Schneider-Brachert W. Postzygotic HRAS and KRAS mutations cause nevus sebaceous and Schimmelpenning syndrome. Nat. Genet. 2012;44:783–787. doi: 10.1038/ng.2316. [DOI] [PubMed] [Google Scholar]
- 25.Kuentz P., St-Onge J., Duffourd Y., Courcet J.B., Carmignac V., Jouan T., Sorlin A., Abasq-Thomas C., Albuisson J., Amiel J. Molecular diagnosis of PIK3CA-related overgrowth spectrum (PROS) in 162 patients and recommendations for genetic testing. Genet. Med. 2017;19:989–997. doi: 10.1038/gim.2016.220. [DOI] [PubMed] [Google Scholar]
- 26.Happle R. Lethal genes surviving by mosaicism: a possible explanation for sporadic birth defects involving the skin. J. Am. Acad. Dermatol. 1987;16:899–906. doi: 10.1016/s0190-9622(87)80249-9. [DOI] [PubMed] [Google Scholar]
- 27.Rivière J.B., Mirzaa G.M., O’Roak B.J., Beddaoui M., Alcantara D., Conway R.L., St-Onge J., Schwartzentruber J.A., Gripp K.W., Nikkel S.M., Finding of Rare Disease Genes (FORGE) Canada Consortium De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Genet. 2012;44:934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirzaa G., Timms A.E., Conti V., Boyle E.A., Girisha K.M., Martin B., Kircher M., Olds C., Juusola J., Collins S. PIK3CA-associated developmental disorders exhibit distinct classes of mutations with variable expression and tissue distribution. JCI Insight. 2016;1:1. doi: 10.1172/jci.insight.87623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gymnopoulos M., Elsliger M.A., Vogt P.K. Rare cancer-specific mutations in PIK3CA show gain of function. Proc. Natl. Acad. Sci. USA. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butler M.G., Dasouki M.J., Zhou X.P., Talebizadeh Z., Brown M., Takahashi T.N., Miles J.H., Wang C.H., Stratton R., Pilarski R., Eng C. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herman G.E., Butter E., Enrile B., Pastore M., Prior T.W., Sommer A. Increasing knowledge of PTEN germline mutations: Two additional patients with autism and macrocephaly. Am. J. Med. Genet. A. 2007;143A:589–593. doi: 10.1002/ajmg.a.31619. [DOI] [PubMed] [Google Scholar]
- 32.Negishi Y., Miya F., Hattori A., Johmura Y., Nakagawa M., Ando N., Hori I., Togawa T., Aoyama K., Ohashi K. A combination of genetic and biochemical analyses for the diagnosis of PI3K-AKT-mTOR pathway-associated megalencephaly. BMC Med. Genet. 2017;18:4. doi: 10.1186/s12881-016-0363-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wouters V., Limaye N., Uebelhoer M., Irrthum A., Boon L.M., Mulliken J.B., Enjolras O., Baselga E., Berg J., Dompmartin A. Hereditary cutaneomucosal venous malformations are caused by TIE2 mutations with widely variable hyper-phosphorylating effects. Eur. J. Hum. Genet. 2010;18:414–420. doi: 10.1038/ejhg.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lalonde E., Ebrahimzadeh J., Rafferty K., Richards-Yutz J., Grant R., Toorens E., Marie Rosado J., Schindewolf E., Ganguly T., Kalish J.M. Molecular diagnosis of somatic overgrowth conditions: A single-center experience. Mol. Genet. Genomic Med. 2019;7:e536. doi: 10.1002/mgg3.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nätynki M., Kangas J., Miinalainen I., Sormunen R., Pietilä R., Soblet J., Boon L.M., Vikkula M., Limaye N., Eklund L. Common and specific effects of TIE2 mutations causing venous malformations. Hum. Mol. Genet. 2015;24:6374–6389. doi: 10.1093/hmg/ddv349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soblet J., Limaye N., Uebelhoer M., Boon L.M., Vikkula M. Variable Somatic TIE2 Mutations in Half of Sporadic Venous Malformations. Mol. Syndromol. 2013;4:179–183. doi: 10.1159/000348327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couto J.A., Vivero M.P., Kozakewich H.P., Taghinia A.H., Mulliken J.B., Warman M.L., Greene A.K. A somatic MAP3K3 mutation is associated with verrucous venous malformation. Am. J. Hum. Genet. 2015;96:480–486. doi: 10.1016/j.ajhg.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Twigg S.R.F., Hufnagel R.B., Miller K.A., Zhou Y., McGowan S.J., Taylor J., Craft J., Taylor J.C., Santoro S.L., Huang T. A Recurrent Mosaic Mutation in SMO, Encoding the Hedgehog Signal Transducer Smoothened, Is the Major Cause of Curry-Jones Syndrome. Am. J. Hum. Genet. 2016;98:1256–1265. doi: 10.1016/j.ajhg.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spencer D.H., Tyagi M., Vallania F., Bredemeyer A.J., Pfeifer J.D., Mitra R.D., Duncavage E.J. Performance of common analysis methods for detecting low-frequency single nucleotide variants in targeted next-generation sequence data. J. Mol. Diagn. 2014;16:75–88. doi: 10.1016/j.jmoldx.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lumbroso S., Paris F., Sultan C., European Collaborative Study Activating Gsalpha mutations: analysis of 113 patients with signs of McCune-Albright syndrome--a European Collaborative Study. J. Clin. Endocrinol. Metab. 2004;89:2107–2113. doi: 10.1210/jc.2003-031225. [DOI] [PubMed] [Google Scholar]
- 41.Theda C., Hwang S.H., Czajko A., Loke Y.J., Leong P., Craig J.M. Quantitation of the cellular content of saliva and buccal swab samples. Sci. Rep. 2018;8:6944. doi: 10.1038/s41598-018-25311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang F., Liu L., Fang E., Zhang G., Chen T., Cao K., Li Y., Li M.M. Molecular Diagnosis of Mosaic Overgrowth Syndromes Using a Custom-Designed Next-Generation Sequencing Panel. J. Mol. Diagn. 2017;19:613–624. doi: 10.1016/j.jmoldx.2017.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.