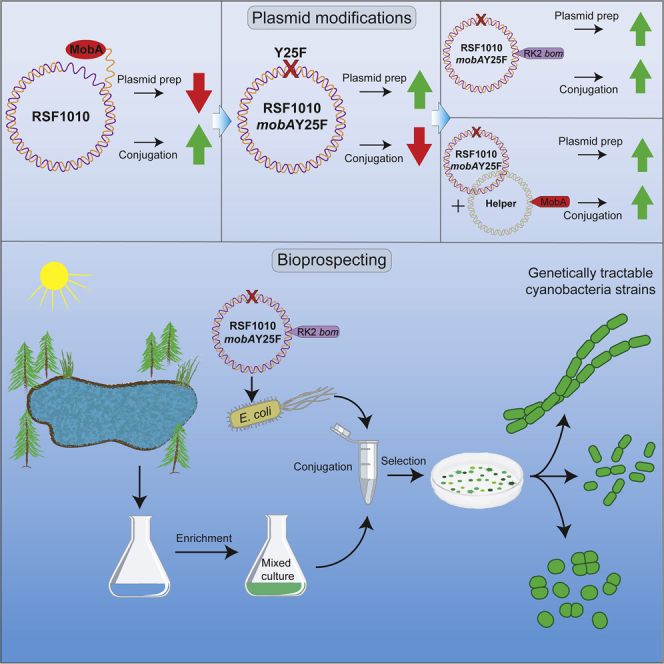
Summary
To facilitate the genetic engineering of diverse cyanobacterial strains, we have modified broad-host-range RSF1010-based plasmids to improve transmissibility, increase copy number, and facilitate cloning. RSF1010-based plasmids replicate in diverse bacterial strains but produce low amounts of useable DNA for cloning. We previously engineered a mobAY25F mutation in RSF1010-based plasmids that improved cloning but decreased conjugation efficiency. Here, we engineered RSF1010-based plasmids to restore conjugation efficiency, which was demonstrated in three diverse laboratory strains of cyanobacteria. We then used an improved RSF1010-based plasmid in mating experiments with cultured samples of wild cyanobacteria. This plasmid, which confers antibiotic resistance and carries a yfp reporter gene, allowed selection of exconjugant cyanobacteria and facilitated the isolation of genetically tractable strains from mixed wild cultures. Improved RSF1010 vectors can be used for bioprospecting genetically tractable strains and are compatible with the CYANO-VECTOR cloning system, a versatile toolbox for constructing plasmids for cyanobacterial genetic engineering.
Subject Areas: Biological Sciences, Molecular Biology, Molecular Microbiology, Bioengineering, Metabolic Engineering, Microbial Biotechnology
Graphical Abstract
Highlights
-
•
An RSF1010 mobAY25F mutation facilitates plasmid cloning but reduces conjugation
-
•
Addition of an RK2-bom site improves conjugation efficiency of mobAY25F vectors
-
•
A helper plasmid carrying the mobA gene improves conjugation efficiency of mobAY25F
-
•
Improved RSF1010-based vectors can be used for bioprospecting of cyanobacteria
Biological Sciences; Molecular Biology; Molecular Microbiology; Bioengineering; Metabolic Engineering; Microbial Biotechnology
Introduction
Cyanobacteria have attracted interest for use as biotechnology production platforms (Ducat et al., 2011, Heidorn et al., 2011, Hays and Ducat, 2015). They grow rapidly, efficiently convert sunlight energy to biomass, and many strains are genetically tractable (Dismukes et al., 2008, Zhu et al., 2008, Heidorn et al., 2011). In addition, without the need for a carbohydrate feedstock, cyanobacteria have the potential to be more cost effective for large-scale production than traditional microorganisms typically used for heterologous gene expression such as Escherichia coli or Saccharomyces cerevisiae (Ducat et al., 2011). As opposed to heterotrophic microorganisms, cyanobacteria are photoautotrophs that use sunlight for energy and CO2 as a carbon source (Quintana et al., 2011). Cyanobacteria are a diverse group of bacteria. They can be found in practically every sunlit environment on the Earth, from marine and freshwater bodies of water, to soil crusts, and even bare rocks (Shih et al., 2013). Cyanobacteria are the only organisms to have evolved oxygenic photosynthesis, which appeared over 2.7 billion years ago, and all higher plants and algae derived their chloroplasts from the cyanobacterial lineage by endosymbiosis (Martin and Kowallik, 1999, Schirrmeister et al., 2015).
In addition to their potential as production platforms, cyanobacteria have a number of advantages over more traditional crops used for the production of biomass as a source of food or fuel. They can utilize concentrated CO2 as a carbon source to scrub greenhouse gas emissions from factories or power plants, and some strains can be grown on brackish and saltwater or wastewater unsuitable for consumption or crop watering (Rawat et al., 2011, Olguín, 2012). It is also worth noting that cyanobacteria drove the Great Oxygenation Event about 2.3 billion years ago, which elevated atmospheric oxygen levels and supported the rise of all animal life (Bekker et al., 2004). Cyanobacteria belong to a widely divergent prokaryotic phylum with a large range of morphological, physiological, and ecological characteristics. However, most research on cyanobacterial genetics has been limited to a handful of well-studied “domesticated” laboratory strains. Improved genetic methods are needed for bioprospecting to identify new diverse genetically tractable strains for the study of their biology and to develop them for biotechnological applications.
The broad-host-range plasmid RSF1010 is a member of the IncQ group of plasmids, which stably replicate in a wide variety of gram-negative and gram-positive bacteria, including E. coli and several strains of cyanobacteria (Trieu-Cuot et al., 1987, Sode et al., 1992, Meyer, 2009). One of the reasons for the broad-host-range characteristic of RSF1010 is that it encodes replication initiation machinery that is host independent (Scherzinger et al., 1991, Meyer, 2009). In addition, although RSF1010 is not self-transmissible, it harbors an origin of transfer and mobilization genes and can co-opt type IV secretion machinery of other self-transmissible plasmids such as RK2 and RP4 (Meyer, 2009). Conjugation from E. coli is a standard method for transferring recombinant plasmids into cyanobacteria because of its efficacy in a wide range of organisms including filamentous strains (Elhai et al., 1997). Natural transformation is used for routine genetic experiments in only two strains of unicellular cyanobacteria, Synechococcus elongatus PCC 7942 and Synechocystis PCC 6803 (Wendt and Pakrasi, 2019), and even for these strains, conjugation is often used when higher efficiency of DNA transfer is required, such as for the construction of genetic libraries (Rubin et al., 2015).
Although the RSF1010 replicon appears well suited for heterologous gene expression, DNA yields are typically low using standard plasmid preparation procedures (Gormley and Davies, 1991, Taton et al., 2014). This is partially due to its maintenance at low copy number, but it is also a side effect of its self-mobility (Frey et al., 1992). One of the RSF1010 proteins, MobA, is a component of the relaxosome that nicks RSF1010 DNA at the bom site (for basis of mobility, also called oriT for origin of transfer), as an initial step in the mobilization process, and the nicked intermediate contains MobA covalently linked to the 5′ end of the cleaved strand (Monzingo et al., 2007). When using standard plasmid preparation techniques that involve heat or alkaline denaturation steps, the nicked DNA can melt to single strands. Some of the RSF1010 DNA isolated from cells will be relaxed nicked molecules with a covalently linked MobA protein or denatured single-stranded molecules that are not recovered in most plasmid preparation protocols and are unsuitable for cloning experiments. To make RSF1010 more tractable for laboratory cloning procedures, a mutation was made in the mobA gene (Taton et al., 2014). This mutation, mobAY25F, produces a defective MobA protein that lacks the ability to cleave RSF1010 DNA at its bom site. Although this RSF1010 mobAY25F variant, in the donor plasmid pCVD047, is more amenable to plasmid preparation and cloning, the loss of MobA functionality reduces the overall conjugation efficiency by two to four orders of magnitude when RP4 is used as the conjugal plasmid (Monzingo et al., 2007, Taton et al., 2014).
A set of modifications were previously made to the RSF1010 plasmid that facilitate its use as a device compatible with the CYANO-VECTOR plasmid assembly platform (Taton et al., 2014). The CYANO-VECTOR platform comprises numerous plasmid devices including functional and selection modules, chromosomal integration sequences, E. coli origins of replication, as well as cyanobacterial and broad-host-range replicons such as the ones based on RSF1010. Each donor plasmid has custom-designed GC-rich sequences that allow for seamless assembly of multiple parts and devices into destination plasmids. Destination plasmids typically contain three or four genetic parts or devices, for example, an E. coli ori, a selectable marker, a cyanobacterial replicon for the target strain, and a functional device such as a reporter-gene construct.
In this study, we describe modifications to the CYANO-VECTOR-compatible plasmid pCVD047 and a conjugation helper plasmid to provide greater quantities of usable plasmid DNA, boost conjugation efficiency, and make the RSF1010 broad-host-range plasmid system more versatile and easier to use. We tested these modifications in three diverse cyanobacterial model strains and showed that they increased conjugation efficiency. In addition, we conjugated one of the modified RSF1010-based plasmids, pAM5404, into mixed cultures derived from wild environmental samples and isolated exconjugant cyanobacterial strains through antibiotic selection and screening for reporter gene expression. These experiments demonstrate a practical application of an efficient broad-host-range conjugation system that enables the isolation of genetically tractable cyanobacteria from a mixed culture. When combined with the wide variety of custom genetic parts and devices available using the CYANO-VECTOR assembly portal, the method described in this work can be a useful tool for bioprospecting for genetically tractable bacterial strains from any environment.
Results
Generation of pCVD047 Variants
RSF1010-based plasmids can be transferred into cyanobacterial strains by conjugation from E. coli. The conjugal donor E. coli strain carries a conjugation plasmid such as pRL443 and an optional helper plasmid such as pRL623. pRL443 is a kanamycin-sensitive mutant of RP4, an IncP-group plasmid with type IV conjugation machinery and transfer functions (tra genes) for self-mobilization (Elhai et al., 1997). The optional helper plasmid, pRL623, carries the ColK mob gene required to mobilize pBR322-based plasmids and also includes three restriction methyltransferases to protect cargo plasmids from restriction activity by Anabaena enzymes AvaI, AvaII, and AvaIII (Elhai et al., 1997). The cargo plasmid is introduced into this background to generate an E. coli donor strain containing all three plasmids. The cargo plasmid may be a “suicide” plasmid designed to recombine into the host chromosome, or a shuttle plasmid that can replicate in E. coli and the cyanobacterial recipient.
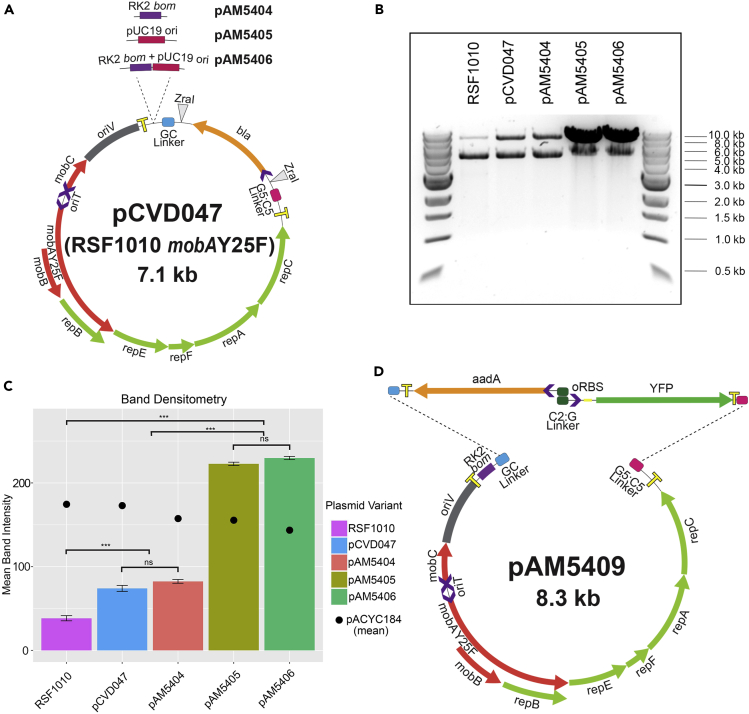
The first strategy used to restore conjugation efficiency and maintain high DNA yields for pCVD047 was to add an RK2-bom site as a second origin of transfer, generating plasmid pAM5404 (Figure 1A). RK2 is an IncP conjugal plasmid very similar to RP4 (Babic et al., 2008). The bom site is a short non-coding region of direct and inverted repeats recognized by the RK2/RP4 TraJ relaxase, which nicks the double-stranded DNA before conjugative transfer (Babic et al., 2008). We anticipated that the TraJ encoded by the RP4 conjugal plasmid in the standard conjugal system described above would recognize and nick the shuttle plasmid DNA at the RK2-bom site more efficiently than at RSF1010's native bom site, increasing its mobilization efficiency. In addition, to increase plasmid DNA yields from E. coli cells for cloning experiments while also retaining the RSF1010 broad-host-range oriV, a high copy number origin of replication from pUC19 was added to pCVD047 and pAM5404 to generate plasmids pAM5405 and pAM5406, respectively (Figure 1A).
Figure 1.
Construction of Improved RSF1010-Based Plasmids
(A) Diagram depicting the CYANO-VECTOR donor plasmid pCVD047, which contains the mobilization-replication region of RSF1010 mobAY25F, with additional components to create plasmids pAM5404 (RK2-bom), pAM5405 (pUC-ori), and pAM5406 (RK2-bom, pUC-ori).
(B) Electrophoresis of total plasmid DNA from plasmid preparations of equal amounts of E. coli cells containing both an RSF1010 variant (upper band) and the control plasmid pACYC184 (lower band), linearized with EcoRV-HF. Outside lanes are 1-kb ladder size markers.
(C) Densitometry of electrophoretic gel bands for RSF1010 variants normalized to the control plasmid pACYC184. Bars represent mean relative band density from triplicate samples; error bars are mean ± SE. Statistical significances were inferred by the Tukey's test (HSD, honestly significant difference); ***p < 0.001; ns, not significant.
(D) Diagram of pAM5404-derived shuttle plasmid pAM5409, which carries the spectinomycin/streptomycin resistance cassette aadA and the oRBS-yfp gene with an optimized ribosome-binding site. Strains and plasmids are listed in Table S2 and oligonucleotides are listed in Table S3.
RSF1010, pCVD047 (mobAY25F), pAM5404 (RK2-bom), pAM5405 (pUC-ori), and pAM5406 (pUC-ori, RK2-bom) were transformed into E. coli DH10B along with a control plasmid, pACYC184. We used densitometry of stained gel images to examine the relative DNA concentrations of the different RSF1010-based plasmids. E. coli cells harboring both plasmids were grown in triplicate, and cultures were standardized by OD600 measurement. Plasmid DNA was extracted from equal numbers of cells, linearized, and separated by agarose gel electrophoresis (Figure 1B). The amounts of the RSF1010 variants and pACYC184 were determined by analyzing relative band intensities. All RSF1010 variants with the mobAY25F substitution produced more useable DNA than wild-type RSF1010 (Figure 1C). The variants containing the pUC origin of replication, pAM5405 and pAM5406, generated much greater amounts of DNA compared with the control plasmid (Figure 1C).
Generation of Shuttle Plasmids and Conjugation into Cyanobacteria
Shuttle plasmids carrying a spectinomycin/streptomycin resistance cassette (aadA) and a PconII-driven yfp gene were generated by seamless assembly (Figure 1D) using the CYANO-VECTOR toolkit. Shuttle plasmids were constructed with the RSF1010 mobilization-replication region from pCVD046 (wild type), pCVD047 (mobAY25F), pAM5404 (RK2-bom), pAM5405 (pUC-ori), and pAM5406 (pUC-ori, RK2-bom). These plasmids were named pAM5407–pAM5411, respectively (Table S2). Each shuttle plasmid was subsequently transformed into the conjugal donor E. coli strain AM1359, which contains the helper plasmid pRL623 and conjugal plasmid pRL443 to generate E. coli cells capable of conjugal transfer of the cargo plasmid. These cells were used in biparental mating experiments to transfer the modified RSF1010 plasmids into cyanobacterial strains.
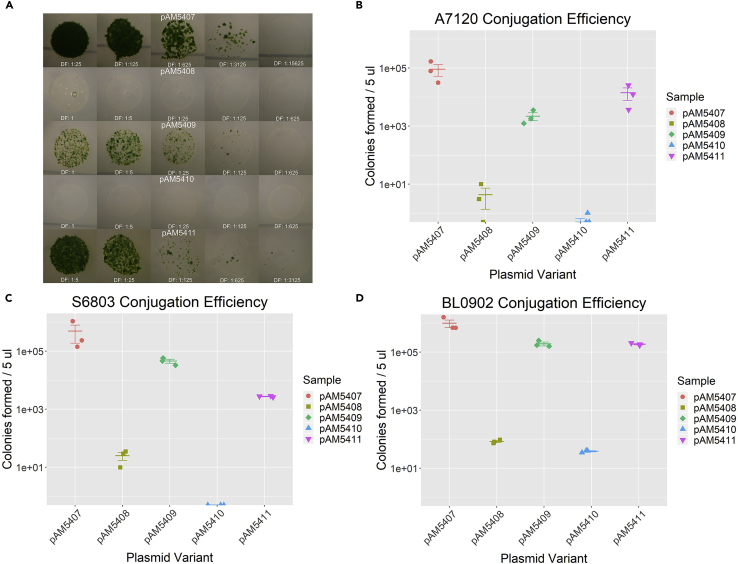
We first performed conjugations into the filamentous nitrogen-fixing cyanobacterial strain Anabaena PCC 7120. The filaments were fragmented to short filaments by sonication to provide better determination of conjugation efficiency, allowed to recover overnight, and mixed in triplicate with cultures of AM1359 E. coli containing pAM5407 or one of the four variants. Serial dilutions of the cell mixtures were spotted onto BG-11 + 5% Luria broth (LB) plates and allowed to conjugate overnight at 30°C. After 18–24 h, plates were underlaid with antibiotic and grown in humidified chambers under medium light (70–100 μmol photons m−2 s−1) at 30°C until colonies formed (Figure 2A). The mean conjugation efficiency for triplicate spots was calculated from the total number of colonies formed per spot multiplied by the dilution factor (Figure 2B). As previously noted, the Y25F mutation in the MobA protein causes the conjugation efficiency of pAM5408 (Y25F) to drop dramatically, by about four orders of magnitude (Taton et al., 2014). However, addition of the RK2-bom site to the plasmid leads to a substantial recovery, greater than two orders of magnitude, of the conjugative efficiency of pAM5409 (RK2-bom) (Figure 2B). pAM5410 (pUC-ori) has about the same conjugation efficiency as pAM5408, which demonstrates that the addition of the pUC origin of replication has little effect on conjugation. Similarly, the pUC origin also does not interfere with the addition of the RK2-bom site, because pAM5411 (pUC-ori, RK2-bom) demonstrated a restoration of conjugative ability similar to that of pAM5409.
Figure 2.
Conjugation Efficiency of RSF1010-Based Plasmids into Three Cyanobacterial Strains
(A) Images of serially diluted 5-μL mating spots of Anabaena PCC 7120 (A7120) harboring one of five RSF1010 plasmid variants.
(B–D) Dot plots of colony counts from triplicate samples of mating plates for Anabaena PCC 7120 (B), Synechocystis PCC 6803 (S6803) (C), and Leptolyngbya BL0902 (BL0902) (D). Each strain was mated with E. coli conjugal donor strains containing plasmids pAM5407, pAM5408, pAM5409, pAM5410, or pAM5411. Dots represent colony counts from each replicate sample; error bars show the mean ± SEM.
These experiments were repeated in two other strains, Synechocystis PCC 6803 and Leptolyngbya BL0902 (Figures 2C and 2D). The results obtained for Anabaena PCC 7120 were largely consistent across strains; the addition of the RK2-bom site restored conjugation efficiency by two to four orders of magnitude. However, we did notice a strain-specific effect of the addition of the pUC replication origin in pAM5410 (pUC-ori) and pAM5411 (pUC-ori, RK2-bom). In Synechocystis PCC 6803, these plasmids displayed a reduced conjugation efficiency relative to their control variant (Figure 2C). pAM5410 (pUC-ori) was less efficient than pAM5408 (Y25F), and pAM5411 (pUC-ori, RK2-bom) was less efficient than pAM5409 (RK2-bom). The negative effect of the pUC origin was much smaller than the positive effect of the RK2-bom site addition, such that pAM5411 (pUC-ori, RK2-bom) still had a substantially increased conjugation efficiency compared with pAM5408 (Y25F). This negative effect of the pUC origin was not seen in Leptolyngbya BL0902, which closely mirrored the conjugation efficiency results obtained for Anabaena PCC 7120.
Addition of the mobA Gene In trans
As an alternative to modifying the RSF1010 plasmid backbone itself by the addition of the pUC origin and RK2-bom, we expressed the RSF1010 mobA gene in trans from a helper plasmid to restore conjugation efficiency lost by the mobAY25F mutation. We cloned the mobA gene into the helper plasmid pRL623 to create plasmid pAM5505. The mobA gene, including the upstream promoters p1, p2, and p3, and a fragment of mobC were cloned into pRL623 at a unique SacI site. Promoters p1 and p3 drive mobA expression, whereas p2, sandwiched between them, faces the opposite direction and drives expression of the mobC gene (Figure 1A). MobC seems to play a role in repression of RSF1010 gene expression, as well as controlling copy number, as an RSF1010 mobC deletion mutant increased plasmid copy number (Frey et al., 1992). To avoid potential confounding effects of expressing a partial MobC polypeptide, we inserted a premature stop codon into the mobC gene fragment.
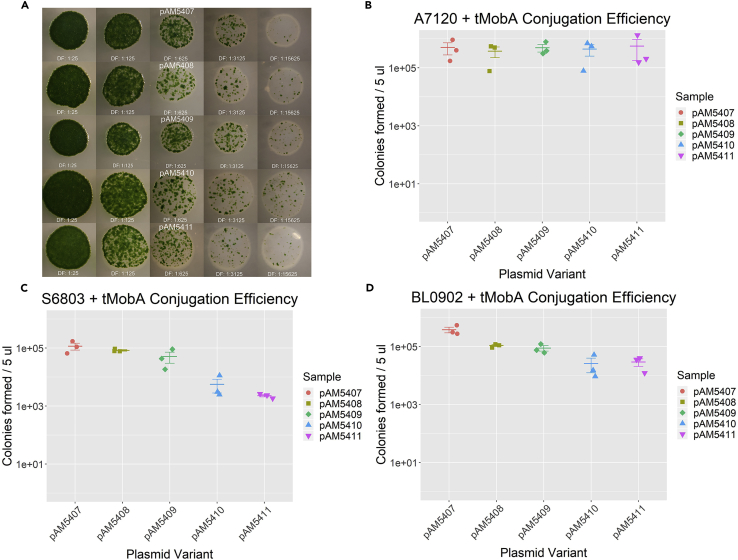
The pAM5505 helper plasmid allowed us to supply a fully functional MobA protein exclusively during mating from the E. coli conjugal donor strain. pCVD047 and derivative plasmids grown in standard E. coli cloning strains would therefore remain un-nicked in supercoiled form, allowing for more efficient purification and better performance in subsequent cloning steps compared with unmodified RSF1010. pAM5505 carrying the mobA gene was co-transformed with conjugal plasmid pRL443 to generate a new strain of conjugal E. coli, AM5501. This E. coli strain was used in conjugal matings with several cyanobacterial strains as described above (Figure 3A). With Anabaena PCC 7120, the addition of MobA in trans completely restored conjugation efficiency of all RSF1010 plasmid variants to wild-type levels (Figure 3B).
Figure 3.
Conjugation Efficiency of RSF1010-Based Plasmids into Three Cyanobacterial Strains, with MobA Supplied In trans
(A) Images of serially diluted 5-μL mating spots of Anabaena PCC 7120 (A7120) harboring one of five RSF1010 variants.
(B–D) Dot plots of colony counts from triplicate samples of mating plates for Anabaena PCC 7120 (B), Synechocystis PCC 6803 (S6803) (C), and Leptolyngbya BL0902 (BL0902) (D). Each strain was mated with E. coli conjugal donor strains containing the helper plasmid pAM5505 producing MobA in trans (tMobA) and cargo plasmid pAM5407, pAM5408, pAM5409, pAM5410, or pAM5411. Dots represent colony counts from each replicate sample; error bars show the mean ± SEM.
Mating experiments were also performed with Leptolyngbya BL0902 and Synechocystis PCC 6803. Interestingly, for Synechocystis PCC 6803 we again saw that the addition of the pUC origin of replication resulted in a strain-specific reduction in conjugation efficiency of one to two orders of magnitude (Figures 2C and 3C). Again, the effect on conjugation efficiency is much smaller than the restorative effect provided by supplying MobA in trans, such that pAM5410 (pUC-ori) and pAM5411 (pUC-ori, RK2-bom) still yield substantially higher conjugation efficiency when MobA is expressed from the pAM5505 helper plasmid. Leptolyngbya BL0902 conjugation efficiencies using the pAM5505 helper plasmid were similar to those of Anabaena PCC 7120, such that each RSF1010 variant produced conjugation efficiencies close to those of the wild-type RSF1010-based plasmid pAM5407 (Figure 3D). Altogether, we consistently observed a rescue of conjugative ability with the pAM5505 helper plasmid. Compared with adding the RK2-bom, the expression of MobA in trans from pAM5505 has the benefit of providing backward compatibility with previously generated pCVD047-derived plasmids. Any pCVD047-derived plasmid should benefit from conjugation with the pAM5505 helper plasmid without needing to make any modifications to the plasmid itself.
Conjugation of pAM5404 (RK2-bom) into Mixed Wild Cultures for Strain Isolation
One of the main objectives of this study was to increase the conjugation efficiency of the pCVD047 broad-host-range plasmid for its transfer into mixed cultures of wild cyanobacteria for the isolation of new genetically tractable strains. Successful conjugation and maintenance of the modified pAM5404 shuttle plasmid would be demonstrated by resistance to spectinomycin (Sp) and streptomycin (Sm) antibiotics and expression of the yellow fluorescent protein (YFP) reporter protein.
To test this approach, water samples were collected from several different freshwater locations in Southern California. Samples were inoculated into liquid BG-11 medium and grown under medium light levels at 30°C until they reached a high density to enrich for cyanobacteria. These mixed wild cultures were then pelleted and used for biparental mating with AM1359 E. coli carrying the pAM5404 shuttle plasmid. Then, 25 μL of the mixed cyanobacteria and E. coli donor cells were spotted onto BG-11 + 5% LB plates. After overnight incubation, spots were scraped from the plates and resuspended in BG-11 liquid medium. Serial dilutions were then spread onto BG-11 plates containing Sp and Sm to allow for greater distribution of cells to obtain isolated colonies. Numerous potential exconjugant cyanobacterial colonies with a blue-green color were formed on the conjugation plates. A total of 48 colonies that had formed 7–14 days after mating were picked for growth in liquid BG-11 medium containing Sp and Sm. The cultures were analyzed by microscopy to determine cell size and morphology and decide whether or not the selected colonies were likely to be exconjugant cyanobacteria versus green algae or diatoms. Twelve isolates containing cyanobacteria were selected for further isolation via serial dilution and streaking onto BG-11 plates containing Sp and Sm and the anti-fungal Benomyl to reduce growth of background fungal contamination.
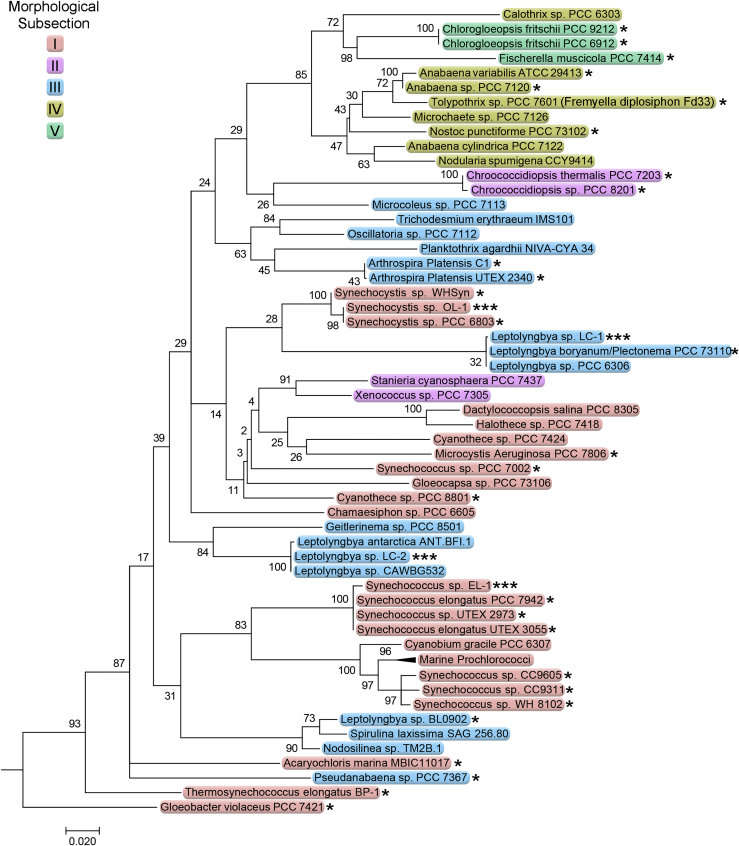
Most of the samples resulted in the isolation of multiple colonies of one morphological type of cyanobacteria, and a single representative of four different types were selected for further isolation and cultivation. Monoalgal strains were then examined by fluorescence microscopy, and the four selected strains expressed the YFP reporter (Figure 4). The 16S rRNA and internal transcribed spacer (ITS) region of each monoalgal strain were amplified by PCR using a set of cyanobacteria-specific PCR primers, as described (Taton et al., 2003). PCR products were sequenced, and the phylogenetic relationships between cyanobacterial strains capable of RSF1010-based vector replication were used to generate a phylogenetic tree (Figure 5). Cyanobacterial strains that have been previously reported to be genetically tractable are marked with an asterisk, and strains identified in this work are marked with three asterisks. Branches with no reported genetically tractable strains were removed or collapsed to condense the tree and highlight those strains capable of genetic manipulation. A complete list of the genetically tractable strains in Figure 5 is given in Table S1.
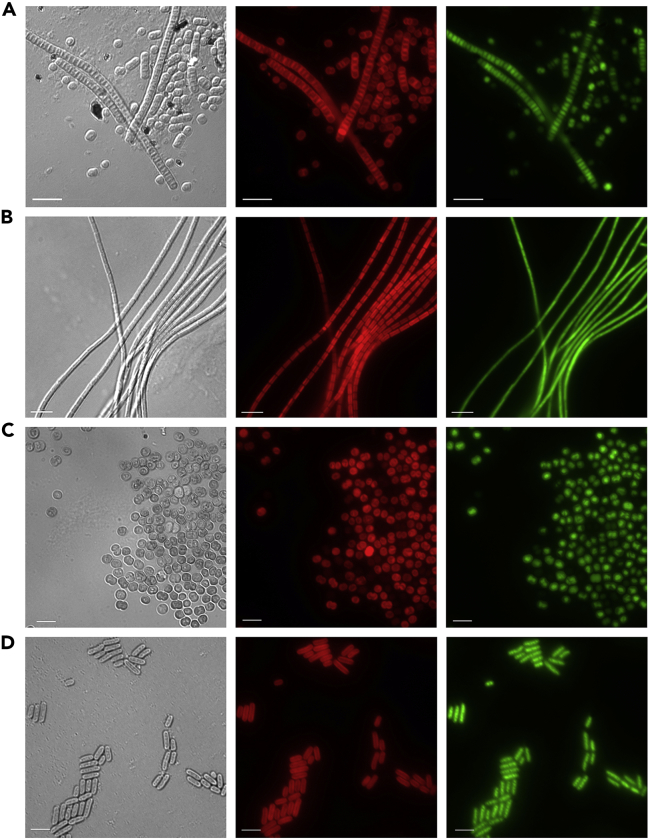
Figure 4.
Micrographs of Exconjugant Cyanobacterial Strains
Four strains exhibiting expression of the aadA gene for spectinomycin/streptomycin resistance and the oRBS-yfp gene for reporter fluorescence were isolated from wild mixed cultures after conjugation with pAM5404. For all images: left panel, white-light differential interference contrast image; middle panel, red autofluorescence of photosynthetic pigments; right panel; green YFP fluorescence. Strains are (A) LC-1, (B) LC-2, (C) OL-1, and (D) EL-1. Scale bars, 5 μm.
Figure 5.
Phylogenetic Analysis of 16S rRNA Gene Sequences by Maximum Likelihood
Evolutionary distances were inferred by using the maximum likelihood method based on the Kimura two-parameter model (Kimura, 1980). The tree with the highest log likelihood (−15732.33) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying neighbor joining and BIONJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood approach and then selecting the topology with superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter = 0.7089]). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 60.75% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 78 nucleotide sequences. It was then trimmed for size and to remove some branches with no genetically tractable strains and to collapse clades. There were a total of 1,281 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016). Cyanobacterial morphological subsections I–V are described in Bergey's Manual of Systematic Bacteriology Vol. 1 (Pierson and Castenholz, 2001). Single asterisks mark strains amenable to genetic manipulations (see also Table S1), and triple asterisks mark the four genetically tractable strains isolated by bioprospecting in this work.
Two filamentous strains, designated LC-1 and LC-2, were isolated from cultures sampled from Lake Cachuma, CA (Figures 4A and 4B). Strain LC-1 cells appeared as nearly round single cells to barrel-shaped cells in short- to medium-length unbranched filaments. Cells in long filaments had a shorter length than width with an average cell length of 1.5 ± 0.4 μm and average cell width of 2.3 ± 0.2 μm. Filaments varied in length, and short filaments or individual cells were easily detached from longer filaments. Cultures grew well on BG-11 agar plates under medium light and formed small colonies, but LC-1 did not grow well in shaking liquid BG-11 cultures. YFP expression varied between cells within each filament, which is potentially due to variations in plasmid copy number. BLASTn analysis of LC-1 16S rRNA gene sequence using the National Center for Biotechnological Information database determined its closest phylogenetic relative to be Leptolyngbya boryana PCC 6306 (Figure 5).
Strain LC-2 cells were in long filaments, with a rectangular cell morphology, and showed no constrictions between cells. Cells were longer than wide with an average cell length of 2.3 ± 0.6 μm and average cell width of 1.0 ± 0.1 μm. LC-2 cultures grew well in liquid and solid media under medium light. On BG-11 agar plates, filaments did not form discrete colonies but instead filaments radiate and spread to cover large areas. Variability of YFP expression was noted along filaments. Based on the LC-1 16S rRNA gene sequence, its closest relatives were identified as being in the polyphyletic genus Leptolyngbya, but in a different cluster than the LC-1 strain (Figure 5).
The same unicellular coccoid strain was isolated from multiple water sources in San Diego County, CA, with identical 16S sequences and cell morphology. One strain, designated OL-1, isolated from Otay Lake, was selected for characterization (Figure 4C). OL-1 has round or oval cell morphology, occurring as predominantly round single cells or elliptical cells in pairs. The typical cell was 1.9 ± .2 μm long and 1.6 ± .3 μm wide. Strain OL-1 grew well in liquid and solid BG-11 medium under medium light levels, and cells formed compact colonies when plated on BG-11 agar. BLASTn analysis of the 16S rRNA gene sequence placed OL-1 into the genus Synechocystis (Figure 5).
A strain, designated EL-1, was isolated from Eastlake, CA, and forms rod-shaped single cells (Figure 4D). Under some growth conditions, EL-1 formed pseudofilamentous cells that occasionally have an asymmetric division that produces a small cell at one end of the filament (not shown). Cells often form stacks of cells that line up side by side. Strain EL-1 grew well in liquid BG-11 medium under medium light levels, and cells formed compact colonies when plated on BG-11 agar. BLASTn analysis of the 16S rRNA gene sequence placed EL-1 in a cluster of the polyphyletic genus Synechococcus that includes S. elongatus PCC 7942 (Figure 5).
Plasmid Stability and Loss of pAM5409
The bioprospecting for genetically tractable strains reported here involved transfer of a modified RSF1010-based plasmid vector by conjugative mating with E. coli followed by selection for antibiotic-resistant cyanobacteria. The consequence of this is that all isolated wild strains contain the pAM5409 plasmid. To determine the stability of the pAM5409 plasmid in the wild strains as well as to obtain the original wild strain lacking the plasmid vector, we designed experiments to monitor loss of pAM5409. Of the wild strains, two were selected for testing plasmid stability when grown without the selective pressure of antibiotics. Strains Synechocystis OL-1 and Synechococcus EL-1 harboring pAM5409 were chosen because they formed compact colonies on plates, which facilitates genetic experiments, and showed robust growth in liquid media.
The OL-1 and EL-1 strains were grown continuously for 30 days with no antibiotics and subcultured into fresh medium about every 7 days when cultures neared stationary phase to ensure that cells were continually dividing. At day 30, samples were removed, serially diluted, and spread onto BG-11 plates to allow formation of individual colonies. Fifty-seven colonies of each strain were picked and patched onto BG-11 plates with and without antibiotics to test for loss of antibiotic resistance. Of the 57 OL-1 colonies, 2 had lost antibiotic resistance and failed to grow under antibiotic selection, but still grew on non-selective media. All the 57 EL-1 colonies grew under antibiotic selection, so liquid cultures of EL-1 were grown with further subculturing, and colony selection and antibiotic testing was done at 60, 90, and 120 days. At 60 and 90 days, 114 colonies were selected, and all grew under antibiotic selection. However, after 120 days of growth, 41 of 114 tested colonies had lost antibiotic resistance.
Cells from antibiotic-sensitive patches on plates were streaked onto fresh plates with and without antibiotic to confirm loss of antibiotic resistance, and new liquid cultures were grown from individual colonies. We further confirmed the lack of the pAM5409 plasmid in the antibiotic-sensitive cultures by PCR (Figure S1). These experiments showed that the plasmid was stably maintained in the majority of cells in a culture population even without antibiotic selection, and also that it is feasible to cure the strains of the exogenous plasmid. These cured wild-type strains can serve as platforms for genetic experiments using any RSF1010-derived plasmid vector. Overall, these experiments show the promise of bioprospecting for diverse cyanobacterial strains possessing a wide range of physiological characteristics that are genetically tractable for further experimentation and potential biotechnological applications.
Discussion
Bioprospecting harnesses the diversity of natural organisms as a source for useful strains and molecules. Many biotechnological applications of cyanobacteria require genetic engineering; therefore, it is necessary to identify strains that are amenable to genetic manipulation. In this work, we present a method for specifically isolating genetically tractable strains from environmental samples by performing conjugal mating of broad-host-range plasmids into mixed cultures enriched for cyanobacteria. Antibiotic selection followed by screening for YFP reporter expression was used for the isolation and identification of genetically tractable strains. The potential applications for this bioprospecting strategy are diverse.
The ability to readily identify genetically tractable strains would facilitate the development of new strains for basic research, investigations of natural products, metabolic engineering, and biomanufacturing. An increasing number of strains have been shown to be amenable to genetic manipulations in recent years (Figure 5). Cyanobacteria are a potent source of genetic diversity for the identification of novel bioactive compounds such as antimicrobial, anti-inflammatory, and anti-cancer agents (Brito et al., 2015, Montalvão et al., 2016). To harness this untapped potential, it is helpful to use genetic approaches to study and modify these organisms. Availability of efficient, easy-to-use broad-host-range plasmids such as those described in this work will make that process much easier. Our bioprospecting approach ensures that the identified strains can replicate a broad-host-range plasmid so that they can be genetically modified for the production of fuels, feeds, or secondary metabolites, as well as to probe novel endogenous biosynthetic pathways.
RSF1010-based plasmids have been used extensively for gene expression in a wide variety of bacteria, including strains of cyanobacteria (Sode et al., 1992, Mermet-Bouvier et al., 1993, Tolonen et al., 2006, Meyer, 2009). The innate transmissibility of RSF1010-based plasmids, although helpful for efficient conjugation, makes it difficult to use these plasmids with standard plasmid preparation and DNA-cloning techniques. Although impairing the nicking activity of the RSF1010 MobA protein facilitates cloning manipulations, it significantly reduces conjugation efficiency. We addressed this problem by editing the plasmid and conjugation system to restore conjugation efficiency. We present two methods of restoring plasmid transmissibility while retaining the ease of use for DNA preparation and recombinant DNA manipulations. One was to circumvent the necessity of the MobA protein by adding a secondary bom/oriT site for the initiation of transfer, allowing pAM5404 (RK2-bom) and pAM5406 (pUC-ori, RK2-bom) to take advantage of the RK2 conjugation system of the standard conjugal E. coli strains used for cyanobacterial genetics. The second method was to move a functional MobA protein onto a helper plasmid in the conjugal donor strain. Expressed in trans, MobA was capable of restoring pCVD047 conjugation efficiency to near-original levels. Limiting the functional mobilization machinery, by either method, to the conjugal donor strain used during the mating process means that the RSF1010-based plasmid vectors are protected from MobA's nicking activity during cloning experiments.
Addition of a second high copy number origin of replication, derived from pUC19, to the pCVD047 replicon donor fragment significantly increased the amount of plasmid DNA generated during plasmid preparation. This will be helpful for all cloning applications. However, there were strain-specific impacts on transmission or replication for plasmids containing this origin. Addition of the pUC19 origin does not seem to affect the number of antibiotic-resistant exconjugant colonies for RSF1010 variants in Leptolyngbya BL0902 or Anabaena PCC 7120, whereas there was a substantial decrease in Synechocystis PCC 6803. Indeed, compared with their counterpart plasmids with and without the RK2-bom site, shuttle plasmids pAM5410 (pUC-ori) and pAM5411 (pUC-ori, RK2-bom) demonstrated significantly lower conjugation efficiency. This decreased conjugation efficiency was seen both with and without MobA in trans. The immediate cause of this effect is unknown; however, we have a few speculations. The addition of the pUC19 origin might interfere with the native plasmid replication process in Synechocystis PCC 6803, or Synechocystis PCC 6803 may possess an innate defense mechanism against high copy number plasmids. Nevertheless, even with the decrease in conjugation efficiency, both addition of the RK2-bom site and supplying mobA in trans from a helper plasmid still restored transmissibility compared with the RSF1010 mobAY25F mutant in Synechocystis PCC 6803.
Cyanobacteria have been genetically modified to express fuel and plastic precursor molecules such as ethanol, isobutanol, isobutyraldehyde, alkanes, hydrogen, and free fatty acids (Schirmer et al., 2010, Ducat et al., 2011, Quintana et al., 2011, Hays and Ducat, 2015, Lee et al., 2017). Cyanobacteria and other microalgae will be valuable platforms for the production of renewable biomass, feedstock, and fertilizer. Traditional agriculture has benefitted from millennia of crop development, genetic manipulation through selective breeding, and more recently, direct genetic modification of agricultural staples such as corn and wheat. These strategies can be adapted to accelerate the process of domesticating algae and cyanobacteria and to engineer them for optimal performance. The broad-host-range-based genetic toolkit described in this work could be an integral part of a future where algae-based agriculture is an efficient and renewable source of a range of materials, nutritional products, and fuel compounds.
Limitations of the Study
For cyanobacterial bioprospecting, there were a few hurdles to overcome during the mating and strain isolation process. BG-11 medium supports the growth of eukaryotic algae, which led to high background levels of algae after conjugation. This may be overcome by treating cultures with eukaryotic-specific antibiotics such as cycloheximide or by raising the pH of growth media to enrich for cyanobacteria, which typically grow well at high pH. Wild cultures also tend to form microbial communities and biofilms. Many microorganisms survived antibiotic treatment, including heterotrophic bacteria, fungi, and eukaryotes, and often formed mats or clumps. Fluorescence microscopy showed that some bacteria in these communities survived antibiotic selection, but did not express the YFP reporter. It is possible that these communities protect sensitive organisms from the antibiotic and that procedures or culture conditions that break up the communities would improve the differential selection of genetically tractable strains.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We would like to thank Kevin Trieu and Neli Razavi for assistance with laboratory research. This work was supported by the United States Department of Energy, Office of Energy Efficiency and Renewable Energy, under Contract Number EE0008246 and the National Institute of General Medical Sciences of the United States National Institutes of Health under Award Number R01GM118815. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author Contributions
Conceptualization, B.B., J.W.G., and A.T.; Methodology, B.B.; Investigation, B.B.; Formal Analysis, B.B. and A.T. Writing – Original Draft, B.B.; Writing – Review & Editing, J.W.G. and A.T.; Resources, J.W.G.; Visualization, B.B. and A.T.; Supervision, J.W.G.
Declaration of Interests
The authors declare no competing interests.
Published: October 25, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.09.002.
Supplemental Information
References
- Babic A., Guérout A.M., Mazel D. Construction of an improved RP4 (RK2)-based conjugative system. Res. Microbiol. 2008;159:545–549. doi: 10.1016/j.resmic.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Bekker A., Holland H.D., Wang P.L., Rumble D., Stein H.J., Hannah J.L., Coetzee L.L., Beukes N.J. Dating the rise of atmospheric oxygen. Nature. 2004;427:117–120. doi: 10.1038/nature02260. [DOI] [PubMed] [Google Scholar]
- Brito Â., Gaifem J., Ramos V., Glukhov E., Dorrestein P.C., Gerwick W.H., Vasconcelos V.M., Mendes M.V., Tamagnini P. Bioprospecting Portuguese Atlantic coast cyanobacteria for bioactive secondary metabolites reveals untapped chemodiversity. Algal Res. 2015;9:218–226. [Google Scholar]
- Dismukes G.C., Carrieri D., Bennette N., Ananyev G.M., Posewitz M.C. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 2008;19:235–240. doi: 10.1016/j.copbio.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Ducat D.C., Way J.C., Silver P.A. Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 2011;29:95–103. doi: 10.1016/j.tibtech.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Elhai J., Vepritskiy A., Muro-Pastor A.M., Flores E., Wolk C.P. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 1997;179:1998–2005. doi: 10.1128/jb.179.6.1998-2005.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey J., Bagdasarian M.M., Bagdasarian M. Replication and copy number control of the broad-host-range plasmid RSF1010. Gene. 1992;113:101–106. doi: 10.1016/0378-1119(92)90675-f. [DOI] [PubMed] [Google Scholar]
- Gormley E.P., Davies J. Transfer of plasmid RSF1010 by conjugation from Escherichia coli to Streptomyces lividans and Mycobacterium smegmatis. J. Bacteriol. 1991;173:6705–6708. doi: 10.1128/jb.173.21.6705-6708.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays S.G., Ducat D.C. Engineering cyanobacteria as photosynthetic feedstock factories. Photosynth. Res. 2015;123:285–295. doi: 10.1007/s11120-014-9980-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidorn T., Camsund D., Huang H.H., Lindberg P., Oliveira P., Stensjö K., Lindblad P. Synthetic biology in cyanobacteria: engineering and analyzing novel functions. Methods Enzymol. 2011;497:539–579. doi: 10.1016/B978-0-12-385075-1.00024-X. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7. 0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.J., Choi J., Lee S.-M., Um Y., Sim S.J., Kim Y., Woo H.M. Photosynthetic CO 2 conversion to fatty acid ethyl esters (FAEEs) using engineered cyanobacteria. J. Agric. Food Chem. 2017;65:1087–1092. doi: 10.1021/acs.jafc.7b00002. [DOI] [PubMed] [Google Scholar]
- Martin W., Kowallik K. Annotated English translation of Mereschkowsky’s 1905 paper ‘Über natur und ursprung der chromatophoren impflanzenreiche’. Eur. J. Phycol. 1999;34:287–295. [Google Scholar]
- Mermet-Bouvier P., Cassier-Chauvat C., Marraccini P., Chauvat F. Transfer and replication of RSF1010-derived plasmids in several cyanobacteria of the genera Synechocystis and Synechococcus. Curr. Microbiol. 1993;27:323–327. [Google Scholar]
- Meyer R. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid. 2009;62:57–70. doi: 10.1016/j.plasmid.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalvão S., Demirel Z., Devi P., Lombardi V., Hongisto V., Perälä M., Hattara J., Imamoglu E., Tilvi S.S., Turan G. Large-scale bioprospecting of cyanobacteria, micro- and macroalgae from the Aegean Sea. New Biotechnol. 2016;33:399–406. doi: 10.1016/j.nbt.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Monzingo A.F., Ozburn A., Xia S., Meyer R.J., Robertus J.D. The structure of the minimal relaxase domain of MobA at 2.1 Å resolution. J. Mol. Biol. 2007;366:165–178. doi: 10.1016/j.jmb.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olguín E.J. Dual purpose microalgae-bacteria-based systems that treat wastewater and produce biodiesel and chemical products within a biorefinery. Biotechnol. Adv. 2012;30:1031–1046. doi: 10.1016/j.biotechadv.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Pierson B.K., Castenholz R.W. The Archaea and the deeply branching and phototrophic bacteria. In: Boone D.R., Castenholz R.W., editors. Volume 1. Springer-Verlag; 2001. pp. 474–486. (Bergey’s Manual of Systematic Bacteriology). [Google Scholar]
- Quintana N., Van Der Kooy F., Van De Rhee M.D., Voshol G.P., Verpoorte R. Renewable energy from Cyanobacteria: energy production optimization by metabolic pathway engineering. Appl. Microbiol. Biotechnol. 2011;91:471–490. doi: 10.1007/s00253-011-3394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat I., Ranjith Kumar R., Mutanda T., Bux F. Dual role of microalgae: phycoremediation of domestic wastewater and biomass production for sustainable biofuels production. Appl. Energy. 2011;88:3411–3424. [Google Scholar]
- Rubin B.E., Wetmore K.M., Price M.N., Diamond S., Shultzaberger R.K., Lowe L.C., Curtin G., Arkin A.P., Deutschbauer A., Golden S.S. The essential gene set of a photosynthetic organism. Proc. Natl. Acad. Sci. U S A. 2015;112:E6634–E6643. doi: 10.1073/pnas.1519220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E., Haring V., Lurz R., Otto S. Plasmid RSF1010 DNA replication in vitro promoted by purified RSF1010 RepA, RepB and RepC proteins. Nucleic Acids Res. 1991;19:1203–1211. doi: 10.1093/nar/19.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer A., Rude M.A., Li X., Popova E., del Cardayre S.B. Microbial biosynthesis of alkanes. Science. 2010;329:559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- Schirrmeister B.E., Gugger M., Donoghue P.C.J. Cyanobacteria and the Great oxidation event: evidence from genes and fossils. Palaeontology. 2015;58:769–785. doi: 10.1111/pala.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih P.M., Wu D., Latifi A., Axen S.D., Fewer D.P., Talla E., Calteau A., Cai F., Tandeau de Marsac N., Rippka R. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U S A. 2013;110:1053–1058. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sode K., Tatara M., Takeyama H., Burgess J.G., Matsunaga T. Conjugative gene transfer in marine cyanobacteria: Synechococcus sp., Synechocystis sp., and Pseudanabaena sp. Appl. Microbiol. Biotechnol. 1992;37:369–373. doi: 10.1007/BF00210994. [DOI] [PubMed] [Google Scholar]
- Taton A., Grubisic S., Brambilla E., De Wit R., Wilmotte A. Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): a morphological and molecular approach. Appl. Environ. Microbiol. 2003;69:5157–5169. doi: 10.1128/AEM.69.9.5157-5169.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taton A., Unglaub F., Wright N.E., Zeng W.Y., Paz-Yepes J., Brahamsha B., Palenik B., Peterson T.C., Haerizadeh F., Golden S.S. Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res. 2014;42:1–16. doi: 10.1093/nar/gku673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolonen A.C., Liszt G.B., Hess W.R. Genetic manipulation of Prochlorococcus strain MIT9313: green fluorescent protein expression from an RSF1010 plasmid and Tn5 transposition. Appl. Environ. Microbiol. 2006;72:7607–7613. doi: 10.1128/AEM.02034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Carlier C., Martin P., Courvalin P. Plasmid transfer by conjugation from Escherichia coli to Gram-positive bacteria. FEMS Microbiol. Lett. 1987;48:289–294. [Google Scholar]
- Wendt K.E., Pakrasi H.B. Genomics approaches to deciphering natural transformation in cyanobacteria. Front. Microbiol. 2019;10:1–7. doi: 10.3389/fmicb.2019.01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.G., Long S.P., Ort D.R. What is the maximum efficiency with which photosynthesis can convert solar energy into biomass? Curr. Opin. Biotechnol. 2008;19:153–159. doi: 10.1016/j.copbio.2008.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.