Abstract
Intellectual disability (ID) is a genetically and clinically heterogeneous disorder, characterized by limited cognitive abilities and impaired adaptive behaviors. In recent years, exome sequencing (ES) has been instrumental in deciphering the genetic etiology of ID. Here, through ES of a large cohort of individuals with ID, we identified two bi-allelic frameshift variants in METTL5, c.344_345delGA (p.Arg115Asnfs∗19) and c.571_572delAA (p.Lys191Valfs∗10), in families of Pakistani and Yemenite origin. Both of these variants were segregating with moderate to severe ID, microcephaly, and various facial dysmorphisms, in an autosomal-recessive fashion. METTL5 is a member of the methyltransferase-like protein family, which encompasses proteins with a seven-beta-strand methyltransferase domain. We found METTL5 expression in various substructures of rodent and human brains and METTL5 protein to be enriched in the nucleus and synapses of the hippocampal neurons. Functional studies of these truncating variants in transiently transfected orthologous cells and cultured hippocampal rat neurons revealed no effect on the localization of METTL5 but alter its level of expression. Our in silico analysis and 3D modeling simulation predict disruption of METTL5 function by both variants. Finally, mettl5 knockdown in zebrafish resulted in microcephaly, recapitulating the human phenotype. This study provides evidence that biallelic variants in METTL5 cause ID and microcephaly in humans and highlights the essential role of METTL5 in brain development and neuronal function.
Keywords: intellectual disability, microcephaly, METTL5, zebrafish, facial dysmorphism, autosomal recessive intellectual disability, m6A methyltransferase
Main Text
Intellectual disability (ID) is characterized by significant impairment in cognitive ability and adaptive behaviors, with a disease onset generally before adulthood.1 With a prevalence of 1%–2% worldwide,2, 3 ID is a complex group of disorders that has high phenotypic variability as well as heterogeneous etiology, encompassing genetic and environmental factors.4 Autosomal-recessive ID (ARID) is estimated to account for more than 50% of genetic causes of ID.4 The identification of novel ARID genes has gained momentum in recent years through implementation of contemporary high-throughput sequencing technologies (e.g., exome sequencing) and study of large consanguineous families.5, 6, 7, 8, 9, 10, 11, 12 However, these molecular studies highlight extreme genetic heterogeneity with an estimate of more than 2,500 genes associated with various forms of ID.13
Here, we illustrate the integration of two large-scale studies, GENCODYS6 and CARID,9 and subsequent functional analyses to identify variants that affect function in METTL5, segregating with ID, microcephaly, and facial dysmorphisms in two large families of Pakistani and Yemenite origin. Written informed consents were obtained for all individuals involved. This study adhered to the World Health Association Declaration of Helsinki (2013) and was approved by the Institutional Review Board at University of Maryland School of Medicine, Baltimore, USA, Center of Excellence in Molecular Biology (CEMB), University of the Punjab, Lahore, Pakistan, the Institutional Review Board Commissie Mensgebonden Onderzoek Regio Arnhem-Nijmegen, the Netherlands, and an ethical votum for MRNET in Erlangen, Germany. Medical and family history, developmental childhood milestones, anthropometric measurements, and findings of physical examination were collected and detailed clinical phenotypes were described with Human Phenotype Ontology (HPO) terms.14 Venous blood samples were collected from research participants for DNA extraction.
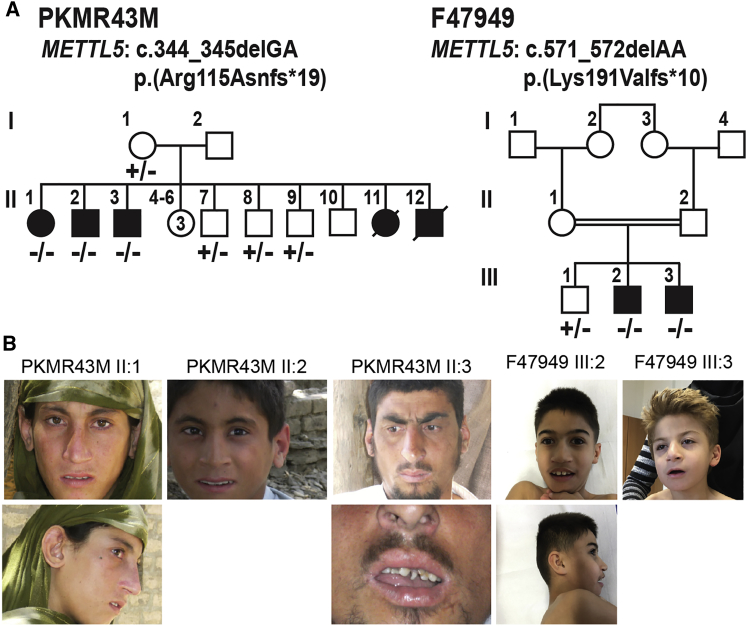
Family 1 (PKMR43M; Figure 1A; Table 1) was enrolled from Dargai, Malakand Agency in Khyber Pakhtoon Khawa province of Pakistan. There are five individuals presenting with severe ID (HP:0010864), of which two were deceased at the time of assessment. The age of affected individuals at the time of evaluation ranged from 14 to 31 years, but the ID was already apparent in early childhood. All three affected individuals had significant delay in childhood milestones including verbal, fine motor, and social skills. There was no history of epilepsy. The apparent behavioral and physical abnormalities noticed in the affected individuals included attention deficit hyperactivity disorder (ADHD) (HP:0007018), aggressive behavior (HP:0000718), microcephaly (HP:0000252), muscular hypotonia (HP:0001252), overhanging nasal tip (HP:0011833), wide nasal base (HP:0012810), and abnormality of dental morphology (HP:0006482) (Figure 1B; Table 1). The affected female had short stature (HP:0004322) and decreased body weight (HP:0004322), i.e., height and weight below 3rd percentile for her age and gender. Evaluation of motor system was unremarkable and gait was normal in all affected individuals with no apparent hearing or vision loss (Table 1). Magnetic resonance imaging (MRI) of the individual II:2 confirmed the measured microcephaly and did not reveal any degeneration of brain tissue (Figure S1).
Figure 1.
Homozygous Frameshift Variants in METTL5 Lead to Intellectual Disabilities Associated with Dysmorphic Features
(A) Pedigrees of Pakistani family PKMR43M and Yemenite family F47949 segregating intellectual disabilities and microcephaly due to different frameshift deletions in METTL5, c.344_345delGA and c.571_572delAA, respectively. The filled symbols represent affected individuals and a double horizontal line connecting parents represents a consanguineous marriage.
(B) Facial appearance of affected individuals. All affected individuals have microcephaly. Various facial dysmorphisms were reported such as large ear shape (PKMR43M II:1, F47949 III:2), dental anomalies (PKMR43M II:3), and large nose (F47949 III:2).
Table 1.
Clinical Findings of Affected Individuals from Families PKMR43M and F47949
|
Family 1, PKMR43M |
Family 2, F47949 |
Family from Hu et al.11 |
|||||
|---|---|---|---|---|---|---|---|
| c.344_345delGA (p.Arg115Asnfs∗19) | c.571_572delAA (p.Lys191Valfs∗10) | c.182G>A (p.Gly61Asp) | |||||
| Individual | II:1 | II:2 | II:3 | III:3 | III:2 | III:3 | III:5 |
| Gender | female | male | male | male | male | male | female |
| Age at evaluation | 16 years | 14 years | 31 years | 7 years | 12 years | 10 years | 5 years |
| Congenital anomalies | no | no | no | fetal tachycardia, hydrops fetalis | ASDII, pulmonic stenosis | not reported | not reported |
| General Features | |||||||
| Mental retardation | severe | severe | severe | severe | moderate to severe | severe | severe |
| Speech delay | yes | yes | yes | yes, no speech | yes, 4–5 word sentences | slurred speech | not reported |
| Weight | 32 kg (−4.9 SD) | 50 kg (−0.1 SD) | 89 kg (1.9 SD) | 16 kg (−3.5 SD) | 26 kg (−2.8 SD) | N/A | N/A |
| Height | 152 cm (−1.6 SD) | 157 cm (−0.8 SD) | 157 cm (−2.7 SD) | 114 cm (−2 SD) | 135 cm (−2.3 SD) | 133 cm (25%ile) | 95 cm (3%ile) |
| Head circumference | 51 cm (−3.1 SD) | 48 cm (−4.2 SD) | 51 cm (−2.8 SD) | 46 cm (−5 SD) | 48 cm (−4.7 SD) | 46 cm (−7 SD) | 44 cm(−8 SD) |
| Head Size | microcephaly | microcephaly | microcephaly | microcephaly | microcephaly | microcephaly | microcephaly |
| Hypotonia | no | mild | no | no | no | N/A | N/A |
| Epilepsy | no | no | no | no | no | no | seizure |
| Spasticity | no | no | no | yes | yes | N/A | N/A |
| Behavioral problem | ADHD | ADHD, aggressive | ADHD, aggressive | hand biting, autistic | hand biting, autistic | short temper, aggressive | short temper, aggressive |
| Eyes | |||||||
| Abnormalities | no | no | no | no | no | strabismus | strabismus |
| Nose | |||||||
| Nose shape anomalies | no | broad nasal base, over hanging nasal tip | over hanging nasal tip | large nose with broad tip | large nose with broad tip | narrow nasal base, broad nasal ridge | narrow nasal base, broad nasal ridge |
| Mouth | |||||||
| Dental anomalies | no | no | dysplastic teeth | no | no | N/A | N/A |
| Shape anomalies | no | no | no | no | no | long philtrum and thin upper lip | long philtrum and thin upper lip |
| Ear | |||||||
| Ear shape anomalies | large | no | large | low-set and posteriorly rotated | low-set and posteriorly rotated | large | no |
| Hearing loss | no | no | no | no | no | not reported | not reported |
| Vestibular deficit | no | no | no | no | no | unbalanced gait | no |
ASDII, atrial septal defect type II; ADHD, attention-deficit/hyperactivity disorder
Family 2 (F47949; Figure 1A; Table 1) had Yemenite origin and was enrolled and evaluated in Germany at the Institutes of Human Genetics in Essen and Düsseldorf. Two affected male individuals were 7 and 12 years old at the time of enrollment and clinical evaluation. Birth history was significant for fetal tachycardia and non-immune hydrops fetalis (HP:0001790) in affected individual III:3, while secundum atrial septal defect (HP:0001684) and pulmonic stenosis (HP:0004415) was observed in affected individual III-2. One affected individual presented severe ID (HP:0010864) and the other one moderate to severe ID (HP:0002342). Both affected individuals had delayed childhood milestones, self-mutilating behavior (HP:0000742), and autism spectrum disorder (HP:0000729). There was no history of epilepsy or motor weakness. Physical examination was significant for microcephaly (HP:0000252), short stature (HP:0004322), low body weight (HP:0004325), wide nasal base (HP:0012810), large nose with broad nasal tip (HP:0000455), low-set and posteriorly rotated ears, and truncal ataxia (HP:0002078) (Figure 1B; Table 1). Overall, the two families presented with a phenotype of ID, speech delay, and microcephaly segregating in an autosomal-recessive inheritance pattern (Figure 1A; Table 1; see Supplemental Note).
Exome sequencing was performed on DNA from one affected individual from each family at Radboudumc, Nijmegen, the Netherlands6 and at the Institute of Human Genetics, Erlangen, Germany.9 Among ≥15 variants that passed our initial filtration criteria (see Riazuddin et al.6 and Reuter et al.9 for complete description), Sanger sequencing revealed segregation of two novel bi-allelic 2-bp deletions, c.344_345delGA and c.571_572delAA, in METTL5 (GenBank: NM_014168.2) with the ID phenotype in families PKMR43M and F47949, respectively (Figure 1A). Both variants are predicted to disrupt the reading frame and cause premature truncation (p.Arg115Asnfs∗19; p.Lys191Valfs∗10) of the encoded METTL5 protein.
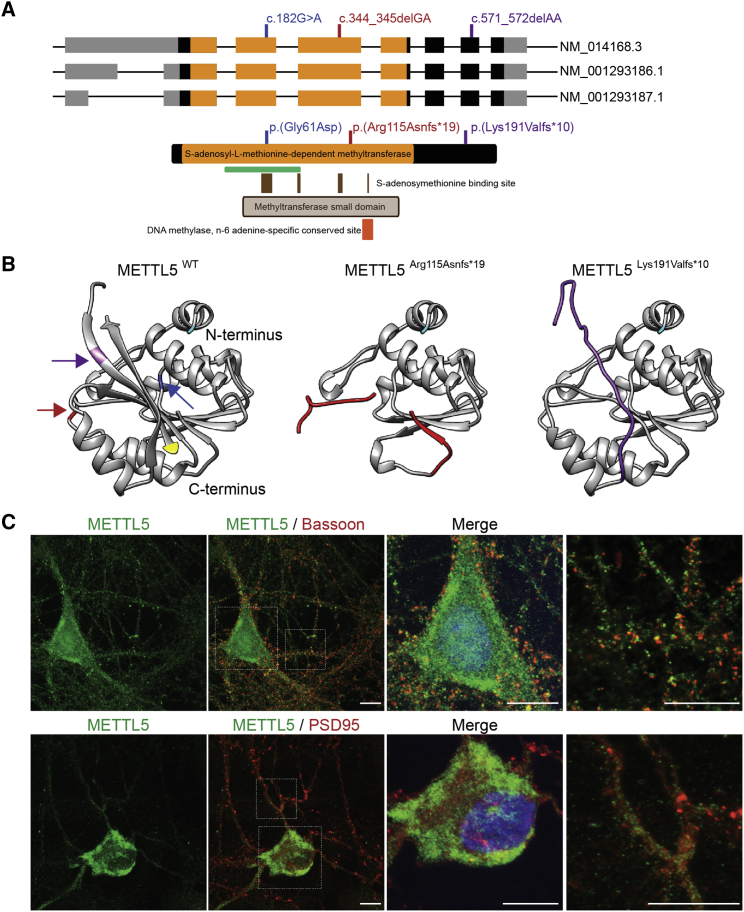
METTL5 is a functionally uncharacterized member of the methyltransferase superfamily, which encompasses 33 METTL proteins with a seven-beta-strand methyltransferase domain. By homology with other family members, it is predicted that METTL5 contains an S-adenosyl-L-methionine-dependent DNA methyltransferase domain and a DNA methylase N6 adenine-specific conserved site. In the event that the mRNA harboring c.344_345delGA escapes the predicted non-mediated decay15 in vivo, the resulting protein will lack the fully functional S-adenosyl-L-methionine-dependent methyltransferase domain and DNA methylase N6 adenine-specific site, whereas the different predicted domains will remain conserved in METTL5L191Vfs∗10 (Figure 2A).
Figure 2.
Frameshift Variants in METTL5 Impact the Predicted Domains and Conformation of the Protein, Ubiquitously Expressed in the Brain
(A) Alternative splicing leads to three isoforms of human METTL5. Non-coding segments and coding regions of exons are denoted by gray and black boxes, respectively. Regions coding for the S-adenosyl-L-methionine-dependent-methyltransferase domain are colored in orange. Variants analyzed in this study are depicted in red (PKMR43M), purple (F47949), and blue.11 The numbering of the position of variants c.182G>A (p.Gly61Asp), c.344_345delGA (p.Arg115Asnfs∗19), and c.571_572delAA (p.Lys191Valfs∗10) is based on accession number GenBank: NM_014167.2. The three isoforms lead to a unique protein, predicted to contain an S-adenosyl-L-methionine-dependent-methyltransferase (orange, amino acids 12–161), a methyltransferase small domain (brown box, amino acids 46–146), S-adenosylmethionine binding sites (dark brown box, amino acids 58L, 59G, 60C, 61G, 62C, 63G, 64V, 81D, 82I, 107C, 108D, 109V, 126V), and a DNA-methylase n-6-adenine-specific conserved site (red box, amino acids 123–129). The localization of the peptide used to produce the METTL5 antibody described in the following analysis is shown as a green bar (Novus Biologicals Cat# NBP1-56640, RRID:AB_11039697, amino acids 35–83).
(B) Protein modeling of the WT and the two novel mutant proteins, using PHYRE2 software,16 shows that the overall conformation of the protein is not affected by the different disease-causing variants. However, due to early termination codon, 2 α helix and 2 β sheet are missing in METTL5R115Nfs∗19 and 2 β sheet are missing in METTL5L191Vfs∗10. The red and purple arrows represent the site of truncation in variant p.Arg115Asnfs∗19 and p.Lys191Valfs∗10, respectively, and the blue arrow highlights the amino acid at position 61. Cyan, N terminus of the protein; yellow, C terminus of the protein.
(C) Representative immunofluorescent labeling images of endogenous METTL5 (green) highlight the partial co-localization of the WT protein with pre-synaptic (Bassoon, Enzo Life Sciences Cat# SAP7F407, RRID:AB_2313990, red) and post-synaptic (PSD95, UC Davis/NIH NeuroMab Facility Cat# 75-028, RRID:AB_2292909, red) markers in rat hippocampal neurons in culture. All images are projection of confocal optical sections stack. Scale bars: 10 μm.
We analyzed the expression profile of METTL5 in the developing and adult human brain (8 pcw to 40 years old), from the RNA-seq data available in the Allen Brain Atlas (see Web Resources). METTL5 is expressed from very early development (8 pcw) and expression persists through adulthood in multiple sub-structures of the human brain, including the cerebellar cortex, hippocampus, and striatum (Table S1). To characterize the cellular localization of METTL5, we used commercially available polyclonal antibody (NBP1-56640, RRID: AB_11039697, Novus Biologicals) (Figure S2) and performed immunostaining and confocal imaging on postnatal (P) day 30 mouse brain. Similar to humans, we found low but ubiquitous expression in the mouse brain (Figure S3). Immunolabeling of cultured rat hippocampal neurons showed an enrichment of METTL5 in the soma and the nucleus as well as in the pre- and post-synaptic regions (Figure 2D). Taken together with the domain structures, these data suggest that METTL5 might have a global epigenetic regulatory role in the brain as well as a synapse-dependent role. Several publications highlighted the importance of synapse-autonomous regulatory mechanisms,17, 18 including regulation involving METTL proteins.19
To evaluate the functional impact of the ID-associated variants on METTL5 protein structure and function, we first performed molecular modeling using PHYRE216 and Chimera20 programs (Figure 2C). As anticipated, both frameshift variants (c.344_345delGA and c.571_572delAA) are predicted to have significant impact on the protein secondary structure and remove the evolutionary conserved two α helix and/or β sheets from the C-terminal region.
To functionally validate the in silico predictions, we next investigated the impact of the variants on the stability and/or targeting of METTL5 in heterologous cells. We included in our study another candidate missense variant (c.182G>A [p.Gly61Asp]) (Figures 2A and 2B) that has been recently reported in an Iranian family with two affected individuals. While clinical features were similar to the phenotypes (Table 1) found in our two families, including severe ID, microcephaly, short temper, aggressive behavior, and various facial dysmorphisms,11 no functional studies were performed to determine the pathogenicity of the p.Gly61Asp variant on the encoded METTL5.
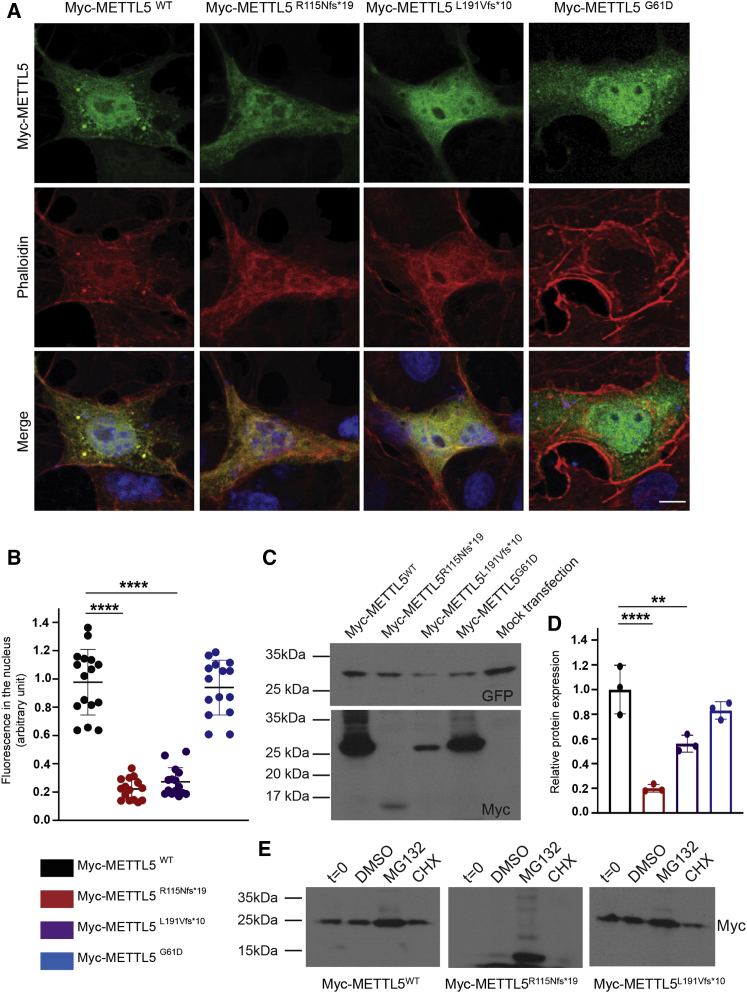
For these studies, full-length METTL5 was amplified from a human liver tissue cDNA library (TakaraBio) and was inserted in pCMV-Myc and peGFP-C2 vectors (TakaraBio). The constructs harboring ID-associated variants (METTL5G61D; METTL5R115Nfs∗19; METTL5L191Vfs∗10) were prepared through site-directed mutagenesis (Agilent Technologies) using the wild-type construct (METTL5WT) as a template. All constructs sequences were validated by Sanger sequencing (primers sequences are available upon request). When transiently expressed in COS7 cells, both Myc- and GFP-tagged METTL5WT as well as mutant proteins were found in the cytoplasm and in slightly higher amounts in the nucleus (Figures 3A and S4A). Similarly, when transfected in cultured rat hippocampal neurons, wild-type and mutant METTL5-GFP tagged proteins were observed in the nucleus, as well as in the neuronal dendrites (Figure S5).
Figure 3.
Disease-Causing Variants Affect the Stability of METTL5
(A) Immunolabeling of Myc-METTL5 fusion proteins for WT and the three variants associated with ID, after transfection in COS7 cells. The subcellular localization shows that METTL5 (green) accumulates in the nucleus (blue) and forms aggregates in the cytoplasm. The two frameshift variants as well as the missense variant do not seem to affect this localization. All images are projection of confocal optical sections stack. Scale bar: 10 μm.
(B) Quantification of the signal intensity for METTL5 in the nucleus in the different conditions show that only the truncating variants affect the level of expression of the protein. COS7 cells were co-transfected with peGFP empty vector and the different pCMV-Myc-METTL5 constructs. The intensity of the METTL5 and GFP signals in the nucleus were measured using ImageJ software and averaged from five areas per nucleus. METTL5 signal was normalized against GFP signal, minimizing the effect of transfection efficiency among cells and experiments (one-way ANOVA analysis followed by a Bonferroni post hoc test, ∗∗∗∗p < 0.0001).
(C and D) Western blot on transfected HEK293T cells with Myc-METTL5 WT or mutant constructs reveals that both frameshift variants decrease the stability of the mutant proteins, but the missense variant does not.
(C) Representative image of western blot assay. An antibody against Myc (Covance Research Products Inc., Cat# MMS-164P-100, RRID:AB_291335) was used to analyze Myc-METTL5 signal. GFP signal was used as a transfection control.
(D) Quantification of the signal intensity for Myc and GFP was measured using ImageJ software. Myc signal intensity was normalized against the GFP signal intensity (one-way ANOVA analysis followed by a Bonferroni post hoc test, ∗∗p < 0.01, ∗∗∗∗p < 0.0001).
(E) Representative image of western blot assay. 48 h after transfection, cells were collected (t = 0) to assess initial expression level of the protein. Remaining cells were treated either with vehicle (0.5% DMSO), MG132 (50 μM), or cyclohexamide (CHX, 0.1 mM) for 4 h. Mutant METTL5 bands intensity increases drastically after treatment with MG132, while the cyclohexamide treatment does not alter differently WT and mutant proteins, suggesting that the disease-causing variants affect the stability of the protein rather than the mRNAs.
In COS7 cells, no apparent difference in the levels and localization of METTL5G61D protein was observed, when compared with wild-type METTL5 (Figure 3B). In contrast, the levels of METTL5R115Nfs∗19 and METTL5L191Vfs∗10 were significantly reduced as compared to wild-type METTL5 (Figure 3B). Western blot analyses using transiently transfected HEK293T cells further confirmed that both truncating variants significantly (∗∗p < 0.01, ∗∗∗p < 0.001) affected the stability of the mutant METTL5 protein (Figures 3C, 3D, and S4). To determine whether the observed decrease in protein expression is due to protein instability, we challenged the stability of Myc-tagged proteins. At 48 h post transfection, cells were treated for 4 h with either MG132 (50 μM, proteasome inhibitor, Sigma-Aldrich) or CHX (cyclohexamide, 0.1 mM, synthesis blocker, Sigma-Aldrich). Post-MG132-treatment western blot analysis revealed a dramatic increase in the levels of truncated proteins while post-CHX treatment did not highlight any remarkable difference between the WT and the mutant proteins, which suggest that c.344_345delGA and c.571_572delAA variants significantly reduce the stability of the corresponding proteins (Figure 3E).
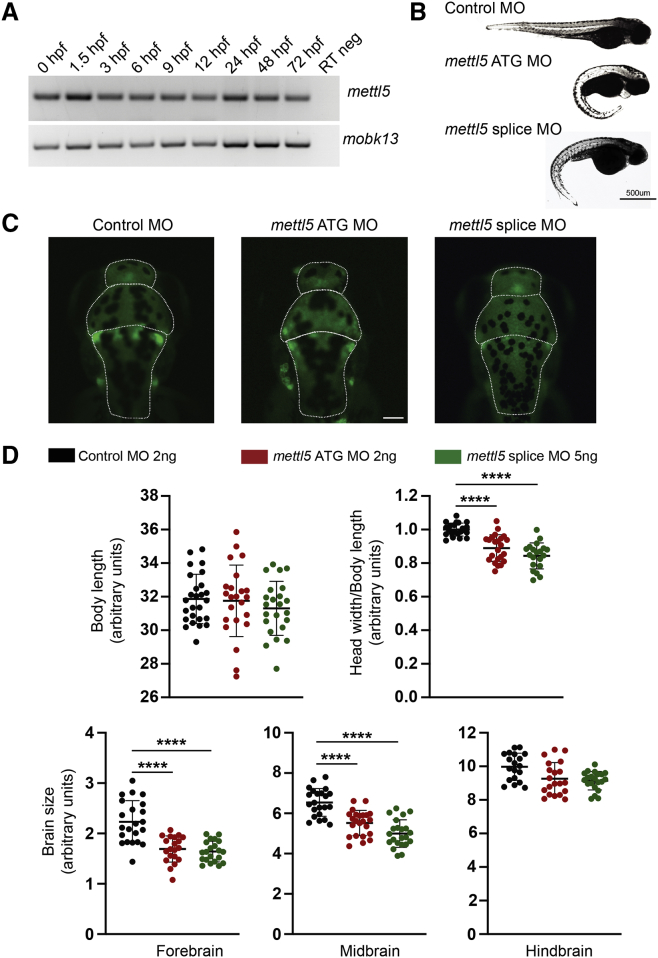
We next investigated the function of Mettl5 in the developing brain and neurons in zebrafish, which express the orthologous mettl5 gene (GenBank: NM_001005949.1) throughout development (Figure 4A).22 Using specific translation blocking morpholino (MO) (5′-GCTCTCCAGCTCTTTCAGCTTCATT-3′; 2 ng) and splice site (exon 3) targeting MO (5′-GGTTAGTGAGTTTTCTTACCCTGGT-3′; 5 ng), we knocked down mettl5 in embryos of NeuroD-eGFP transgenic zebrafish strain.23 At 72 h post-fertilization (hpf) mettl5 morphants had reduced head size, mimicking the human microcephaly, and curved tails, while the control morpholino (5′- CCTCTTACCTCAGTTACAATTTATA-3′)-injected embryos had normal growth (Figures 4B, 4C, and S6). To rule out MO toxicity, we used various dilutions and also blocked the p53 pathway via co-injection of a p53-targeting MO.24 The phenotype of the co-injected embryos was identical to that of those injected with the Mettl5 ATG blocker or splice-targeting MO alone. Thus, morphant developmental deficits appear to be specific to the knockdown of mettl5 expression. Next, we further examined the brain structure in mettl5 morphants and controls embryos. Although no statistically significant difference in hindbrain size was observed, both the forebrain and the midbrain were adversely altered with a 24%–26% and 16%–23% size reduction, respectively (p < 0.0001), which further confirms the essential role of mettl5 in brain development.
Figure 4.
mettl5 Knock-down in zebrafish Reproduces the Microcephaly Phenotype
(A) mettl5 is expressed as early as 20 min after fertilization in zebrafish embryos. mobk13 was used as a loading control. Hpf, hours post fertilization.
(B) Representative images of dorsal view of zebrafish morphology from control, ATG blocker mettl5 MO, and splice-targeting mettl5 MO, injected larvae, at 72 hpf. Scale bar: 500 μm.
(C) Representative top images of injected zebrafish’s brain, at 72 hpf. Brain area were measured according to the white dotted line. Scale bar: 10 μm.
(D) Zebrafish were imaged from a dorsal and lateral view, at 72 hpf. The total body length as well as the brain maximum width were measured using ImageJ software. The brain width/body length ratio was calculated and normalized against the mean for the fish injected with the Control MO and defined as the “microcephaly index,” as previously described.21 The larvae injected with the mettl5 ATG MO and splice-targeting MO show a significant microcephaly phenotype compared to the fish injected with the control MO (n = 20/condition, data are represented as the mean ± SD, one-way ANOVA analysis followed by a Bonferroni post hoc test, ∗∗∗∗p < 0.0001).
(E) Each brain area was measured using ImageJ software and normalized against the mean for the Control MO-injected fish (n = 20/condition, data are represented as the mean ± SD, one-way ANOVA analysis followed by a Bonferroni post hoc test, ∗∗∗∗p < 0.0001).
Even though the precise function of METTL5 is currently uncharacterized, our study highlights its essential role in human and zebrafish brain development and cognitive function. In this study, we have identified two bi-allelic frameshift variants: c.344_345delGA and c.571_572delAA. Despite a low pLI score for METTL5 (pLi = 0), no loss-of-function variants in a homozygous state was found in the gnomAD database, indicating the in-tolerance to bi-allelic truncating variants. Our functional analyses revealed that both c.344_345delGA and c.571_572delAA variants drastically impact the expression and the conformation of the protein (Figures 2 and3), and thus taken together with the genetic and in silico analysis, both these variants can be classified as “pathogenic” according to the ACMG/AMP guidelines25 (Table S2). The candidate missense variant p.Gly61Asp reported previously,11 replacing highly evolutionary conserved amino acid (Figure S7), is predicted to be pathogenic by several in silico algorithms (Table S3). However, our 3D modeling (Figure S7) as well as expression and localization studies (Figure 3) failed to demonstrate a functional impact on the encoded protein, and thus classified here as “variant of uncertain significance” (Table S2).
In humans, variants in METTL proteins have been associated with various disorders, such as sclerosis (METTL126 [MIM: 404466]), liver cancer (METTL327 [MIM: 612472]), breast cancer (METTL3,28 METTL629), colon cancer (METTL8 [MIM: 609525] and METTL1630), pancreatic cancer (METTL1331 [MIM: 617987]), otoprotection/hearing loss (MIM: 605429) (METTL1332), and osteoporosis (METTL21C33 [MIM: 615259]). Furthermore, variants in other methyltransferase-like genes associated with ID have been reported previously.34, 35, 36, 37 For instance, truncating alleles of METTL23 (MIM: 615262) have been identified in families with mild non-syndromic ARID or cognitive dysfunction with mild dysmorphic features (MIM: 615942). Similarly, micro-duplications (351 kb and 432 kb genomic region), both containing METTL4 among other genes, were associated with ID and mild dysmorphic features.36, 37 In addition to these METTL proteins, various methyltransferases, including histone methyltransferases (EHMT138 [MIM: 607001], KMT2A39 [MIM: 159555]), DNA methyltransferase (DNMT3A40 [MIM: 602769]), tRNA (TRMT141 [MIM: 611669], NSUN242, 43 [MIM: 610916]), and rRNA methyltransferases (FTSJ144 [MIM: 300499]), have been implicated in ID, highlighting the importance of methyltransferases in the brain development and cognitive functions.
A growing body of evidence shows that methyltransferases and other epigenetic regulators play an important role in neurodevelopment and/or neuroplasticity.45, 46 The presence of putative DNA methylase N6 adenine-specific conserved site as well as the accumulation of METTL5 in the nucleus tend to suggest an important role of METTL5 as an epigenetic regulator. In silico and homology analyses predicted METTL5 interaction with DNA; however, there is only experimental evidence showing that it interacts with RNA.47 N6-adenine RNA methylation has been implicated in the regulation of gene expression, translation efficiency, mRNA stability,48 as well as in neuropsychiatric disorders,49 although the mechanism and impact are not fully understood yet.50, 51, 52 Among others, METTL3 and METTL14 (MIM: 616504) are of particular interest as they belong to the METTL family and form a complex which is important for N6-adenine RNA methylation.53 A recent study, using Mettl14 transgenic mice, showed that Mettl14 regulates the size of the brain and more particularly the size of the cerebral cortex, via altered histone modifications.54 Knocking out Mettl14 specifically in the neural stem cells leads to microcephaly.54 Similarly, the ablation of Mettl3 in mice leads to developmental defects associated with depletion of m6A and dramatic change in the expression of apoptosis and cerebellum development-related genes.55 In addition to their roles in the regulation of brain development, both Mettl3 and Mettl14 are key molecules that can alter learning abilities.56, 57 METTL3-METTL14 complex is one of the main components, but it does not account for all the N6-methyladenosine (m6A) modifications. The identification of METTL16, another methyltransferase-like protein, as a new m6A methyltransferase58 suggests that other methyltransferase-like family members like METTL5 may also carry m6A methyltransferase activity.
In summary, METTL5 variants underlie autosomal-recessive ID with microcephaly in humans. METTL5 is expressed in the developing and aging brain, accumulates in the nucleus and the synapses of neurons, and shares several domains structure with other m6A modifiers. Therefore, we speculate that human METTL5 participates in the epigenetic regulation of DNA/RNA in the neurons. Our studies reveal that, as in humans, zebrafish Mettl5 is critical for proper brain development and possibly neuronal function.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We are very thankful to the affected individuals and their families who have participated to this study. We would like to thank Dr. Zaghloul for generously sharing the NeuroD-eGFP zebrafish line with us. We would like to thank Muhammad A. Usmani and Drs. R. Yousaf, S. Yousaf, and A.P.J. Giese for their technical assistance. Confocal images were acquired thanks to the University of Maryland, School of Medicine, CIBR Confocal microscopy core facility and Department of Physiology (Baltimore, Maryland, USA).
This work was supported by NIH R01NS107428 (to Saima Riazuddin), the EU FP7 Large-Scale Integrating Project Genetic and Epigenetic Networks in Cognitive Dysfunction (241995) (to H.v.B.), the DFG (AB 3939 / 2-2) (to R.A.J.), and NRPU Projects from Higher Education commission of Pakistan (to M.S. and J.A.). D.L.P. is recipient of a CAPES Fellowship (99999.013311/2013-01).
Published: September 26, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.09.007.
Contributor Information
Sheikh Riazuddin, Email: riazuddin@aimrc.org.
Saima Riazuddin, Email: sriazuddin@som.umaryland.edu.
Web Resources
Allen Brain Atlas, https://portal.brain-map.org/
CADD, https://cadd.gs.washington.edu/
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/variation/689363/; https://www.ncbi.nlm.nih.gov/clinvar/variation/689379/
ExAC Browser, http://exac.broadinstitute.org/
GenBank, https://www.ncbi.nlm.nih.gov/genbank/
gnomAD Browser, https://gnomad.broadinstitute.org/
InterPro, https://www.ebi.ac.uk/interpro/
MutationTaster, http://www.mutationtaster.org/
OMIM, https://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PROVEAN, http://provean.jcvi.org
Supplemental Data
Reference
- 1.Schalock R.L., Luckasson R.A., Shogren K.A., Borthwick-Duffy S., Bradley V., Buntinx W.H., Coulter D.L., Craig E.M., Gomez S.C., Lachapelle Y. The renaming of mental retardation: understanding the change to the term intellectual disability. Intellect. Dev. Disabil. 2007;45:116–124. doi: 10.1352/1934-9556(2007)45[116:TROMRU]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 2.Maulik P.K., Mascarenhas M.N., Mathers C.D., Dua T., Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res. Dev. Disabil. 2011;32:419–436. doi: 10.1016/j.ridd.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 3.McKenzie K., Murray A.L. Evaluating the use of the Child and Adolescent Intellectual Disability Screening Questionnaire (CAIDS-Q) to estimate IQ in children with low intellectual ability. Res. Dev. Disabil. 2015;37:31–36. doi: 10.1016/j.ridd.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Vissers L.E., Gilissen C., Veltman J.A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 5.Harripaul R., Noor A., Ayub M., Vincent J.B. The Use of Next-Generation Sequencing for Research and Diagnostics for Intellectual Disability. Cold Spring Harb. Perspect. Med. 2017;7:7. doi: 10.1101/cshperspect.a026864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riazuddin S., Hussain M., Razzaq A., Iqbal Z., Shahzad M., Polla D.L., Song Y., van Beusekom E., Khan A.A., Tomas-Roca L., UK10K Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol. Psychiatry. 2017;22:1604–1614. doi: 10.1038/mp.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Najmabadi H., Hu H., Garshasbi M., Zemojtel T., Abedini S.S., Chen W., Hosseini M., Behjati F., Haas S., Jamali P. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 8.Anazi S., Maddirevula S., Faqeih E., Alsedairy H., Alzahrani F., Shamseldin H.E., Patel N., Hashem M., Ibrahim N., Abdulwahab F. Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol. Psychiatry. 2017;22:615–624. doi: 10.1038/mp.2016.113. [DOI] [PubMed] [Google Scholar]
- 9.Reuter M.S., Tawamie H., Buchert R., Hosny Gebril O., Froukh T., Thiel C., Uebe S., Ekici A.B., Krumbiegel M., Zweier C. Diagnostic Yield and Novel Candidate Genes by Exome Sequencing in 152 Consanguineous Families With Neurodevelopmental Disorders. JAMA Psychiatry. 2017;74:293–299. doi: 10.1001/jamapsychiatry.2016.3798. [DOI] [PubMed] [Google Scholar]
- 10.Trujillano D., Bertoli-Avella A.M., Kumar Kandaswamy K., Weiss M.E., Köster J., Marais A., Paknia O., Schröder R., Garcia-Aznar J.M., Werber M. Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur. J. Hum. Genet. 2017;25:176–182. doi: 10.1038/ejhg.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu H., Kahrizi K., Musante L., Fattahi Z., Herwig R., Hosseini M., Oppitz C., Abedini S.S., Suckow V., Larti F. Genetics of intellectual disability in consanguineous families. Mol. Psychiatry. 2019;24:1027–1039. doi: 10.1038/s41380-017-0012-2. [DOI] [PubMed] [Google Scholar]
- 12.Harripaul R., Vasli N., Mikhailov A., Rafiq M.A., Mittal K., Windpassinger C., Sheikh T.I., Noor A., Mahmood H., Downey S. Mapping autosomal recessive intellectual disability: combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol. Psychiatry. 2018;23:973–984. doi: 10.1038/mp.2017.60. [DOI] [PubMed] [Google Scholar]
- 13.Musante L., Ropers H.H. Genetics of recessive cognitive disorders. Trends Genet. 2014;30:32–39. doi: 10.1016/j.tig.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Robinson P.N., Köhler S., Bauer S., Seelow D., Horn D., Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis B.P., Green R.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutton M.A., Schuman E.M. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 18.Kelleher R.J., 3rd, Govindarajan A., Tonegawa S. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron. 2004;44:59–73. doi: 10.1016/j.neuron.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Merkurjev D., Hong W.T., Iida K., Oomoto I., Goldie B.J., Yamaguti H., Ohara T., Kawaguchi S.Y., Hirano T., Martin K.C. Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 2018;21:1004–1014. doi: 10.1038/s41593-018-0173-6. [DOI] [PubMed] [Google Scholar]
- 20.Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jobst-Schwan T., Schmidt J.M., Schneider R., Hoogstraten C.A., Ullmann J.F.P., Schapiro D., Majmundar A.J., Kolb A., Eddy K., Shril S. Acute multi-sgRNA knockdown of KEOPS complex genes reproduces the microcephaly phenotype of the stable knockout zebrafish model. PLoS ONE. 2018;13:e0191503. doi: 10.1371/journal.pone.0191503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thisse B., Heyer V., Lux A., Alunni V., Degrave A., Seiliez I., Kirchner J., Parkhill J.P., Thisse C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–519. doi: 10.1016/s0091-679x(04)77027-2. [DOI] [PubMed] [Google Scholar]
- 23.Obholzer N., Wolfson S., Trapani J.G., Mo W., Nechiporuk A., Busch-Nentwich E., Seiler C., Sidi S., Söllner C., Duncan R.N. Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. J. Neurosci. 2008;28:2110–2118. doi: 10.1523/JNEUROSCI.5230-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langheinrich U., Hennen E., Stott G., Vacun G. Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signaling. Curr. Biol. 2002;12:2023–2028. doi: 10.1016/s0960-9822(02)01319-2. [DOI] [PubMed] [Google Scholar]
- 25.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alcina A., Fedetz M., Fernández O., Saiz A., Izquierdo G., Lucas M., Leyva L., García-León J.A., Abad-Grau Mdel.M., Alloza I. Identification of a functional variant in the KIF5A-CYP27B1-METTL1-FAM119B locus associated with multiple sclerosis. J. Med. Genet. 2013;50:25–33. doi: 10.1136/jmedgenet-2012-101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M., Wei L., Law C.T., Tsang F.H., Shen J., Cheng C.L., Tsang L.H., Ho D.W., Chiu D.K., Lee J.M. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 28.Cai X., Wang X., Cao C., Gao Y., Zhang S., Yang Z., Liu Y., Zhang X., Zhang W., Ye L. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–19. doi: 10.1016/j.canlet.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 29.Gatza M.L., Silva G.O., Parker J.S., Fan C., Perou C.M. An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nat. Genet. 2014;46:1051–1059. doi: 10.1038/ng.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeon S.Y., Jo Y.S., Choi E.J., Kim M.S., Yoo N.J., Lee S.H. Frameshift Mutations in Repeat Sequences of ANK3, HACD4, TCP10L, TP53BP1, MFN1, LCMT2, RNMT, TRMT6, METTL8 and METTL16 Genes in Colon Cancers. Pathol. Oncol. Res. 2018;24:617–622. doi: 10.1007/s12253-017-0287-2. [DOI] [PubMed] [Google Scholar]
- 31.Liu S., Hausmann S., Carlson S.M., Fuentes M.E., Francis J.W., Pillai R., Lofgren S.M., Hulea L., Tandoc K., Lu J. METTL13 Methylation of eEF1A Increases Translational Output to Promote Tumorigenesis. Cell. 2019;176:491–504.e21. doi: 10.1016/j.cell.2018.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yousaf R., Ahmed Z.M., Giese A.P., Morell R.J., Lagziel A., Dabdoub A., Wilcox E.R., Riazuddin S., Friedman T.B., Riazuddin S. Modifier variant of METTL13 suppresses human GAB1-associated profound deafness. J. Clin. Invest. 2018;128:1509–1522. doi: 10.1172/JCI97350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J., Hsu Y.H., Mo C., Abreu E., Kiel D.P., Bonewald L.F., Brotto M., Karasik D. METTL21C is a potential pleiotropic gene for osteoporosis and sarcopenia acting through the modulation of the NF-κB signaling pathway. J. Bone Miner. Res. 2014;29:1531–1540. doi: 10.1002/jbmr.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernkopf M., Webersinke G., Tongsook C., Koyani C.N., Rafiq M.A., Ayaz M., Müller D., Enzinger C., Aslam M., Naeem F. Disruption of the methyltransferase-like 23 gene METTL23 causes mild autosomal recessive intellectual disability. Hum. Mol. Genet. 2014;23:4015–4023. doi: 10.1093/hmg/ddu115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiff R.E., Ali B.R., Baron B., Yu T.W., Ben-Salem S., Coulter M.E., Schubert C.R., Hill R.S., Akawi N.A., Al-Younes B. METTL23, a transcriptional partner of GABPA, is essential for human cognition. Hum. Mol. Genet. 2014;23:3456–3466. doi: 10.1093/hmg/ddu054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kashevarova A.A., Nazarenko L.P., Skryabin N.A., Salyukova O.A., Chechetkina N.N., Tolmacheva E.N., Sazhenova E.A., Magini P., Graziano C., Romeo G. Array CGH analysis of a cohort of Russian patients with intellectual disability. Gene. 2014;536:145–150. doi: 10.1016/j.gene.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 37.Kashevarova A.A., Nazarenko L.P., Skryabin N.A., Nikitina T.V., Vasilyev S.A., Tolmacheva E.N., Lopatkina M.E., Salyukova O.A., Chechetkina N.N., Vorotelyak E.A. A mosaic intragenic microduplication of LAMA1 and a constitutional 18p11.32 microduplication in a patient with keratosis pilaris and intellectual disability. Am. J. Med. Genet. A. 2018;176:2395–2403. doi: 10.1002/ajmg.a.40478. [DOI] [PubMed] [Google Scholar]
- 38.Kleefstra T., Brunner H.G., Amiel J., Oudakker A.R., Nillesen W.M., Magee A., Geneviève D., Cormier-Daire V., van Esch H., Fryns J.P. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am. J. Hum. Genet. 2006;79:370–377. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lebrun N., Giurgea I., Goldenberg A., Dieux A., Afenjar A., Ghoumid J., Diebold B., Mietton L., Briand-Suleau A., Billuart P., Bienvenu T. Molecular and cellular issues of KMT2A variants involved in Wiedemann-Steiner syndrome. Eur. J. Hum. Genet. 2018;26:107–116. doi: 10.1038/s41431-017-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatton-Brown K., Seal S., Ruark E., Harmer J., Ramsay E., Del Vecchio Duarte S., Zachariou A., Hanks S., O’Brien E., Aksglaede L., Childhood Overgrowth Consortium Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet. 2014;46:385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davarniya B., Hu H., Kahrizi K., Musante L., Fattahi Z., Hosseini M., Maqsoud F., Farajollahi R., Wienker T.F., Ropers H.H., Najmabadi H. The Role of a Novel TRMT1 Gene Mutation and Rare GRM1 Gene Defect in Intellectual Disability in Two Azeri Families. PLoS ONE. 2015;10:e0129631. doi: 10.1371/journal.pone.0129631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan M.A., Rafiq M.A., Noor A., Hussain S., Flores J.V., Rupp V., Vincent A.K., Malli R., Ali G., Khan F.S. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012;90:856–863. doi: 10.1016/j.ajhg.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abbasi-Moheb L., Mertel S., Gonsior M., Nouri-Vahid L., Kahrizi K., Cirak S., Wieczorek D., Motazacker M.M., Esmaeeli-Nieh S., Cremer K. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012;90:847–855. doi: 10.1016/j.ajhg.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freude K., Hoffmann K., Jensen L.R., Delatycki M.B., des Portes V., Moser B., Hamel B., van Bokhoven H., Moraine C., Fryns J.P. Mutations in the FTSJ1 gene coding for a novel S-adenosylmethionine-binding protein cause nonsyndromic X-linked mental retardation. Am. J. Hum. Genet. 2004;75:305–309. doi: 10.1086/422507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kleefstra T., Schenck A., Kramer J.M., van Bokhoven H. The genetics of cognitive epigenetics. Neuropharmacology. 2014;80:83–94. doi: 10.1016/j.neuropharm.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 46.Iwase S., Bérubé N.G., Zhou Z., Kasri N.N., Battaglioli E., Scandaglia M., Barco A. Epigenetic Etiology of Intellectual Disability. J. Neurosci. 2017;37:10773–10782. doi: 10.1523/JNEUROSCI.1840-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franke C., Gräfe D., Bartsch H., Bachmann M.P. Use of Nonradioactive Detection Method for North- and South-Western Blot. Methods Mol. Biol. 2015;1314:63–71. doi: 10.1007/978-1-4939-2718-0_8. [DOI] [PubMed] [Google Scholar]
- 48.Parashar N.C., Parashar G., Nayyar H., Sandhir R. N6-adenine DNA methylation demystified in eukaryotic genome: From biology to pathology. Biochimie. 2018;144:56–62. doi: 10.1016/j.biochi.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 49.Yao B., Cheng Y., Wang Z., Li Y., Chen L., Huang L., Zhang W., Chen D., Wu H., Tang B., Jin P. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun. 2017;8:1122. doi: 10.1038/s41467-017-01195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lesbirel S., Wilson S.A. The m6A-methylase complex and mRNA export. Biochim. Biophys. Acta. Gene Regul. Mech. 2019;1862:319–328. doi: 10.1016/j.bbagrm.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer K.D. m6A-mediated translation regulation. Biochim. Biophys. Acta. Gene Regul. Mech. 2019;1862:301–309. doi: 10.1016/j.bbagrm.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malla S., Melguizo-Sanchis D., Aguilo F. Steering pluripotency and differentiation with N6-methyladenosine RNA modification. Biochim. Biophys. Acta. Gene Regul. Mech. 2019;1862:394–402. doi: 10.1016/j.bbagrm.2018.10.013. [DOI] [PubMed] [Google Scholar]
- 53.Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y., Li Y., Yue M., Wang J., Kumar S., Wechsler-Reya R.J., Zhang Z., Ogawa Y., Kellis M., Duester G., Zhao J.C. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 2018;21:195–206. doi: 10.1038/s41593-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang C.X., Cui G.S., Liu X., Xu K., Wang M., Zhang X.X., Jiang L.Y., Li A., Yang Y., Lai W.Y. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018;16:e2004880. doi: 10.1371/journal.pbio.2004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koranda J.L., Dore L., Shi H., Patel M.J., Vaasjo L.O., Rao M.N., Chen K., Lu Z., Yi Y., Chi W. Mettl14 Is Essential for Epitranscriptomic Regulation of Striatal Function and Learning. Neuron. 2018;99:283–292.e5. doi: 10.1016/j.neuron.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Z., Wang M., Xie D., Huang Z., Zhang L., Yang Y., Ma D., Li W., Zhou Q., Yang Y.G., Wang X.J. METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 2018;28:1050–1061. doi: 10.1038/s41422-018-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warda A.S., Kretschmer J., Hackert P., Lenz C., Urlaub H., Höbartner C., Sloan K.E., Bohnsack M.T. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–2014. doi: 10.15252/embr.201744940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.