Abstract
Age-related hearing impairment (ARHI) is the most common sensory impairment in the aging population; a third of individuals are affected by disabling hearing loss by the age of 65. It causes social isolation and depression and has recently been identified as a risk factor for dementia. The genetic risk factors and underlying pathology of ARHI are largely unknown, meaning that targets for new therapies remain elusive, yet heritability estimates range between 35% and 55%. We performed genome-wide association studies (GWASs) for two self-reported hearing phenotypes, using more than 250,000 UK Biobank (UKBB) volunteers aged between 40 and 69 years. Forty-four independent genome-wide significant loci (p < 5E−08) were identified, considerably increasing the number of established trait loci. Thirty-four loci are novel associations with hearing loss of any form, and only one of the ten known hearing loci has a previously reported association with an ARHI-related trait. Gene sets from these loci are enriched in auditory processes such as synaptic activities, nervous system processes, inner ear morphology, and cognition, while genetic correlation analysis revealed strong positive correlations with multiple personality and psychological traits for the first time. Immunohistochemistry for protein localization in adult mouse cochlea implicate metabolic, sensory, and neuronal functions for NID2, CLRN2, and ARHGEF28. These results provide insight into the genetic landscape underlying ARHI, opening up novel therapeutic targets for further investigation. In a wider context, our study also highlights the viability of using self-report phenotypes for genetic discovery in very large samples when deep phenotyping is unavailable.
Introduction
ARHI is characterized by a non-syndromic bilateral, sensorineural hearing loss that progresses with increasing age and is an established risk factor for depression1, 2, 3 and dementia.4, 5, 6, 7 Hearing loss was ranked fourth in the latest study into the Global Burden of Diseases,8 yet hearing amplification devices are the only treatment option currently available for ARHI, representing an area of unmet clinical need. The economic burden of the condition is increasing due to the growing size of the aging population; a recent review highlighted the excess medical costs due to hearing impairment as $3.3–$12.8 billion in the USA alone.9
ARHI is expected to be a highly genetically heterogeneous trait given that more than 150 genetic loci have been identified in non-syndromic congenital hearing loss alone (see Hereditary Hearing Loss Homepage in Web Resources). Previous GWASs of ARHI-related phenotypes have only identified a small number of promising candidate genes, though there has been poor replication of findings to date, possibly reflecting varied phenotyping methods and limited sample sizes.10, 11, 12, 13, 14, 15, 16, 17, 18, 19 Alternatively, it might suggest that the genetic contribution to ARHI has been overestimated. To date, five genomic loci have been significantly associated with ARHL in previous GWASs,15, 16, 18 only two of which were replicated in independent population samples. Understanding the genetic etiology of ARHI sheds light on the pathophysiology of the condition and ultimately facilitates the development of novel therapeutic or preventative interventions.
Here, we conducted two large hearing GWASs with sample sizes of more than 250,000 individuals using the self-reported hearing difficulty and hearing aid use of UKBB participants and refined our results using a combination of conditional analysis and replication analysis. In addition, we performed in silico functional annotation and in vivo expression analysis (see Figure 1 for study design) to understand the role of gene variants in the biological mechanisms of ARHI.
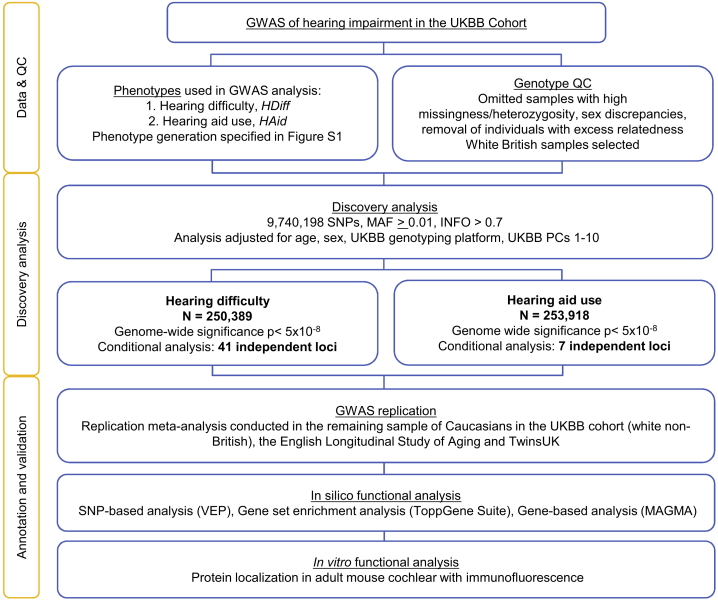
Figure 1.
Workflow Schematic for Discovery and Validation of Associated Loci
N, sample size; QC, quality control; PC, principal components; MAF, minor allele frequency; INFO, quality metric, combination of imputation score and dosage confidence.
Our aim was to identify the genetic components of adult hearing impairment in the UK population and provide insight into the pathology of ARHI and associated traits.
Subjects and Methods
Participants
The sample used for this study consisted of individuals who participated in the UKBB study. The UKBB is a national resource, initially set up to study lifestyle and genetic factors affecting aging traits with the aim of understanding and improving healthy aging at a population level. More than 500,000 volunteers attended 23 assessment centers across the UK between 2007 and 2013 where they donated samples for genotyping, completed lifestyle questionnaires, and had standard measurements taken. The UKBB resource is described extensively elsewhere.20
The cohort used for discovery association analysis consisted of UKBB participants with “white British” ancestry. The UKBB sample classification white British is derived from both principal component (PC) analysis and self-declared ethnicity.21 Samples with excess heterozygosity, excess relatedness, and sex discrepancies were identified and removed prior to analysis, resulting in samples sizes of n = 250,389 and n = 253,918 for hearing difficulty (HDiff) and hearing aid (HAid) use, respectively.
For replication analysis, we used the UKBB ethnic group “Caucasians” (white non-British Europeans). To assign participants into discrete ancestry clusters, we used the 1st and 2nd PC vectors provided by UKBB. A k-means clustering algorithm was applied to generate clusters for each PC. We then combined cluster indices for the PCs (1.1, 1.2, …, 5.5), compared them against self-reported ancestry, and assigned the ancestry group accordingly. If contradictory, the pairwise clusters took precedence over the self-report grouping.
Two samples were used for replication analysis, the English Longitudinal Study of Aging (ELSA) and TwinsUK. They were selected as they comprise predominantly Northern European ancestry samples and include relevant questionnaire data. ELSA is a longitudinal study, consisting of around 12,000 respondents from the Health Survey for England. Eight waves of data collection have been completed since 2002.22 TwinsUK is the largest adult twin registry in the UK and comprises more than 13,000 healthy twin volunteers aged 16–98. Collection of data and biological materials commenced in 1992 and is ongoing. During study participation, twins regularly complete health and lifestyle questionnaires and visit for clinical evaluation.23
Phenotype Definitions
Two phenotypes were derived for this study; a phenotype representing self-reported hearing difficulty (HDiff) and a phenotype representing self-reported hearing aid use (HAid). Participants in the UKBB study completed a touchscreen questionnaire during their visit to the assessment center, which included questions regarding hearing status. Participants were assigned case/control status based on their responses to questionnaire measures regarding hearing difficulty and hearing aid use. HDiff case subjects responded “Yes” to both of the questions “Do you have any difficulty with your hearing?” and “Do you find it difficult to follow a conversation if there is background noise (such as TV, radio, children playing)?” HDiff control subjects were selected if their response to both of these questions was “No.” Participants with any other combination of responses were removed. In addition, HDiff control subjects aged <50 were removed from analysis, as were any control subjects that responded “Yes” to the question “Do you use a hearing aid most of the time?” HAid case subjects responded “Yes” to “Do you use a hearing aid most of the time?” and control subjects responded “No.” Details of how the UKBB phenotype was derived are displayed in Figure S1.
If participants answered the questionnaire twice, i.e., attended an assessment center for a repeat visit, the answer at the second time point was used in analysis, in order to increase the mean age of the sample. To reduce the likelihood of including congenital forms of deafness, participants who selected “I am completely deaf” in the UKBB questionnaire were excluded from analysis. Note that a further, objective measure of hearing, the speech reception threshold using the “Digits in Noise” (DIN) protocol, was obtained from 160,955 of the UK Biobank participants.24, 25 Preliminary heritability assessment of the DIN did not yield clear heritability or association with age and therefore it was not considered suitable for the present study.
Questionnaire responses for the ELSA and TwinsUK replication samples were derived to obtain comparable phenotypes to the UKBB phenotype (Figure S1). For the ELSA sample, case/control phenotypes were derived from responses to questionnaire measures collected during study Wave 7. The HDiff phenotype was derived using responses from two questions: “Do you ever have any difficulties with your hearing?” and “Do you find it difficult to follow a conversation if there is background noise, such as TV, radio or children playing (using a hearing aid as usual)?” Case subjects were defined as participants who responded “Yes” to both questions, and control subjects who responded “No” to both questions. As in the UKBB analysis, control subjects who reported hearing aid use or age <50 were removed, as were any cochlear implant users in the case or control samples. The HAid phenotype was derived using the question “Whether ever wears a hearing aid;” case subjects responded “Yes most of the time,” or “Yes some of the time” while control subjects responded “No.” During ELSA data processing, age was capped at 90 years, and thus individuals aged >90 were reported to be 90 years of age. Resulting ELSA samples sizes for association analysis of these traits were HDiff = 3,545 and HAid = 4,482.
The TwinsUK phenotypes were likewise derived from responses to questions. HDiff case subjects responded either “Yes, diagnosed by doctor or health professional” or “Yes, not diagnosed by health professional” to the question “Do you suffer from hearing loss?” while control subjects responded “No.” HAid case subjects responded or indicated “Yes” to either of “Do you wear a hearing aid?” and “Wearing a hearing aid.” HAid control subjects responded “No.” As TwinsUK is a longitudinal study, a number of participants gave responses to the same questions on multiple occasions. The most recent response was included in analysis, unless the latest response indicated that hearing had improved. In this scenario, the participant was excluded. TwinsUK recruits adult twins aged over 18 years. Twins aged <40 were removed from analysis so that the lower age limit was comparable to the UKBB cohort. In order to retain the size of the TwinsUK sample and thus optimize power, controls aged <50 were not removed (as in the discovery HDiff UKBB analysis). Resulting Twins UK samples sizes for association analysis of these traits were HDiff = 3,636 and HAid = 3,435.
Genotyping and Imputation
The ∼500,000 samples in UKBB were genotyped on one of two arrays; 50,000 samples were genotyped on the Affymetrix UK BiLEVE Axiom array while the remaining ∼450,000 were genotyped on the Affymetrix UK Biobank Axiom array. The two arrays shared 95% coverage resulting in >800,000 genotyped SNPs. Imputation was carried out centrally by UKBB, primarily using the HRC reference panel and IMPUTE2.26 SNPs which do not feature on this panel were imputed with the UK 10K and 1000G panel. Analysis in this study was conducted with version 3 of the UKBB imputed data with 487,409 samples imputed and available for analysis following UKBB centrally performed QC filters.
ELSA samples were genotyped at UCL Genomics in two batches using the Illumina HumanOmni 2.5M platform. Imputation was carried out centrally by ELSA with IMPUTE2, using the 1000 Genomes phase I dataset27 (see Web Resources).
Genotyping of TwinsUK was conducted with a combination of Illumina arrays; HumanHap300, HumanHap610Q, 1M-Duo, and 1.2MDuo 1M. The imputation reference was 1000G Phase3 v5 (GRCh37).
Association Analysis
Discovery association analysis was performed using a linear mixed-effects model approach to test for association between imputed SNP dosages and the two traits. BOLT-LMM v.228 was used for the association analysis, which corrects for population stratification and within-sample relatedness. In addition, the analysis was adjusted for age, sex, UKBB genotyping platform, and UKBB PCs1-10. For quality control, SNPs were filtered based on two thresholds: (1) minor allele frequency (MAF) ≥ 0.01 and (2) INFO score > 0.7. By implementing a MAF cutoff of 0.01, we reduced the likelihood of including participants with forms of congenital deafness, as we only detected variants that occur at least in 1/100 participants, a higher frequency of variants than the frequency of congenital deafness. Individuals with <98% genotype call rate were removed.
Conditional and Joint Analysis
Conditional and joint SNP analysis was performed to identify independent signals within highly associated regions, using GCTA-COJO.29 This analysis requires the linkage disequilibrium reference sample, which was obtained by random selection of 10,000 individuals from the UKBB cohort with white British ancestry. The reference sample size was selected to maximize power based on previous data simulations.29 The distance assumed for complete linkage equilibrium was 10 Mb and a cut off value of R2 = 0.9 was used to check for collinearity between the selected SNPs and those to be tested. Alleles with a frequency difference > 0.2 between the reference sample and GWAS sample were excluded. Independent SNPs identified with GCTA-COJO were mapped to the nearest protein coding gene using variant effect predictor (VEP),30 genome build GRCh37. VEP was used to establish whether the SNP was in an exonic, intronic, or intergenic region and also the functional consequence of the variant at that position.
LD Score Regression
Univariate linkage disequilibrium score regression (LDSC) was used to calculate whether inflated test statistics were likely due to the polygenic nature of the trait or confounding bias, by analyzing the relationship between test statistic and LD.31
We also performed genetic correlation analysis between the HDiff trait and 765 traits available for correlation analysis on LD hub.31, 32 To filter our results, we calculated a conservative significance threshold with a multiple-test correction (0.05/764, p = 6.5E−5) and selected those with a correlation (rg) > 0.3 or < −0.3 to report in this study and grouped the remaining traits into five categories.
Heritability Estimates
SNP heritability estimates for the two traits were calculated with BOLT-LMM (h2g). As HDiff and HAid are both qualitative traits, these estimates were recalculated to the liability scale. Sample and population prevalence were specified as the case prevalence in the analyzed sample; HDiff at 0.35 and HAid at 0.052. SNP heritability was also calculated using a region-based approach with Heritability Estimation from Summary Statistics (HESS).33 SNP heritability was partitioned into 1702 approximately independent loci (see Web Resources). The EUR 1000G reference panel was used for LD estimation (see Web Resources).
Replication Association Analysis
The lead SNPs for each locus identified with conditional analysis (Table 1) were tested for association with HDiff and HAid phenotypes in each of the three cohorts UKBB (white non-British Europeans), TwinsUK, and ELSA. The UKBB white non-British sample was examined using the same protocol as the white British dataset described above, under the linear mixed-effects models method with BOLT-LMM adjusting for age, sex, UKBB PCs 1-10, and genotyping platform. As BOLT-LMM is unsuitable for analysis of samples with N < 5,000, alternative software was used for the TwinsUK and ELSA association analysis. The TwinsUK sample was analyzed using a linear mixed-effects model regression adjusting for age and sex with GEMMA,34 the most suitable software for twins as it can control for family structure. For the ELSA sample, one of each pair of related individuals was excluded from analysis during central quality control checks at ELSA (relatedness was estimated in PLINK 1.935), and PLINK2 logistic regression was used to test for association in the ELSA sample, adjusting for age and sex.
Table 1.
Independent SNPs Significantly Associated (p < 5 × 10−8) with the Two Phenotypes Regarding Hearing Ability in the UK Biobank Discovery Sample
| Chr | SNP | EA | EAF | INFO | β | SE | p Value | pJ-Value | Nearest Gene | Distance to Gene (bp) | Other Genes within 100 kb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hearing Difficulty GWAS | |||||||||||
| 22 | rs36062310 | G | 0.96 | 1.000 | −0.0315 | 0.003 | 1.90E−22 | 1.92E−22 | KLHDC7B | 0 | SYCE3, ADM2, ARSA,aCHKB, CPT1B, LMF2, MAPK8IP2, MIOX, NCAPH2, ODF3B, SBF1, SCO2, SYCE3, TYMP |
| 5 | rs6453022 | C | 0.50 | 1.000 | −0.0126 | 0.001 | 1.70E−21 | 2.07E−12 | ARHGEF28 | 0 | – |
| 6 | rs759016271 | AGTAGTCCACTTTTCTTCTTTGCCTG | 0.39 | 0.997 | −0.0127 | 0.001 | 6.10E−21 | 6.16E−21 | ZNF318 | 0 | CRIP3, SLC22A7, CUL9, DNPH1, TTBK1 |
| 5 | rs6890164 | A | 0.51 | 0.993 | 0.0119 | 0.001 | 3.30E−19 | 4.15E−10 | ARHGEF28 | 6177 | – |
| 11 | rs7951935 | G | 0.62 | 0.996 | −0.0114 | 0.001 | 7.80E−17 | 7.85E−17 | TYR | 1472 | NOX4 |
| 6 | rs35186928 | G | 0.62 | 0.991 | −0.0109 | 0.001 | 1.70E−15 | 1.69E−15 | HLA-DQA1 | 13352 | HLA-DRB1, HLA-DRB3, HLA-DRB5, HLA-DRB6 |
| 6 | rs9493627 | G | 0.68 | 1.000 | −0.0104 | 0.001 | 1.40E−13 | 1.41E−13 | EYA4a | 0 | – |
| 22 | rs132929 | G | 0.59 | 0.999 | −0.0098 | 0.001 | 2.20E−13 | 4.61E−13 | BAIAP2L2a | 0 | SLC16A8, PICK1, PLA2G6, POLR2F |
| 22 | rs5756795 | T | 0.54 | 1.000 | −0.0092 | 0.001 | 5.10E−12 | 1.09E−11 | TRIOBPa | 0 | GALR3, GCAT, GGA1,aH1F0, LGALS1, NOL12, PDXP, SH3BP1 |
| 14 | rs1566129 | T | 0.41 | 1.000 | 0.0091 | 0.001 | 1.40E−11 | 1.37E−11 | NID2 | 0 | GNG2, RTRAF |
| 4 | rs35414371 | T | 0.87 | 0.998 | −0.0131 | 0.002 | 1.60E−11 | 1.64E−11 | CLRN2a | 1965 | LAP3, MED28, QDPR |
| 3 | 3:182069497_TA_T | TA | 0.84 | 0.989 | −0.0118 | 0.002 | 4.10E−11 | 4.07E−11 | ATP11B | 441791 | – |
| 11 | rs12225399 | G | 0.65 | 0.989 | −0.009 | 0.001 | 8.60E−11 | 8.67E−11 | PHLDB1 | 0 | ARCN1, IFT46, KMT2A, TMEM25, TREH, TTC36 |
| 11 | rs55635402 | A | 0.81 | 0.996 | 0.0105 | 0.002 | 2.90E−10 | 2.94E−10 | TUBa | 0 | EIF3F, NLRP10, OR10A3, RIC3 |
| 16 | rs62033400 | A | 0.61 | 0.999 | 0.0085 | 0.001 | 2.90E−10 | 2.95E−10 | FTO | 0 | RPGRIP1L |
| 8 | rs13277721 | G | 0.49 | 0.992 | −0.0083 | 0.001 | 3.30E−10 | 3.35E−10 | AGO2 | 0 | PTK2 |
| 2 | rs62188635 | C | 0.45 | 0.988 | 0.0083 | 0.001 | 4.70E−10 | 4.72E−10 | KLF7 | 50519 | – |
| 6 | rs2236401 | C | 0.49 | 0.997 | −0.0081 | 0.001 | 9.30E−10 | 9.38E−10 | SYNJ2a | 0 | SERAC1,aGTF2H5 |
| 7 | rs4947828 | T | 0.23 | 0.999 | −0.0096 | 0.002 | 1.00E−09 | 1.02E−09 | GRB10 | 0 | – |
| 10 | rs6597883 | T | 0.84 | 0.989 | 0.0111 | 0.002 | 1.00E−09 | 1.05E−09 | CTBP2 | 0 | – |
| 5 | rs34442808 | T | 0.49 | 0.992 | −0.008 | 0.001 | 1.30E−09 | 1.32E−09 | MCTP1, SLF1 | 0 | – |
| 10 | rs835267 | A | 0.53 | 0.996 | 0.008 | 0.001 | 1.60E−09 | 1.58E−09 | EXOC6 | 0 | CYP26A1, CYP26C1 |
| 10 | rs4948502 | T | 0.57 | 0.995 | 0.0081 | 0.001 | 1.70E−09 | 5.63E−10 | ARID5B | 0 | – |
| 10 | rs10824108 | G | 0.42 | 0.999 | −0.0079 | 0.001 | 3.00E−09 | 1.24E−08 | ADK | 0 | AP3M1, VCL |
| 1 | rs12027345 | G | 0.57 | 0.995 | 0.0079 | 0.001 | 3.60E−09 | 3.64E−09 | MAST2 | 12668 | GPBP1L1, MAST2, TMEM69, TMA16P2, GPBP1L1 |
| 6 | rs217289 | G | 0.56 | 0.992 | −0.0078 | 0.001 | 4.90E−09 | 4.92E−09 | SNAP91 | 0 | – |
| 3 | rs13093972 | A | 0.55 | 0.992 | −0.0078 | 0.001 | 5.50E−09 | 5.56E−09 | ZBTB20 | 121137 | – |
| 15 | rs62015206 | C | 0.41 | 1.000 | −0.0078 | 0.001 | 7.70E−09 | 7.76E−09 | MAPK6 | 15613 | BCL2L10, GNB5 |
| 5 | rs10475169 | A | 0.88 | 1.000 | −0.0117 | 0.002 | 9.30E−09 | 9.37E−09 | IRX2 | 190445 | – |
| 17 | rs17671352 | T | 0.38 | 0.999 | 0.0078 | 0.001 | 1.00E−08 | 1.43E−08 | ACADVL | 0 | DVL2,aDLG4, ASGR1, CLDN7, CTDNEP1, EIF5A, ELP5, GABARAP, GPS2, NEURL4, PHF23, SLC2A4, YBX2 |
| 1 | rs7525101 | C | 0.56 | 1.000 | −0.0075 | 0.001 | 1.50E−08 | 1.45E−08 | LMX1Aa | 61973 | – |
| 17 | rs12938775 | G | 0.50 | 1.000 | 0.0075 | 0.001 | 1.60E−08 | 2.25E−08 | PAFAH1B1 | 0 | CLUH, RAP1GAP2 |
| 8 | rs76837345 | A | 0.93 | 0.997 | −0.0146 | 0.003 | 1.90E−08 | 1.95E−08 | CHMP4C | 0 | IMPA1, SLC10A5, SNX16, ZFAND1 |
| 6 | rs9366417 | G | 0.26 | 0.993 | 0.0085 | 0.002 | 2.10E−08 | 2.12E−08 | SOX4 | 291019 | – |
| 8 | rs3890736 | G | 0.63 | 0.993 | −0.0077 | 0.001 | 2.20E−08 | 2.22E−08 | GFRA2 | 15676 | – |
| 10 | rs143282422 | G | 0.99 | 1.000 | −0.0349 | 0.006 | 2.40E−08 | 3.02E−08 | CDH23a | 0 | C10orf105 |
| 7 | rs9691831 | A | 0.42 | 0.995 | −0.0074 | 0.001 | 3.10E−08 | 3.11E−08 | TMEM213 | 0 | ATP6V0A4,aKIAA1549 |
| 11 | rs141403654 | A | 0.98 | 0.878 | −0.0313 | 0.006 | 3.50E−08 | 3.53E−08 | AGBL2 | 0 | C1QTNF4, FNBP4, MTCH2, NUP160 |
| 18 | rs4611552 | T | 0.78 | 0.995 | −0.0089 | 0.002 | 3.60E−08 | 3.56E−08 | CCDC68 | 9362 | – |
| 13 | rs12552 | A | 0.44 | 0.994 | 0.0073 | 0.001 | 4.80E−08 | 4.86E−08 | OLFM4 | 0 | – |
| 1 | rs10927035 | C | 0.35 | 0.995 | −0.0075 | 0.001 | 4.90E−08 | 4.89E−08 | AKT3 | 0 | SDCCAG8 |
| Hearing Aid GWAS | |||||||||||
| 5 | rs4597943 | G | 0.51 | 0.989 | −0.0042 | 0.001 | 2.10E−11 | 2.09E−11 | ARHGEF28 | 0 | – |
| 2 | rs9677089 | A | 0.75 | 0.989 | −0.0046 | 0.001 | 2.00E−10 | 1.98E−10 | SPTBN1a | 0 | – |
| 6 | rs9321402 | G | 0.68 | 0.999 | −0.0042 | 0.001 | 3.00E−10 | 3.02E−10 | EYA4a | 0 | – |
| 14 | rs1566129 | T | 0.41 | 1.000 | 0.0037 | 0.001 | 2.50E−09 | 2.53E−09 | NID2 | 0 | RTRAF |
| 3 | rs3915060 | C | 0.27 | 0.983 | 0.004 | 0.001 | 9.70E−09 | 9.70E−09 | ILDR1a | 0 | CD86, SLC15A2 |
| 10 | rs10901863 | C | 0.73 | 0.934 | −0.004 | 0.001 | 2.60E−08 | 2.65E−08 | CTBP2 | 0 | – |
| 8 | rs7823971 | C | 0.80 | 0.991 | −0.0043 | 0.001 | 2.70E−08 | 2.68E−08 | RP11-1102P16.1 | 0 | – |
Definition of terms: Chr., chromosome; SNP, single-nucleotide polymorphism; EA, effect allele; EAF, frequency of effect allele in BOLT-LMM; INFO, quality metric, combination of imputation score and dosage confidence; β, effect size from BOLT-LMM approximation to infinitesimal mixed model; SE, standard error of the effect size; p value, infinitesimal mixed-effects model association test p value; pJ-value, p value from a joint analysis of all the selected SNPs; nearest gene, protein-coding gene in closest proximity to SNP; distance to gene (bp), distance in base pairs between SNP and nearest gene, a value of 0 indicates the SNP lies within the gene; other genes within 100 kb, list of genes within 100 kb of the SNP.
Genes previously linked to hearing phenotypes in mice or humans. Two SNPs reached genome-wide significance in the HAid analysis that are in close proximity to HLA-DQA1 on chr 6 (Figure 2) but were not present in conditional analysis results.
For SNPs significantly associated with hearing difficulty in the discovery, a fixed-effect inverse-variance weighted meta-analysis was conducted using METAL36 version 2011-03-25 with the three samples: white non-British UKBB, ELSA, and TwinsUK. BOLT-LMM does not report analyzed sample size per SNP, so to obtain the weight of the UKBB replication sample per SNP, sample size was calculated from PLINK linear regression.
A power calculation37 was performed for each independent locus analyzed in the replication analysis (p < 0.05 for both traits and p < 0.0012 for HDiff and p < 0.00714 for HAid, Tables S1 and S2).
Gene Prioritization, Pathway, and Tissue Enrichment Analysis
Summary statistics from the UKBB HDiff trait were input for Functional Mapping and Annotation of Genome-wide Association Studies (FUMA38). First, the SNP2GENE function was used to identify lead SNPs from HDiff analysis that reached a suggestive level of significance and were independent at r2 < 0.1. These identified lead SNPs were used to perform gene set enrichment analysis with ToppGene Suite.39
Second, gene-set analysis was performed with MAGMA40 using all of the SNPs that were analyzed in the HDiff discovery association analysis. Here, SNPs were mapped to protein coding genes that were within 10 kb of the SNP location. The effects of multiple SNPs were combined to calculate the significance of an association of a gene with HDiff. These significantly associated genes were analyzed for differential expression in 30 general tissue types.
Mouse Tissue Preparation
Adult mouse cochleae were collected at p28–p30 from C57BL/6 mice, bred in an in-house facility. Mice were euthanized according to Schedule 1 procedures as described in United Kingdom legislation outlined in the Animals (Scientific Procedures) Act 1986. Dissected inner ears were fixed in 4% paraformaldehyde diluted in PBS for 1 h at room temperature before being washed several times in PBS. They were then decalcified in 10% EDTA overnight at 4°C, before being separated from the vestibular system. Cochlea were mounted in 4% low-melting point agarose and sectioned on a Vibratome (1000 plus system, Intracel) at 200-μm intervals.
Immunofluorescence
Antibodies used to identify protein localization in the organ of Corti were: anti-nidogen-2 (NID2) at a 1:750 dilution (Ab14513, Abcam), anti-clarin-2 (CLRN2) at 1:1,000 (HPA042407, Atlas Antibodies), and anti-rho guanine nucleotide exchange factor 28 (ARHGEF28) at 1:1,000 (HPA037602, Atlas Antibodies). All were detected using an isotype-specific Alexa Fluor 488 goat anti-rabbit secondary antibody (Santa Cruz Biotechnology). Antibodies were diluted in a goat blocking solution (4% triton, 8% goat serum, 1 g BSA, 50 mL PHEM buffer) and sections were stained with primary antibodies overnight at 4°C. Following PBS washes, sections were incubated with the secondary antibody at 1:1,000 in darkness at room temperature for 2 h. Phalloidin-Atto 647N to f-actin (Sigma-Aldrich) and DAPI were added to the secondary antibody incubations at 1:1,000 to stain hair cell stereocilia and DNA, respectively. Samples were imaged using a Zeiss LSM 880 Airyscan 20× objective.
Results
Phenotype Definition
UKBB participants were categorized using a case-control design based on responses to questions regarding hearing difficulty (HDiff, n = 498,281) and hearing aid use (HAid, n = 316,629) (see Subjects and Methods and Figure S1 for details). Following quality control filters and selection of white British participants (described in Subjects and Methods), the final sample sizes used for association analyses were n = 250,389 (87,056 case subjects and 163,333 control subjects) for HDiff, and n = 253,918 (13,178 case subjects and 240,740 control subjects) for HAid.
Genome-wide Association Analysis
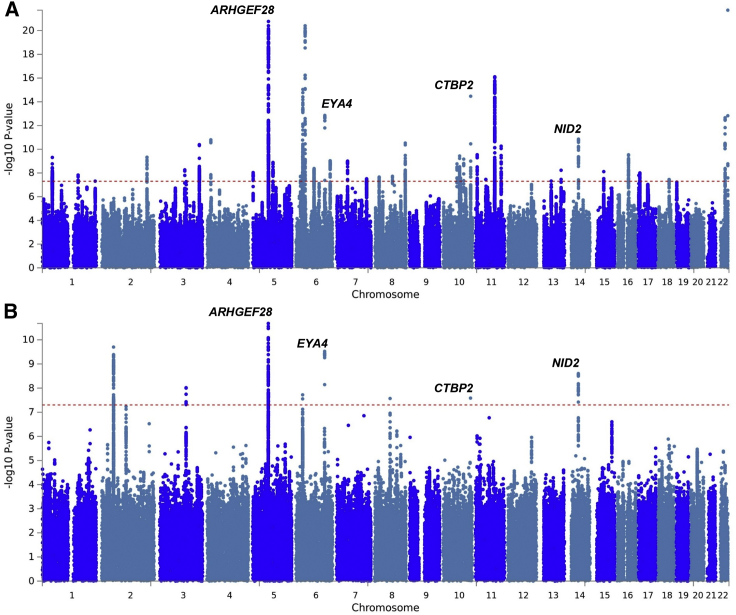
A linear mixed-effects model was used to test for association between 9,740,198 SNPs and the two hearing traits, using BOLT-LMM v.2,28 which corrects for population stratification and within sample relatedness. The studies identified 2,080 and 240 SNPs at genome-wide significance (p < 5E−08) for HDiff and HAid analysis, respectively (Figures 2 and S2). Conditional and joint analysis using GCTA-COJO29 identified that these SNPs represent 41 and 7 independent loci associated with HDiff and HAid, respectively, including four loci common to both traits, thus resulting in 44 independent loci overall.
Figure 2.
Manhattan Plots Displaying GWAS Results for Hearing Difficulty and Hearing Aid Use Phenotypes
Shown are (A) hearing difficulty and (B) hearing aid use. The Manhattan plots display the p values of all SNPs tested in discovery analysis. The threshold for genome wide significance (p < 5 × 10−8) is indicated by a red dotted line. Loci that reached genome-wide significance in both phenotypes are annotated with gene symbol.
SNP heritability estimates for the two traits calculated with BOLT-LMM (h2g) were 0.117 ± 0.001 for HDiff and 0.029 ± 0.001 for HAid. Estimates recalculated to the liability scale are 0.19 and 0.13 for HDiff and HAid, respectively. SNP heritability was also calculated using a region-based approach (Figure S3); one local heritability estimate was significant (Bonferroni corrected p < 0.05 for 1,702 loci) for HDiff, chr5 base positions 71240456–73759326, p = 1.16E–05. No regions were significant in HAid (Figure S3). The Variant Effect Predictor (VEP)30 was used to map independent lead SNPs to the nearest protein coding genes, using the GRCh37 genomic reference. Of 41 independent SNPs associated with HDiff, 6 variants lie in exons, 4 of which result in missense mutations in EYA4 (MIM: 603550), CDH23 (MIM: 605516), KLHDC7B, and TRIOBP (MIM: 609761), 21 SNPs lie within introns and 14 are intergenic (Table 1). Six of the independent SNPs associated with HAid reside in intronic regions and 1 is intergenic. Two highly significant independent associations with HDiff are found within 100 kb of the ARHGEF28 (MIM: 612790) gene, this locus is also highly associated with the HAid phenotype. Other gene loci common to both traits are NID2 (MIM: 605399), CTBP2 (MIM: 602619), and EYA4 (see Figure S4 for locus plots).
Replication Analysis
Replication was attempted for the lead SNPs (41 HDiff and 7 HAid) by meta-analyzing three independent samples: the remaining Caucasians in the UKBB cohort (white, non-British Europeans), TwinsUK, and the English Longitudinal Study of Aging (ELSA), totalling HDiff n = 30,765 and HAid n = 35,004. Two intronic SNPs, rs759016271 in ZNF318 (MIM: 617512) and rs1566129 in NID2, reached significance in the HDiff replication analysis (Bonferroni correction 0.05/41 = 0.0012, p < 0.0012), and a further intronic SNP, rs4597943 in ARHGEF28, replicated in HAid analysis at the significance threshold (0.05/7 = 0.00714, p < 0.00714). An additional 14 SNPs reached nominal significance and the power to detect each variant is also shown (Tables S1 and S2).
We investigated whether any of the candidate genes identified in adult hearing in previously published genetic association studies were replicated within the discovery white British sample (Table 2) and found only two previous variant associations located in close proximity to ISG20 (MIM: 604533) and within TRIOBP, which were identified in a GWAS performed with data from electronic health records.18
Table 2.
Summary Statistics from HDiff and HAid GWAS Analysis, at SNPs Highlighted in Previous Adult Hearing Loss GWAS
|
Variant Highlighted in Previous Study |
Summary Statistics from HDiff and HAid Analysis in the UKBB Cohort |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Citation | Gene | SNP | CHR | BP | A1 | A0 | INFO | UKBB Phenotype | A1FREQ | BETA | SE | p |
| Friedman et al.14 | GRM7 | rs11928865 | 3 | 7155702 | T | A | 0.989 | HDiff | 0.741 | 0.0016 | 0.0015 | 0.28 |
| HAid | 0.742 | −0.0014 | 0.0007 | 0.05 | ||||||||
| Van Laer et al.12 | IQGAP2 | rs457717 | 5 | 75920972 | A | G | 0.986 | HDiff | 0.326 | 0.0013 | 0.0014 | 0.34 |
| HAid | 0.325 | −0.0006 | 0.0007 | 0.37 | ||||||||
| GRM7 | rs161927 | 3 | 7838242 | G | A | 0.988 | HDiff | 0.134 | 0.0038 | 0.0019 | 0.05 | |
| HAid | 0.136 | −0.0002 | 0.0009 | 0.86 | ||||||||
| Girotto et al.17 | DCLK1 | rs248626 | 5 | 141097725 | A | G | 1.000 | HDiff | 0.251 | 0.0018 | 0.0015 | 0.23 |
| HAid | 0.252 | −0.0003 | 0.0007 | 0.71 | ||||||||
| KCNMB2 | rs4603971 | 3 | 177902467 | G | A | 0.992 | HDiff | 0.934 | −0.0015 | 0.0027 | 0.58 | |
| HAid | 0.934 | 0.0006 | 0.0012 | 0.63 | ||||||||
| CMIP | rs898967 | 16 | 81566780 | C | T | 0.981 | HDiff | 0.476 | 0.0010 | 0.0013 | 0.45 | |
| HAid | 0.476 | 0.0002 | 0.0006 | 0.76 | ||||||||
| GRM8 | rs2687481 | 7 | 125869122 | G | T | 0.998 | HDiff | 0.811 | −0.0018 | 0.0017 | 0.28 | |
| HAid | 0.810 | 0.0012 | 0.0008 | 0.14 | ||||||||
| Nolan et al.19 | ESSRG | rs2818964 | 1 | 216682448 | G | A | 0.978 | HDiff | 0.366 | −0.0015 | 0.0014 | 0.27 |
| HAid | 0.366 | 0.0004 | 0.0006 | 0.55 | ||||||||
| Wolber et al.15 | SIK3 | rs681524 | 11 | 116748314 | T | C | 0.992 | HDiff | 0.927 | −0.0010 | 0.0026 | 0.71 |
| HAid | 0.928 | 0.0018 | 0.0012 | 0.13 | ||||||||
| Vuckovic et al.16 | PCDH20 | rs78043697 | 13 | 62467039 | T | C | 0.995 | HDiff | 0.928 | 0.0000 | 0.0025 | 1.00 |
| HAid | 0.928 | 0.0010 | 0.0012 | 0.38 | ||||||||
| SLC28A3 | rs7032430 | 9 | 86714002 | C | A | 0.959 | HDiff | 0.782 | −0.0013 | 0.0016 | 0.43 | |
| HAid | 0.783 | −0.0001 | 0.0008 | 0.91 | ||||||||
| Fransen et al.10 | ACVR1B | rs2252518 | 12 | 52381026 | C | A | 0.996 | HDiff | 0.739 | −0.0010 | 0.0015 | 0.50 |
| HAid | 0.739 | 0.0001 | 0.0007 | 0.85 | ||||||||
| CCBE1 | rs34175168 | 18 | 57180682 | G | A | 0.990 | HDiff | 0.986 | 0.0112 | 0.0056 | 0.04 | |
| HAid | 0.986 | −0.0009 | 0.0026 | 0.74 | ||||||||
| Hoffman et al.18 | ISG20 | rs4932196 | 15 | 89253268 | T | C | 1.000 | HDiff | 0.809 | 0.0085 | 0.0017 | 4.60E−07 |
| HAid | 0.809 | 0.0039 | 0.0008 | 6.40E−07 | ||||||||
| TRIOBP | rs5756795a | 22 | 38122122 | T | C | 1 | HDiff | 0.539 | −0.0092 | 0.0013 | 5.10E−12 | |
| HAid | 0.538 | −0.0027 | 0.0006 | 1.60E−05 | ||||||||
Definition of terms: study, publication of previous finding; gene, gene highlighted in the referenced publication as the lead SNP is either located in the gene region or in close proximity; SNP, single-nucleotide polymorphism; CHR, chromosome; BP, base position; A1, effect allele in analysis; A0, reference allele; INFO, quality metric, combination of imputation score and dosage confidence; UKBB phenotype, phenotype used in this study; A1FREQ, frequency of effect allele in analysis sample; BETA, effect size from BOLT-LMM approximation to infinitesimal mixed model; SE, standard error of the effect size; p value, infinitesimal mixed model association test p value.
This study did not analyze SNP rs58389158, but analyzed rs5756795 which is in complete LD with this SNP in the British population, and referenced in the previous study.
ISG20 is a novel association with hearing, but mutations in TRIOBP cause a form of autosomal-recessive non-syndromic deafness, Deafness, autosomal-recessive 28 (DFNB28 [MIM: 609823]).41, 42 No other lead variants from previous ARHI genetic studies were replicated at nominal level in our analysis, including the first reported ARHI-associated gene variant in GRM711, 13 (MIM: 604101).
Gene Prioritization, Pathway, and Tissue Enrichment Analysis
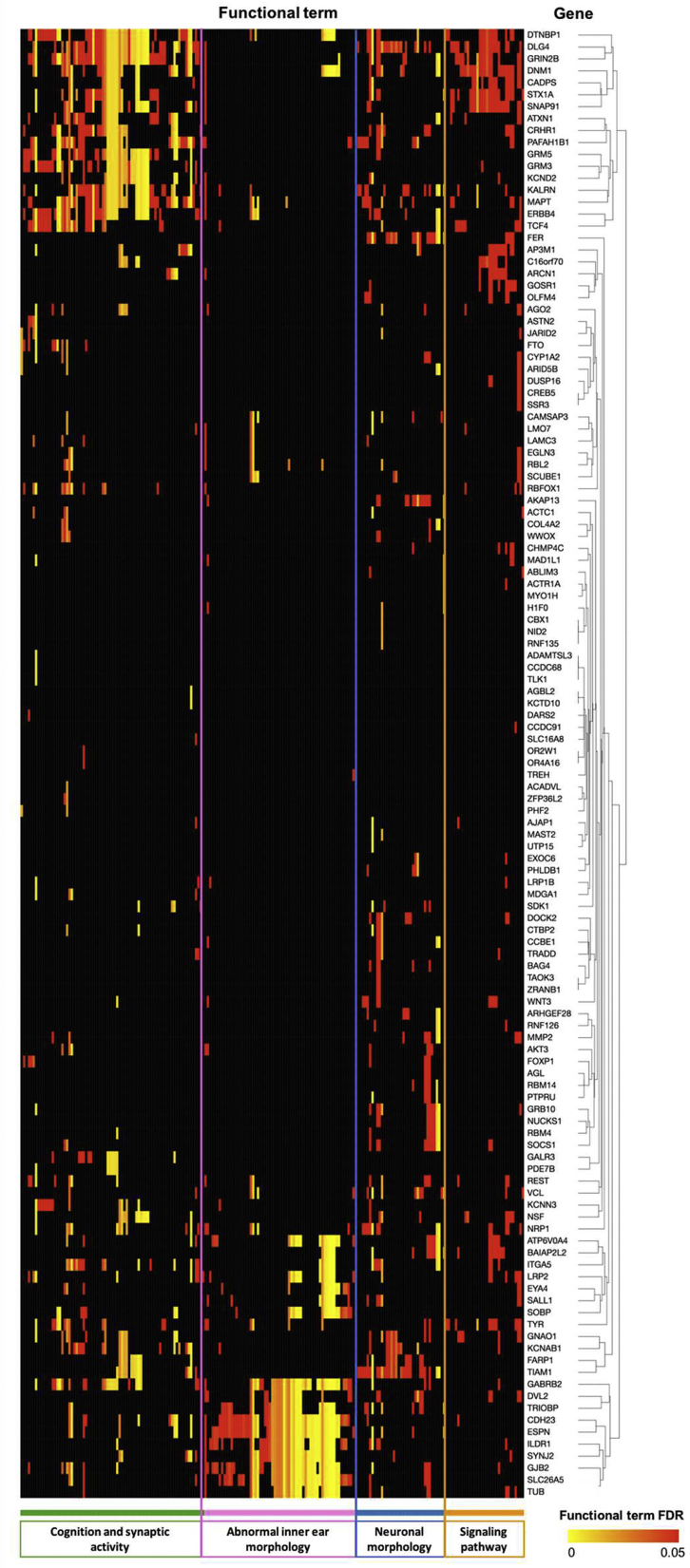
Functional gene annotation was undertaken with genes mapped from SNPs associated at a suggestive level (p < 1E−05) in the HDiff association analysis (Figure 3). Genes were significantly enriched in a number of processes required for auditory function: synaptic activities, trans-synaptic signaling, nervous system processes, modulation of chemical synaptic transmission, positive dendritic spine morphogenesis, and inner ear morphology as well as cognition, learning, or memory. These genes were also significantly enriched with mouse phenotype ontologies, mostly relating to inner ear abnormalities and abnormal auditory brainstem response, and were significant at FDR 0.05 (Figure 3). As well as suggesting pathogenic pathways, this finding demonstrates the shared genetic pathology in mouse and human auditory systems, supporting the use of mouse models to study human auditory function.
Figure 3.
Heatmap of the Enriched Functional Terms for Genes Mapped to Lead SNP at Suggestive Level (HDiff Analysis) via ToppGene Suite
Genes and functional terms were grouped using clustering of the strength of the enrichment of genes for respective functional terms. Functional terms include GO Biological Process, GO Molecular Function, GO Cellular Component, Mouse Phenotype, Pathway, and Disease.
In silico tissue-specific gene expression analysis undertaken with MAGMA40 indicates a significant association between HDiff genes and transcription levels of genes in the brain (p = 5.4E−04; Figure S5).
LD Score Regression
The LD score regression intercepts for the two analyses were 1.032 for HDiff and 1.03 for HAid. The ratio (intercept − 1)/(mean(χ2) − 1) for HDiff was 8% and represents the proportion of inflation in the χ2 statistic that the intercept attributes to alternative explanations than polygenicity. The ratio for HAid was 5%. We also performed genetic correlation analysis between HDiff and the 764 available traits on LDSC.31 After removing the 3 traits that were used to create the HDiff and HAid phenotypes used in this study, 153 traits were significantly correlated with HDiff. Here we have highlighted 41 traits (significant correlation with HDiff and an rg < −0.3 or rg > 0.3) and grouped these into five categories: Hearing, Low mood/depression, Pain, Breathing difficulties, and Health report/Subjective wellbeing (Table S3). Regarding hearing traits, a strong positive correlation was observed between HDiff and self-reported tinnitus “now or most of the time” (rg = 0.6, SE 0.056, p = 1.40E−26). Traits in the low mood/depression group included “frequency of tiredness/lethargy in last 2 weeks” (rg = 0.41, SE 0.029, p = 2.79E−45), neuroticism score (rg = 0.31, SE 0.026, p = 1.94E−34), miserableness (rg = 0.33, SE 0.027, p = 2.69E−33), and whether “seen doctor (GP) for nerves, anxiety, tension or depression” (rg = 0.35, SE 0.03, p = 5.90E−31).
Protein Localization of Putative Novel Hearing Genes
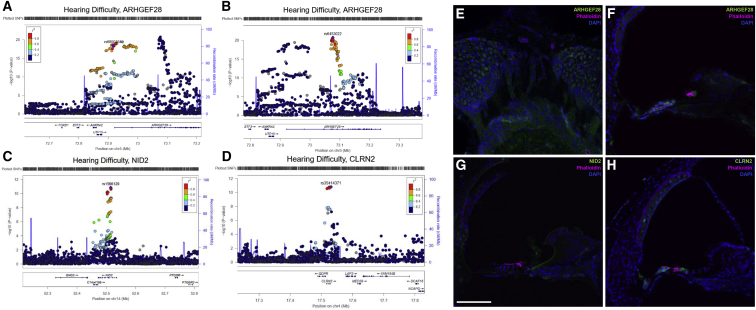
We investigated expression of three putative novel hearing genes—NID2, ARHGEF28, and CLRN2—in adult mouse cochlea using immunohistochemistry based on a number of factors. The lead SNP in NID2 in both HDiff and HAid is located in intron 5 and was also replicated in the HDiff meta-analysis. Two independent lead SNPs were identified at the ARHGEF28 locus in the HDiff analysis, along with a third SNP in the HAid analysis which replicated in the meta-analysis. The lead independent SNP at the CLRN2 locus in the HDiff analysis is within 2 kb of CLRN2, although several other genes are within 100 kb. Because CLRN1 (MIM: 606397), a paralog of CLRN2, is expressed in hair cells and mutations in CLRN1 cause autosomal-recessive Usher syndrome type 3 (USH3 [MIM: 276902]) with progressive sensorineural hearing loss,43, 44 we investigated whether clarin-2 is also expressed in the inner ear.
Immunostaining for nidogen-2, a basement membrane component encoded by NID2, was most prominent in the epithelial lining of the inner spiral sulcus between the tectorial membrane and the inner hair cell (Figures 4 and S6), as well as localizing to nerve fibers and blood vessel basement membranes. Similar to clarin-1, clarin-2 immunostaining localized to the inner and outer hair cells, the primary sensory cells of sound detection (Figures 4 and S6). ARHGEF28 encodes Rho Guanine Nucleotide Exchange Factor 28, for which immunostaining was observed in both hair cells and the spiral ganglion neuron cell bodies and axons (Figures 4 and S6).
Figure 4.
Cochlear Expression of Three Putative Hearing Genes Identified in HDiff and HAid GWASs
(A–D) Locus zoom plots of associated loci, generated with HDiff summary statistics. Four associated loci are plotted which have lead SNPs in or in proximity to ARHGEF28 (A and B), NID2 (C), and CLRN2 (D). Purple indicates lead independent SNP generated from GCTA-COJO conditional analysis. Coloring of remaining SNPs is based on linkage disequilibrium (LD) with the lead SNP. The genes within the region are annotated, and the direction of the transcripts is shown by arrows. Two independent regions were identified within the ARHGEF28 locus; both are shown.
(E–H) Immunofluorescence images of adult mouse cochlea, spiral ganglion neurons (E) and organ of Corti (F–H). Vibratome sections stained with the three proteins of interest in mouse inner ear; DAPI (blue) and Phalloidin (magenta) were also used for staining of actin and nuclei, respectively.
(E) Anti-ARHGEF28 (green) staining is observed in the neuronal cell bodies and axons.
(F) Anti-ARHGEF28 (green) staining is mainly observed in outer and inner hair cells.
(G) Anti-NID2 (green) staining is observed lining blood vessels and the epithelial lining of the inner spiral sulcus.
(H) Anti-CLRN2 (green) staining is observed in outer and inner hair cells, in addition to the stria vascularis.
The scale bar in (G) represents 100 μm. The scale is consistent for all images in this figure.
Discussion
Using data from UKBB, we have performed a GWAS with a sample size of more than 250,000 individuals, identifying 2,320 genome-wide significant SNP associations, representing 44 independent associations with self-reported adult hearing loss in participants aged 40–69 years. Only one of these loci, TRIOBP, has previously been associated with ARHI. Thirty-four of the loci are novel to hearing function while ten loci have previously been linked to some form of hearing loss either in mouse models or humans (Table 1). Two independent associations were found within 100 kb of the ARHGEF28 locus. The findings represent a more than 400-fold increase in the number of reported genome-wide significant SNP associations and a 9-fold increase in independent genomic loci for ARHI-related phenotypes and provide a major breakthrough in revealing the genetic architecture of ARHI.
Pure tone audiometry, the gold standard measure of hearing, requires an audiologist, a quiet environment, and significant clinical time and is therefore challenging to collect in the large samples needed for GWASs. Due to this limitation, previous GWASs in well-characterized audiometric cohorts have included fewer than 5,000 case subjects. The most recent study, by Hoffmann et al.,18 utilized ARHI-related diagnoses in electronic health records to perform a larger study with 6,527 case subjects and 45,882 control subjects and identified 2 genome-wide significant SNPs. Our study is an order of magnitude larger than previous reported work using 87,056 (HDiff) and 13,178 (HAid) case subjects with total sample sizes greater than 250,000 for each trait. The increased sample size is achieved by utilizing self-reported hearing data and self-report hearing aid use rather than using objective audiometric measures. The benefit of the increased power of this study is highlighted by the ability to detect a large number of associations by using self-report measures, as was previously demonstrated with another sensory trait.45
Hearing aid users in the UK will have received a diagnosis of hearing impairment following a pure-tone audiometry test, and so this phenotype provides a good surrogate of abnormal pure tone audiometry. HDiff is a more subjective measure than would be provided by pure-tone audiometry and may well be influenced by psycho-social elements as well as hearing ability. However, it is known that a pure tone-audiometry test does not diagnose all hearing loss including “hidden hearing loss” (a key symptom of which is difficulty hearing with background noise present), meaning that HDiff may be more representative of “real world” hearing impairment. Therefore, different forms of hearing loss may be defined by the two phenotypes, which may have subtle differences in pathology and genetic risk variants. In addition, within the UKBB sample hearing aid use is correlated with socio-economic status and education level,46 providing evidence that environmental factors are associated with and may influence the UKBB phenotypes. These factors were not included as covariates for association analysis as the interaction with hearing loss is not currently fully understood.
A long-standing question in ARHI has been whether the susceptibility genes for ARHI would be variants in known congenital deafness genes or completely novel genes. Approximately one quarter of the genetic loci identified in the UKBB GWAS are known hearing loss genes while the rest are novel. For four of the ten established hearing loci, BAIAP2L247 (MIM: 617536), TUB48 (MIM: 601197), SYNJ249 (MIM: 609410), and SPTBN150 (MIM: 182790), this study provides the first link to hearing pathology in humans, as they have previously only been linked to hearing function in animal models. In addition, a number of these ten loci have existing links to early-onset, congenital deafness. CDH23, which encodes cadherin-23, is a component of the stereocilia tip link, deflection of which results in opening of the mechano-transduction necessary for sound detection.51 It has long been proposed as a candidate human ARHI gene since an exon skipping mutation of cdh23 in the commonly used C57BL/6 strain of mice causes an accelerated age-related hearing loss.52 However, previous mutations in humans have been associated with a form of early-onset recessive hearing loss, Deafness, autosomal-recessive 12 (DFNB12 [MIM: 601386]), and Usher’s syndrome type ID (USHID [MIM: 601067]), which causes early-onset deafness and blindness.53 This study provides a link between this locus and common adult hearing impairment, suggestive of multiple variants being present at the same loci but displaying different types of hearing phenotypes.
Four significant gene loci are common to both analyses: EYA4, NID2, ARHGEF28, and CTBP2. Variants within EYA4 have been reported in autosomal-dominant non-syndromic hearing loss,54, 55, 56 deafness autosomal-dominant 10 (DFNA10 [MIM: 601316]), while NID2 and ARHGEF28 are new associations with hearing impairment. CTBP2, though not previously linked to genetic risk of ARHI, encodes C-terminal Binding Protein 2, a critical protein component of the inner ear hair cell pre-synaptic ribbon.57 The remaining loci were distinct between the two studies.
SNP-based heritability recalculated to the liability scale for HDiff (19%) and HAid (13%) are at the mid-lower end of previous heritability estimates for ARHL.58, 59, 60 This may be because this method accounts only for the additive genetic effects of the SNPs that were included in our analysis.61 This sample is, however, larger than previous samples used for a heritability estimate of common adult hearing loss and the phenotype measures do differ from those used for previous estimates.
Genetic correlation analysis revealed a link between genetic data for common adult hearing loss and depression-associated traits and pain-related traits. Studies have reported numerous epidemiological links between these depressive symptoms and hearing loss,1, 2, 62 but there is no evidence of a common genetic etiology. This correlation and significance does not account for genetic confounding, meaning that intermediate factors could contribute or be causal of the correlations reported. Due to the nature of these significantly correlated traits, the correlation may be confounded by general wellbeing. In addition, sampling bias may be present due to variability between studies such as trait and covariate categorization and inclusion.32
Gene set enrichment analysis using ToppGene contributed further evidence that this trait is highly polygenic and has a heterogeneous pathology. Genes and terms associated with HDiff were enriched for abnormal inner ear abnormality, synaptic function, cognition, and abnormal neuron morphology. These findings indicate that genetic factors have a role in the common dysfunction of a range of mechanisms within the auditory system. Connecting pathogenic genetic variants to distinct mechanisms within the auditory system will enable us to better understand the biological relevance of an individual’s genetic risk for hearing loss, and also how individual subsystems are dysregulated in the aging population. With the knowledge that certain gene groups have a role in particular cell or tissue types, studies can be targeted to a likely pathogenic location.
Analysis via ToppGene also revealed HDiff gene sets overlap with those involved in “cognition,” providing evidence of a possible genetic role to the link between hearing loss and cognition phenotypes. However, the finding from MAGMA analysis of a significant association between HDiff genes and transcription levels of genes in brain tissue does not necessarily confirm this finding or that these genes cause pathology in higher auditory systems with age. The results from these expression studies may simply reflect the fact that sensory cells of the inner ear are of neural origin, or that a substantial amount of neuronal tissue expression data are available in comparison to the limited datasets derived from cochlear tissue.
Suggestive level associations between HDiff and mouse ontologies is a promising finding for the field. Phenotype-driven screens in mice have been critically important in identifying deafness genes and are the model organism of choice for the study of hearing genetics, primarily due to the similarity of audiology systems and convergent evolution. However, there is less evidence that similar genetic predisposition to auditory aging is comparable between mice and humans. These results, together with the identification of CDH23 as a human ARHI loci, supports the use of mouse models in genetic studies of ARHI.
While Nidogen-2 is an established component of the basement membrane protein complex, anti-NID2 staining was not observed in the basilar membrane of the organ of Corti. It was, however, observed staining the blood vessel basement membranes, as has been noted in other tissues previously,63 providing evidence of antibody specificity in our sample. Anti-NID2 staining was most prominent in a restricted region of the epithelial lining between the inner spiral sulcus and spiral limbus, suggesting it may have a role in maintaining the structure of the epithelia here in close proximity to the attachment site of the tectorial membrane.
Similar to clarin-1 (L. Dunbar et al., 2019, ARO Midwinter Meeting, abstract), clarin-2 immunostaining localized to the inner and outer hair cells, the primary sensory cells of sound detection, suggesting that it may have a similar role in hearing (Figures 4 and S5). During preparation of the manuscript, recent evidence from concurrent studies suggests that mutations in clrn2 can cause progressive hearing loss in mouse and zebrafish and that mutations in CLRN2 underlie a recessive form of congenital hearing loss in an Iranian family (L. Dunbar et al., 2019, ARO Midwinter Meeting, abstract; Gopal et al., 2019, ARO Midwinter Meeting, abstract). In addition to observing anti-ARHGEF28 staining in the sensory cells, anti-ARHGEF28 stains the lining of the nerve fibers in the SGN region. Previous reports demonstrate a role for ARHGEF28 in regulation of neurofilaments66, 67 and axon growth and branching.67 It has also been implicated in the pathogenesis of motor neuron disease through formation of neurofilament and ARHGEF28 aggregates,68, 69 perhaps implying a role in neuronal function or maintenance.
Our study should be received in the context of its limitations; first, there is currently a lack of adequately powered studies with which to replicate our results. Despite meta-analyzing three cohorts, the replication sample remains an order of magnitude smaller than the discovery set. It is therefore unsurprising that only three genes, ZNF318, NID2, and ARHGEF28, were replicated. This replication rate likely reflects sample heterogeneity as well as “winner’s curse.”70 This heterogeneity includes varied questionnaire measures used to derive the phenotypes in the three replication samples and the different demographics of the individual cohorts. The lack of samples with consistent phenotype measures is a key limitation in genetic hearing research. One of the three cohorts included in our replication meta-analysis is a subset of the UKBB cohort and therefore is not a truly independent sample from the discovery cohort. However, the identification of known hearing genes, gene annotation analysis, and the results of in vivo expression provide support and putative mechanisms for involvement of these genes in hearing loss.
Second, we could not confirm the age of onset of hearing difficulty or hearing aid prescription, making an accurate diagnosis of ARHI a challenge. Some of the associations, for example, may be driven by individuals with congenital hearing impairment due to highly penetrant variants. We reduced the likelihood of this by implementing a minor allele frequency cut-off of 0.01 (i.e., higher than the known frequency of variants involved in congenital deafness) and by excluding participants who selected “I am completely deaf” in the UKBB questionnaire.
In summary, we have conducted a GWAS on adult hearing using more than 250,000 samples and have identified 44 associated independent loci. Several genes identified are known to have a role in congenital deafness, are already known to have an important role in hearing, or have been identified in mouse models. One locus has been associated with human ARHI traits previously and 34 of the 44 loci identified have not previously been associated with hearing loss phenotypes in humans or mice. Interestingly, these genes have a range of biological functions and are suggestive of multiple pathogenic pathways underlying ARHI and are not restricted to only one of sensory or neural or metabolic forms of hearing loss. For three genes we demonstrate different localized cell-specific expression within the mouse adult cochlea. Importantly, this study demonstrates that self-reported hearing loss in adults is suitable for use in association studies using large cohorts such as the UKBB. Our results present a framework for further study into the auditory pathways influenced by the genomic loci identified and provide therapeutic opportunities in ARHI and possibly dementia.
Declaration of Interests
D.R.M. is a Scientific Advisor for hearX Ltd., Otonomy Inc. The other authors declare no competing interests.
Acknowledgments
The research was carried out using the UK Biobank Resource under application number 11516. H.R.R.W. is funded by a PhD Studentship Grant, S44, from Action on Hearing Loss. The study was also supported by funding from NIHR UCLH BRC Deafness and Hearing Problems Theme, a grant from MED_EL, and the NIHR Manchester Biomedical Research Centre. The English Longitudinal Study of Aging is jointly run by University College London, Institute for Fiscal Studies, University of Manchester, and National Centre for Social Research. Genetic analyses have been carried out by UCL Genomics and funded by the Economic and Social Research Council and the National Institute on Aging. Data governance was provided by the METADAC data access committee, funded by ESRC, Wellcome, and MRC (2015-2018: Grant Number MR/N01104X/1 2018-2020: Grant Number ES/S008349/1). TwinsUK is funded by the Wellcome Trust, Medical Research Council, European Union, the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility, and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London. We would like to thank all the participants of UK Biobank, English Longitudinal Study of Aging, and TwinsUK.
Published: September 26, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.09.008.
Contributor Information
Sally J. Dawson, Email: sally.dawson@ucl.ac.uk.
Frances M.K. Williams, Email: frances.williams@kcl.ac.uk.
Data Availability
Data that support the findings of this study are publicly available upon successful application from the UK Biobank, the English Longitudinal Study of Aging, and TwinsUK. All ELSA GWAS data have been deposited in the European Genome-phenome Archive (Database: EGAS00001001036).
Derived data from the UK Biobank data fields and the discovery GWAS summary statistics that support the findings of this study will be made available as part of the UK Biobank Returns Catalogue following the publication of this manuscript; see Web Resources.
Web Resources
ELSA GWAS QC & Imputation Analysis, https://www.ucl.ac.uk/drupal/site_iehc/sites/iehc/files/elsa_gwas_qc_imputation_analysis.pdf
Hereditary Hearing Loss Homepage, https://hereditaryhearingloss.org/
Region-based heritability estimates genome partition map, https://bitbucket.org/nygcresearch/ldetect-data/downloads/
Region-based heritability reference panel, EUR 1000G, https://www.internationalgenome.org/data/
UK Biobank datashowcase, http://biobank.ctsu.ox.ac.uk/crystal/
Supplemental Data
References
- 1.Brewster K.K., Ciarleglio A., Brown P.J., Chen C., Kim H.O., Roose S.P., Golub J.S., Rutherford B.R. Age-Related Hearing Loss and Its Association with Depression in Later Life. Am. J. Geriatr. Psychiatry. 2018;26:788–796. doi: 10.1016/j.jagp.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rutherford B.R., Brewster K., Golub J.S., Kim A.H., Roose S.P. Sensation and psychiatry: Linking age-related hearing loss to late-life depression and cognitive decline. Am. J. Psychiatry. 2018;175:215–224. doi: 10.1176/appi.ajp.2017.17040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han J.H., Lee H.J., Jung J., Park E.C. Effects of self-reported hearing or vision impairment on depressive symptoms: a population-based longitudinal study. Epidemiol. Psychiatr. Sci. 2019;28:343–355. doi: 10.1017/S2045796018000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jayakody D.M.P., Friedland P.L., Martins R.N., Sohrabi H.R. Impact of aging on the auditory system and related cognitive functions: A narrative review. Front. Neurosci. 2018;12:125. doi: 10.3389/fnins.2018.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei J., Hu Y., Zhang L., Hao Q., Yang R., Lu H., Zhang X., Chandrasekar E.K. Hearing impairment, mild cognitive impairment, and dementia: A meta-analysis of cohort studies. Dement. Geriatr. Cogn. Disord. Extra. 2017;7:440–452. doi: 10.1159/000485178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gurgel R.K., Ward P.D., Schwartz S., Norton M.C., Foster N.L., Tschanz J.T. Relationship of hearing loss and dementia: a prospective, population-based study. Otol. Neurotol. 2014;35:775–781. doi: 10.1097/MAO.0000000000000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin F.R., Metter E.J., O’Brien R.J., Resnick S.M., Zonderman A.B., Ferrucci L. Hearing loss and incident dementia. Arch. Neurol. 2011;68:214–220. doi: 10.1001/archneurol.2010.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vos T., Barber R.M., Bell B., Bertozzi-Villa A., Biryukov S., Bolliger I., Charlson F., Davis A., Degenhardt L., Dicker D. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386:743–800. doi: 10.1016/S0140-6736(15)60692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huddle M.G., Goman A.M., Kernizan F.C., Foley D.M., Price C., Frick K.D., Lin F.R. The economic impact of adult hearing loss: A systematic review. JAMA Otolaryngol. Head Neck Surg. 2017;143:1040–1048. doi: 10.1001/jamaoto.2017.1243. [DOI] [PubMed] [Google Scholar]
- 10.Fransen E., Bonneux S., Corneveaux J.J., Schrauwen I., Di Berardino F., White C.H., Ohmen J.D., Van de Heyning P., Ambrosetti U., Huentelman M.J. Genome-wide association analysis demonstrates the highly polygenic character of age-related hearing impairment. Eur. J. Hum. Genet. 2015;23:110–115. doi: 10.1038/ejhg.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman D.L., Fisher L.M., Ohmen J., Parody R., Fong C.T., Frisina S.T., Mapes F., Eddins D.A., Robert Frisina D., Frisina R.D., Friedman R.A. GRM7 variants associated with age-related hearing loss based on auditory perception. Hear. Res. 2012;294:125–132. doi: 10.1016/j.heares.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Laer L., Huyghe J.R., Hannula S., Van Eyken E., Stephan D.A., Mäki-Torkko E., Aikio P., Fransen E., Lysholm-Bernacchi A., Sorri M. A genome-wide association study for age-related hearing impairment in the Saami. Eur. J. Hum. Genet. 2010;18:685–693. doi: 10.1038/ejhg.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo H., Yang T., Jin X., Pang X., Li J., Chai Y., Li L., Zhang Y., Zhang L., Zhang Z. Association of GRM7 variants with different phenotype patterns of age-related hearing impairment in an elderly male Han Chinese population. PLoS ONE. 2013;8:e77153. doi: 10.1371/journal.pone.0077153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman R.A., Van Laer L., Huentelman M.J., Sheth S.S., Van Eyken E., Corneveaux J.J., Tembe W.D., Halperin R.F., Thorburn A.Q., Thys S. GRM7 variants confer susceptibility to age-related hearing impairment. Hum. Mol. Genet. 2009;18:785–796. doi: 10.1093/hmg/ddn402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolber L.E., Girotto G., Buniello A., Vuckovic D., Pirastu N., Lorente-Cánovas B., Rudan I., Hayward C., Polasek O., Ciullo M. Salt-inducible kinase 3, SIK3, is a new gene associated with hearing. Hum. Mol. Genet. 2014;23:6407–6418. doi: 10.1093/hmg/ddu346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vuckovic D., Dawson S., Scheffer D.I., Rantanen T., Morgan A., Di Stazio M., Vozzi D., Nutile T., Concas M.P., Biino G. Genome-wide association analysis on normal hearing function identifies PCDH20 and SLC28A3 as candidates for hearing function and loss. Hum. Mol. Genet. 2015;24:5655–5664. doi: 10.1093/hmg/ddv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girotto G., Pirastu N., Sorice R., Biino G., Campbell H., d’Adamo A.P., Hastie N.D., Nutile T., Polasek O., Portas L. Hearing function and thresholds: a genome-wide association study in European isolated populations identifies new loci and pathways. J. Med. Genet. 2011;48:369–374. doi: 10.1136/jmg.2010.088310. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann T.J., Keats B.J., Yoshikawa N., Schaefer C., Risch N., Lustig L.R. A Large Genome-Wide Association Study of Age-Related Hearing Impairment Using Electronic Health Records. PLoS Genet. 2016;12:e1006371. doi: 10.1371/journal.pgen.1006371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nolan L.S., Maier H., Hermans-Borgmeyer I., Girotto G., Ecob R., Pirastu N., Cadge B.A., Hübner C., Gasparini P., Strachan D.P. Estrogen-related receptor gamma and hearing function: evidence of a role in humans and mice. Neurobiol. Aging. 2013;34:2077.e1–2077.e9. doi: 10.1016/j.neurobiolaging.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. Genome-wide genetic data on ∼500,000 UK Biobank participants Supplementary Material. bioRxiv. 2017 [Google Scholar]
- 22.Steptoe A., Breeze E., Banks J., Nazroo J. Cohort profile: the English longitudinal study of ageing. Int. J. Epidemiol. 2013;42:1640–1648. doi: 10.1093/ije/dys168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moayyeri A., Hammond C.J., Hart D.J., Spector T.D. The UK adult twin registry (twinsUK resource) Twin Res. Hum. Genet. 2013;16:144–149. doi: 10.1017/thg.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawes P., Fortnum H., Moore D.R., Emsley R., Norman P., Cruickshanks K., Davis A., Edmondson-Jones M., McCormack A., Lutman M., Munro K. Hearing in middle age: a population snapshot of 40- to 69-year olds in the United Kingdom. Ear Hear. 2014;35:e44–e51. doi: 10.1097/AUD.0000000000000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore D.R., Edmondson-Jones M., Dawes P., Fortnum H., McCormack A., Pierzycki R.H., Munro K.J. Relation between speech-in-noise threshold, hearing loss and cognition from 40-69 years of age. PLoS ONE. 2014;9:e107720. doi: 10.1371/journal.pone.0107720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loh P.R., Tucker G., Bulik-Sullivan B.K., Vilhjálmsson B.J., Finucane H.K., Salem R.M., Chasman D.I., Ridker P.M., Neale B.M., Berger B. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J., Ferreira T., Morris A.P., Medland S.E., Madden P.A.F., Heath A.C., Martin N.G., Montgomery G.W., Weedon M.N., Loos R.J., Genetic Investigation of ANthropometric Traits (GIANT) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44 doi: 10.1038/ng.2213. 369–375, S1–S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng J., Erzurumluoglu A.M., Elsworth B.L., Kemp J.P., Howe L., Haycock P.C., Hemani G., Tansey K., Laurin C., Pourcain B.S., Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., Duncan L., Perry J.R.B., Patterson N., Robinson E.B., ReproGen Consortium. Psychiatric Genomics Consortium. Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi H., Kichaev G., Pasaniuc B. Contrasting the Genetic Architecture of 30 Complex Traits from Summary Association Data. Am. J. Hum. Genet. 2016;99:139–153. doi: 10.1016/j.ajhg.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang C.C., Chow C.C., Tellier L.C.A.M., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkp427. W305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shahin H., Walsh T., Sobe T., Abu Sa’ed J., Abu Rayan A., Lynch E.D., Lee M.K., Avraham K.B., King M.-C., Kanaan M. Mutations in a novel isoform of TRIOBP that encodes a filamentous-actin binding protein are responsible for DFNB28 recessive nonsyndromic hearing loss. Am. J. Hum. Genet. 2006;78:144–152. doi: 10.1086/499495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riazuddin S., Khan S.N., Ahmed Z.M., Ghosh M., Caution K., Nazli S., Kabra M., Zafar A.U., Chen K., Naz S. Mutations in TRIOBP, which encodes a putative cytoskeletal-organizing protein, are associated with nonsyndromic recessive deafness. Am. J. Hum. Genet. 2006;78:137–143. doi: 10.1086/499164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fields R.R., Zhou G., Huang D., Davis J.R., Möller C., Jacobson S.G., Kimberling W.J., Sumegi J. Usher syndrome type III: revised genomic structure of the USH3 gene and identification of novel mutations. Am. J. Hum. Genet. 2002;71:607–617. doi: 10.1086/342098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adato A., Vreugde S., Joensuu T., Avidan N., Hamalainen R., Belenkiy O., Olender T., Bonne-Tamir B., Ben-Asher E., Espinos C. USH3A transcripts encode clarin-1, a four-transmembrane-domain protein with a possible role in sensory synapses. Eur. J. Hum. Genet. 2002;10:339–350. doi: 10.1038/sj.ejhg.5200831. [DOI] [PubMed] [Google Scholar]
- 45.Kiefer A.K., Tung J.Y., Do C.B., Hinds D.A., Mountain J.L., Francke U., Eriksson N. Genome-wide analysis points to roles for extracellular matrix remodeling, the visual cycle, and neuronal development in myopia. PLoS Genet. 2013;9:e1003299. doi: 10.1371/journal.pgen.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawyer C.S., Armitage C.J., Munro K.J., Singh G., Dawes P.D. Correlates of Hearing Aid Use in UK Adults: Self-Reported Hearing Difficulties, Social Participation, Living Situation, Health, and Demographics. Ear Hear. 2019;40:1061–1068. doi: 10.1097/AUD.0000000000000695. [DOI] [PubMed] [Google Scholar]
- 47.Bowl M.R., Simon M.M., Ingham N.J., Greenaway S., Santos L., Cater H., Taylor S., Mason J., Kurbatova N., Pearson S., International Mouse Phenotyping Consortium A large scale hearing loss screen reveals an extensive unexplored genetic landscape for auditory dysfunction. Nat. Commun. 2017;8:886. doi: 10.1038/s41467-017-00595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohlemiller K.K., Hughes R.M., Mosinger-Ogilvie J., Speck J.D., Grosof D.H., Silverman M.S. Cochlear and retinal degeneration in the tubby mouse. Neuroreport. 1995;6:845–849. doi: 10.1097/00001756-199504190-00005. [DOI] [PubMed] [Google Scholar]
- 49.Manji S.S.M., Williams L.H., Miller K.A., Ooms L.M., Bahlo M., Mitchell C.A., Dahl H.H.M. A mutation in synaptojanin 2 causes progressive hearing loss in the ENU-mutagenised mouse strain Mozart. PLoS ONE. 2011;6:e17607. doi: 10.1371/journal.pone.0017607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y., Qi J., Chen X., Tang M., Chu C., Zhu W., Li H., Tian C., Yang G., Zhong C. Critical role of spectrin in hearing development and deafness. Sci. Adv. 2019;5:v7803. doi: 10.1126/sciadv.aav7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richardson G.P., Petit C. Hair-Bundle Links: Genetics as the Gateway to Function. Cold Spring Harb. Perspect. Med. 2019 doi: 10.1101/cshperspect.a033142. Published online January 7, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noben-Trauth K., Zheng Q.Y., Johnson K.R. Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat. Genet. 2003;35:21–23. doi: 10.1038/ng1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schultz J.M., Bhatti R., Madeo A.C., Turriff A., Muskett J.A., Zalewski C.K., King K.A., Ahmed Z.M., Riazuddin S., Ahmad N. Allelic hierarchy of CDH23 mutations causing non-syndromic deafness DFNB12 or Usher syndrome USH1D in compound heterozygotes. J. Med. Genet. 2011;48:767–775. doi: 10.1136/jmedgenet-2011-100262. [DOI] [PubMed] [Google Scholar]
- 54.Schönberger J., Wang L., Shin J.T., Kim S.D., Depreux F.F.S., Zhu H., Zon L., Pizard A., Kim J.B., Macrae C.A. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat. Genet. 2005;37:418–422. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- 55.Makishima T., Madeo A.C., Brewer C.C., Zalewski C.K., Butman J.A., Sachdev V., Arai A.E., Holbrook B.M., Rosing D.R., Griffith A.J. Nonsyndromic hearing loss DFNA10 and a novel mutation of EYA4: evidence for correlation of normal cardiac phenotype with truncating mutations of the Eya domain. Am. J. Med. Genet. A. 2007;143A:1592–1598. doi: 10.1002/ajmg.a.31793. [DOI] [PubMed] [Google Scholar]
- 56.Pfister M., Tóth T., Thiele H., Haack B., Blin N., Zenner H.-P., Sziklai I., Nürnberg P., Kupka S. A 4-bp insertion in the eya-homologous region (eyaHR) of EYA4 causes hearing impairment in a Hungarian family linked to DFNA10. Mol. Med. 2002;8:607–611. [PMC free article] [PubMed] [Google Scholar]
- 57.Sheets L., Trapani J.G., Mo W., Obholzer N., Nicolson T. Ribeye is required for presynaptic Ca(V)1.3a channel localization and afferent innervation of sensory hair cells. Development. 2011;138:1309–1319. doi: 10.1242/dev.059451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bedin E., Franzè A., Zadro C., Persico M.G., Ciullo M., Hladnik U., Dolcetta D., Grasso D.L., Riccardi P., Nutile T. Age-related hearing loss in four Italian genetic isolates: an epidemiological study. Int. J. Audiol. 2009;48:465–472. doi: 10.1080/14992020902822039. [DOI] [PubMed] [Google Scholar]
- 59.Bogo R., Farah A., Johnson A.C., Karlsson K.K., Pedersen N.L., Svartengren M., Skjönsberg Å. The role of genetic factors for hearing deterioration across 20 years: a twin study. J. Gerontol. A Biol. Sci. Med. Sci. 2015;70:647–653. doi: 10.1093/gerona/glu245. [DOI] [PubMed] [Google Scholar]
- 60.Gates G.A., Couropmitree N.N., Myers R.H. Genetic associations in age-related hearing thresholds. Arch. Otolaryngol. Head Neck Surg. 1999;125:654–659. doi: 10.1001/archotol.125.6.654. [DOI] [PubMed] [Google Scholar]
- 61.Zaitlen N., Kraft P. Heritability in the genome-wide association era. Hum. Genet. 2012;131:1655–1664. doi: 10.1007/s00439-012-1199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keidser G., Seeto M., Rudner M., Hygge S., Rönnberg J. On the relationship between functional hearing and depression. Int. J. Audiol. 2015;54:653–664. doi: 10.3109/14992027.2015.1046503. [DOI] [PubMed] [Google Scholar]
- 63.Kohfeldt E., Sasaki T., Göhring W., Timpl R. Nidogen-2: a new basement membrane protein with diverse binding properties. J. Mol. Biol. 1998;282:99–109. doi: 10.1006/jmbi.1998.2004. [DOI] [PubMed] [Google Scholar]
- 66.Cañete-Soler R., Wu J., Zhai J., Shamim M., Schlaepfer W.W. p190RhoGEF Binds to a destabilizing element in the 3′ untranslated region of light neurofilament subunit mRNA and alters the stability of the transcript. J. Biol. Chem. 2001;276:32046–32050. doi: 10.1074/jbc.M104104200. [DOI] [PubMed] [Google Scholar]
- 67.Volkening K., Leystra-Lantz C., Strong M.J. Human low molecular weight neurofilament (NFL) mRNA interacts with a predicted p190RhoGEF homologue (RGNEF) in humans. Amyotroph. Lateral Scler. 2010;11:97–103. doi: 10.3109/17482960902995584. [DOI] [PubMed] [Google Scholar]
- 68.Rico B., Beggs H.E., Schahin-Reed D., Kimes N., Schmidt A., Reichardt L.F. Control of axonal branching and synapse formation by focal adhesion kinase. Nat. Neurosci. 2004;7:1059–1069. doi: 10.1038/nn1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Droppelmann C.A., Wang J., Campos-Melo D., Keller B., Volkening K., Hegele R.A., Strong M.J. Detection of a novel frameshift mutation and regions with homozygosis within ARHGEF28 gene in familial amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Frontotemporal Degener. 2013;14:444–451. doi: 10.3109/21678421.2012.758288. [DOI] [PubMed] [Google Scholar]
- 70.Kraft P., Zeggini E., Ioannidis J.P.A. Replication in genome-wide association studies. Stat. Sci. 2009;24:561–573. doi: 10.1214/09-STS290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study are publicly available upon successful application from the UK Biobank, the English Longitudinal Study of Aging, and TwinsUK. All ELSA GWAS data have been deposited in the European Genome-phenome Archive (Database: EGAS00001001036).
Derived data from the UK Biobank data fields and the discovery GWAS summary statistics that support the findings of this study will be made available as part of the UK Biobank Returns Catalogue following the publication of this manuscript; see Web Resources.