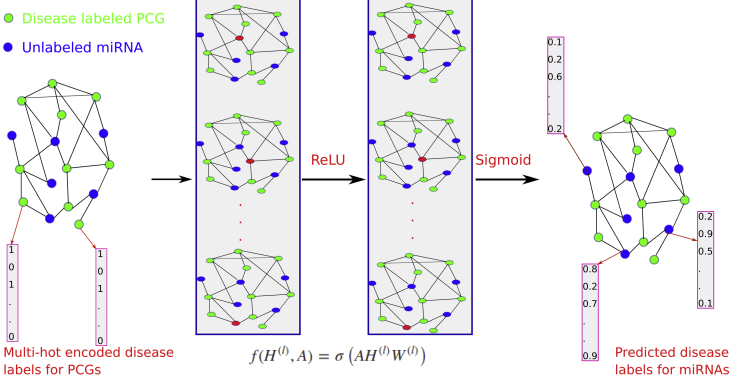
Figure 1.
The Flowchart of DimiG with Two-Layer GCN
Each node (gene) is represented as a vector of expression values across tissues with its sum across tissues from GTEx, and the network is constructed from PCG-PCG and PCG-miRNA interactions. When doing forward propagation, the embedding of the red node in each network is the weighted sum of the embedding of its neighbors, where all nodes in the network are updated simultaneously. The label is a multi-hot vector indicating the presence of diseases. In the end, DimiG can infer the probability between diseases and unlabeled miRNAs.