Summary
Circular RNAs (circRNAs), an important class of regulatory RNAs, have been shown to be the most prevalent in the brain compared with other tissues. However the processes governing their biogenesis in neurons are still elusive. Moreover, little is known about whether and how different biogenesis factors work in synchrony to generate neuronal circRNAs. To address this question, we pharmacologically inhibited the spliceosome and profiled rat neuronal circRNAs using RNA sequencing. We identified over 100 circRNAs that were up-regulated and a few circRNAs that were down-regulated upon spliceosome inhibition. Bioinformatic analysis revealed that up-regulated circRNAs possess significantly longer flanking introns compared with the un-changed circRNA population. Moreover, the flanking introns of up-regulated circRNAs harbor a higher number of distinct repeat sequences and more reverse complementary motifs compared with the unchanged circRNAs. Taken together, our data demonstrate that the biogenesis of circRNAs containing distinct intronic features becomes favored under conditions of limited spliceosome activity.
Subject Areas: Biological Sciences, Molecular Biology, Cell Biology, Omics
Graphical Abstract
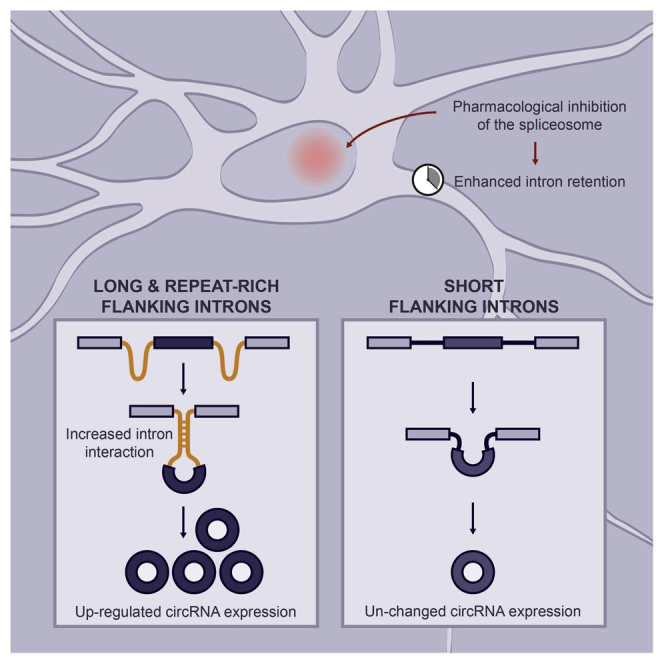
Highlights
-
•
RNA-seq reveals changes of the neuronal circRNA landscape after spliceosome inhibition
-
•
Hundreds of circRNAs are up-regulated under conditions of reduced spliceosome activity
-
•
Long and repeat-rich flanking introns facilitate up-regulation of cognate circRNAs
Biological Sciences; Molecular Biology; Cell Biology; Omics
Introduction
Covalently closed circular RNA molecules (circRNAs) have reemerged as an important class of regulatory RNA. Originally viewed as aberrant splicing by-products with little functional potential (Cocquerelle et al., 1993, Nigro et al., 1991, Pasman et al., 1996), recent studies have uncovered their widespread expression (Barrett et al., 2015, Hansen et al., 2013, Ivanov et al., 2015, Jeck et al., 2013, Memczak et al., 2013, Salzman et al., 2012, Wang et al., 2014, Westholm et al., 2014). Many circRNAs are stable, highly conserved between species (Cocquerelle et al., 1993, Guo et al., 2014, Jeck et al., 2013, Rybak-Wolf et al., 2015, Veno et al., 2015), and show distinct tissue- and cell-type-specific expression (Memczak et al., 2013, Salzman et al., 2013).
CircRNAs are the most abundant in the brain, where a distinct expression pattern is found across different brain regions (Rybak-Wolf et al., 2015, Westholm et al., 2014, You et al., 2015). Several observations suggest a regulatory role of neuronal circRNAs for synaptic response and functions: circRNAs are highly enriched at synapses and often derived from genes with neuronal and synaptic function (Rybak-Wolf et al., 2015, You et al., 2015). The expression of circRNAs is regulated throughout neuronal development (Rybak-Wolf et al., 2015, Westholm et al., 2014), with an increase coinciding with the onset of synaptogenesis in hippocampus (You et al., 2015). Moreover, a dynamic change in neuronal circRNA expression is observed upon induction of homeostatic plasticity (You et al., 2015). Importantly, these findings also highlight that circRNA biogenesis must be under tight control in the brain. However, the mechanisms of neuronal circRNA biogenesis and the conditions under which neuronal circRNA formation is favored or suppressed are still elusive. Although the spliceosome has been implicated as a critical element of circRNA biogenesis (Ashwal-Fluss et al., 2014, Liang et al., 2017, Liang and Wilusz, 2014, Starke et al., 2015, Stegeman et al., 2018), alongside other features such as the presence of complementary sequences in circRNA-flanking introns (Dong et al., 2017, Ivanov et al., 2015, Kramer et al., 2015, Liang and Wilusz, 2014, Zhang et al., 2014) and RNA-binding proteins (Ashwal-Fluss et al., 2014, Conn et al., 2015, Errichelli et al., 2017, Ivanov et al., 2015, Kristensen et al., 2018, Li et al., 2017, Liang et al., 2017), studies performed on primary neurons are missing. Moreover, the interplay between circRNA biogenesis factors and their impact on the circRNA population as a whole has not yet been addressed.
We address this issue here using pharmacological inhibition of the spliceosome and RNA sequencing (RNA-seq) in rat primary hippocampal neurons. We observed a shift in the circRNA landscape following spliceosome inhibition, with a large population of circRNAs becoming up-regulated in their expression. We analyzed the up-regulated population of circRNAs and identified several features that could contribute to their preferential formation under reduced spliceosome activity. Our findings shed new light on the mechanisms generating neuronal circRNAs and point to a complex, multilayered regulation of circRNA biogenesis.
Results
Profiling circRNA after Spliceosome Inhibition in Rat Hippocampal Neurons
To investigate how splicing affects neuronal circRNA biogenesis, we inhibited spliceosome assembly in primary rat hippocampal neurons using Isoginkgetin, a pre-mRNA splicing inhibitor (Figure 1A) (O'Brien et al., 2008). Neurons were incubated with 33μM Isoginkgetin for 24 h, a treatment that resulted in the most effective spliceosome inhibition (Figure S1A). Control neurons were treated with DMSO for 24 h, and rRNA-depleted total RNA was then deep sequenced (see Transparent Methods). From three biological replicates for each condition, we obtained a minimum of 22 million and a maximum of 79 million reads, with the mappable reads ranging from 91.7% to 96.1% (Table S1). The replicates of the same conditions correlated well with one another (Figure S1B).
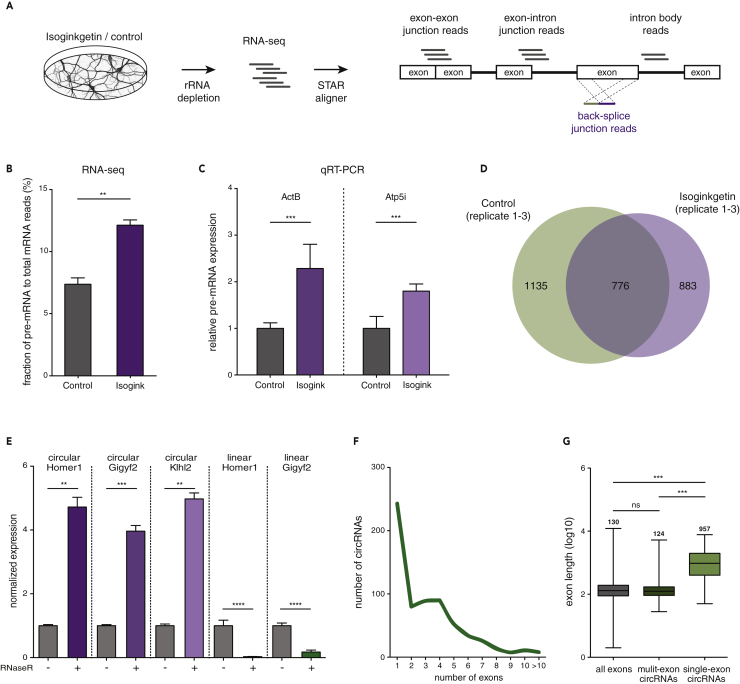
Figure 1.
Profiling circRNA after Spliceosome Inhibition in Primary Rat Hippocampal Neurons
(A) Experiment and analysis pipeline.
(B) The abundance of pre-mRNA in a total mRNA sample was quantified after Isoginkgetin (purple) or control (gray) treatment. Assessed by RNA-seq, Isoginkgetin treatment led to a significant increase in the pre-mRNA population (**p < 0.01, unpaired t test, two-tailed, n = 3).
(C) Relative pre-mRNA expression for two candidate ActB and ATP5I transcripts under control and Isoginkgetin treatment conditions assessed with qRT-PCR (***p < 0.001, unpaired t test, two-tailed, n = 3).
(D) Number of circRNAs identified in all replicates in the Isoginkgetin and control conditions.
(E) Validation of circularity using RNase R treatment followed by qRT-PCR. CircRNAs increased in expression, whereas linear transcripts were depleted after RNase R incubation (****p < 0.0001, ***p < 0.001, **p < 0.01, unpaired t test, two tailed, n = 4).
(F) Numbers of exons that comprise the population of 776 circRNAs.
(G) Length distribution of back-spliced exons. A median length of 957 and 124 bp was found for single- and multi-exon circRNAs, respectively (***p < 0.0001, Kruskal-Wallis test followed by Dunn's multiple comparisons test). Error bars represent SD (B, C, and E). Box-whisker plots show minimum, first quartile, median, third quartile, and maximum of a set of data (G).
We first validated the efficiency of spliceosome inhibition by Isogingketin in primary neurons using two independent methods. As a readout of spliceosome inhibition, we assessed the percentage of unspliced pre-mRNA in the total mRNA population. For this we determined the relative abundance of exon-intron junction reads (pre-mRNA) to the sum of exon-exon junction reads (mature mRNA) and exon-intron junction reads (pre-mRNA). As reads that map to the body of exons cannot be clearly assigned either to pre- or mature mRNA they were not considered. We found a significant 73.5% increase in pre-mRNA expression after Isoginkgetin treatment (6.8% pre-mRNA in control condition versus 11.8% pre-mRNA in Isoginkgetin, Figure 1B). Second, using qRT-PCR we validated the pre-mRNA expression of candidate transcripts using primers amplifying an exon-intron junction (see Transparent Methods). For ActB (β-actin) and ATP5l (ATP synthase membrane subunit E), we observed a 2.3- and 1.8-fold increase in pre-mRNA levels after Isoginkgetin treatment, respectively (Figure 1C).
It has previously been found that Isoginkgetin can also partially inhibit transcription (Boswell et al., 2017). Therefore, we investigated the expression of total mRNA, pre-mRNA, mature mRNA, and introns in Isoginkgetin versus control condition by using exon-exon reads, exon-intron junction reads, and intron-body reads, respectively, for differential expression analysis (see Transparent Methods). We identified 11,052 Isoginkgetin-enriched and 10,166 control-enriched transcript features (Figure S1C). Although the total mRNA expression showed minor differences between Isoginkgetin and control (median logFC [fold change] of −0.16), we observed a larger reduction in mature mRNA levels (median logFC of −0.33) and the strongest difference in pre-mRNA and intron levels (median logFC of 0.62 and 0.92) after spliceosome inhibition (Figure S1D), indicating that Isoginkgetin treatment primarily affects splicing rather than transcription. Together, these results validate the use of Isoginkgetin as a tool to reduce spliceosome activity in primary neurons.
To identify and quantify circRNA expression, we developed a pipeline to identify reads that map to the back-splice junction and are unique for circRNAs. For this, we used the detection of chimeric alignments in STAR, a high-performance aligner for RNA-sequencing data (Dobin et al., 2013). A chimeric alignment consists of two “segments” that map to the genome in a non-canonical order. We searched for back-splice junction reads by filtering all chimeric alignments for those wherein both segments originate from the same chromosome and strand and the 5′ segment aligns downstream of the 3′ segment. We furthermore required that the segments possess splice junctions and be at least 20 bp long. These reads were then realigned to the rat genome and, when originating from the same gene, were used to determine the position of the back-splice junction and quantified. CircRNAs that mapped with at least one unique back-splice junction read and were consistently found in all replicates and conditions were kept. Using this pipeline we identified on average 13,917 neuronal circRNAs in each sample (Table S1). Of these, 1,659 and 1,911 circRNAs were detected in all Isoginkgetin-treated and control samples, respectively. A total of 776 circRNAs overlapped between the Isoginkgetin and control conditions (Figure 1D, Data S1). We verified the circularity of three candidate circRNAs using RNase R treatment followed by qRT-PCR. Primers amplifying the back-splice junction were used to quantify circRNA levels. For the quantification of the parent mRNA transcript, we used a primer set targeting the exon-exon junctions up- or downstream of the exons involved in the back-splice junction formation (see Transparent Methods). As expected, all tested circRNAs showed a strong enrichment after RNase R treatment, whereas the linear parent transcripts were depleted (Figure 1E).
The group of 776 robustly expressed circRNAs was used for subsequent analyses. We investigated the exon distribution of the identified circRNAs and found that 37.2% of them were generated by the circularization of a single exon (Figure 1F). In addition, we found that exons of “single-exon” circRNAs were significantly longer (median of 957 bp) compared with the average rat exon or exons from circRNAs formed by several exons (“multi-exon” circRNAs; median of 124 bp) (Figure 1G). Taken together, we successfully inhibited the spliceosome in primary rat hippocampal neurons and identified hundreds of neuronal circRNAs, many of which are generated by a single, long exon.
Neuronal circRNAs Are Up- and Down-regulated after Spliceosome Inhibition
We next asked whether and how neuronal circRNA and parent mRNA expression are affected by reduced spliceosome activity. Using the back-splice junction reads of the 776 robustly expressed circRNAs and the exon-exon junction reads of their parent mRNAs, we performed a differential expression analysis in Isoginkgetin versus control condition (see Transparent Methods). Looking at the FC differences, we observed a general increase in the expression of circRNAs after spliceosome inhibition compared with their parent mRNAs (Figure 2A). A total of 142 circRNAs were significantly altered between Isoginkgetin and control conditions. The majority of circRNAs was unchanged after spliceosome inhibition, whereas we observed 134 up-regulated circRNAs. Interestingly, we also identified few circRNAs that showed significantly reduced expression after Isoginkgetin treatment (Figures 2A and S2A, Data S1). Our observation indicates that reduced spliceosome activity favors the production of a large subset of neuronal circRNAs, whereas in a small minority of cases it can also inhibit circRNA processing. For almost all the up-regulated circRNAs, the parent mRNA expression was either unchanged or decreased, suggesting that an increase in circRNA production can occur at the cost of processing the parent mRNA. Interestingly, in 11 cases both the circRNA and mRNA expression levels were up-regulated (Figures 2A and S2A, Data S1). The fraction of multi-exon circRNAs was higher in the regulated group compared with the unchanged circRNA population (Figure S2B), whereas there was no significant difference in exon length between regulated and unchanged circRNAs (Figure S2C).
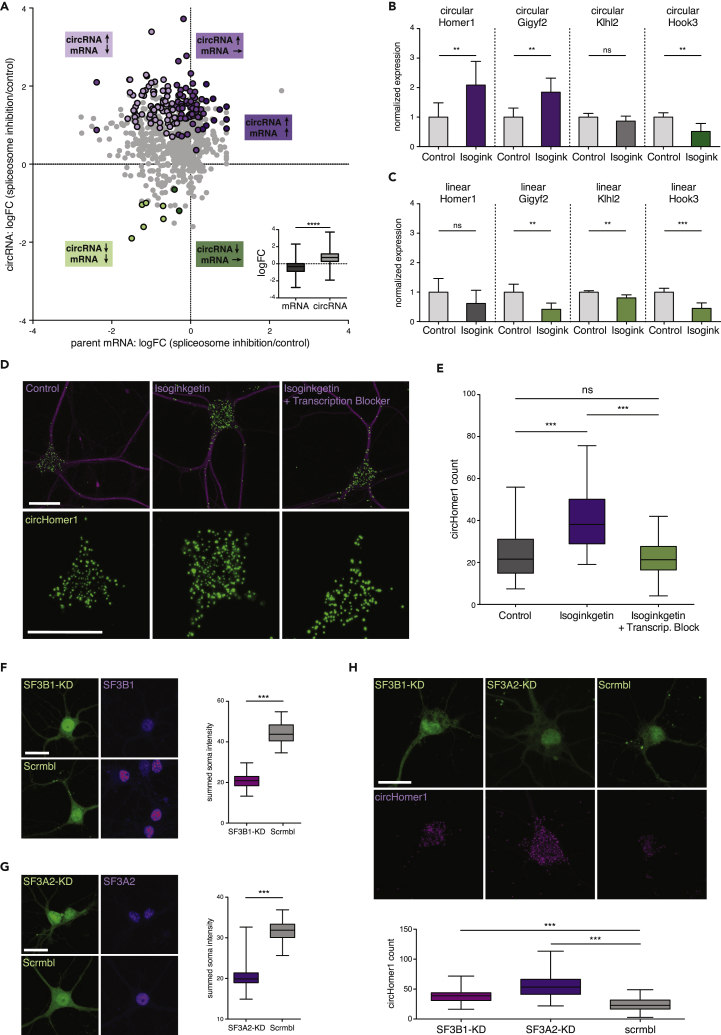
Figure 2.
Spliceosome Inhibition Leads to Expression Changes of circRNAs and Their Parent mRNAs
(A) Scatterplot showing the log2 fold change (FC) of 776 robustly expressed circRNA (y axis) and their parent mRNA (x axis) between Isoginkgetin and control conditions. Magenta dots indicate circRNAs that are significantly up-regulated in Isoginkgetin. The shade of magenta (from light to dark) indicates whether the parent mRNA was down-regulated, unchanged, or up-regulated, respectively. Green dots represent significantly down-regulated circRNAs, with the parent mRNA being also down-regulated (light green dots) or unchanged (dark green dots). Gray dots indicate circRNAs that are unchanged. Inset shows that the general abundance of circRNAs was higher than that of the parent mRNA after Isoginkgetin treatment (****p < 0.0001, unpaired t test, two-tailed).
(B) qRT-PCR validation of circHomer1, circGigyf2, circKlhl2, and circHook3 expression after Isogingketin treatment (**p < 0.01, unpaired t test, two-tailed, n = 3).
(C) qRT-PCR validation of parent Homer1, Gigyf2, Klhl2, and Hook3 mRNA expression (***p < 0.001, **p < 0.01, unpaired t test, two-tailed, n = 3).
(D) Validation of circRNA expression changes using high-resolution in situ hybridization for circHomer1 in control, Isoginkgetin, and Isoginkgetin and transcription-inhibitor treated neurons. Neuronal somata and dendrites were identified using anti-MAP2 immunostaining. Scale bar, 25 μm.
(E) Somatic circHomer1 expression was significantly up-regulated in Isoginkgetin-treated neurons compared with control. The inhibition of transcription prevented the Isoginkgetin-induced up-regulation of circHomer1 (***p < 0.0001, Kruskal-Wallis test followed by Dunn's multiple comparisons test, n = 73, 76, and 63).
(F and G) CircHomer1 expression after RNAi-mediated depletion of (F) SF3B1, (G) SF3A2, or scrambled control. The expression of SF3B1 or SF3A2 (magenta) in positively transfected cells (green) was validated after 4 days (***p < 0.0001, Mann-Whitney U test, n = 46, 39, 52, and 25). Scale bar, 25 μm.
(H) CircHomer1 fluorescence in situ hybridization after SF3B1 or SF3A2 depletion. Somatic circHomer1 was significantly up-regulated in SF3B1 and SF3A2 knockdown cells compared with scrambled control (***p < 0.0001, Mann-Whitney U test, n = 37, 63, and 57). Scale bar, 25 μm. Error bars represent SD. (B and C). Boxplot whiskers show minimum, first quartile, median, third quartile, and maximum (A, E, F, G, and H).
To validate the expression changes of circRNAs and parent mRNAs after Isoginkgetin treatment we performed qRT-PCR on four candidates (Figure S2D). Consistent with our RNA-seq data, we observed for previously reported circHomer1, a circRNA derived from the synaptic scaffolding molecule Homer1 transcript (Brakeman et al., 1997), and circGigyf2, a circRNA derived from a tyrosine kinase receptor signaling regulator (Giovannone et al., 2009), a 2.1- and 1.8-fold increased expression after Isoginkgetin treatment, respectively, whereas the parent mRNA level was either unchanged or down-regulated (Figures 2B and 2C). In addition, we confirmed the down-regulation of circHook3 and the unchanged expression of circKlhl2 using qRT-PCR (Figure 2B). Furthermore, qRT-PCR investigation of expression changes of the linear variants of our candidate transcripts also matched the results of the RNA-seq data (Figure 2C). Taken together, qRT-PCR validation experiments yielded results highly consistent with our RNA-seq results for both circRNA and parent mRNA transcripts, strengthening the validity of our sequencing approach.
In addition, we directly visualized circRNAs in cultured hippocampal neurons using high-resolution in situ hybridization optimized for the detection of circRNAs (Figures 2D and S2E; You et al., 2015). We observed circHomer1 particles in both the cell body and dendrite of neurons (Figure 2D). Consistent with our RNA-seq and qRT-PCR data, the number of circHomer1 particles in the cell body increased significantly after Isoginkgetin treatment. Interestingly, when we treated the neurons simultaneously with Isoginkgetin and a cocktail of transcription inhibitors (see Transparent Methods), circHomer1 expression remained at levels similar to that observed in control conditions (Figures 2D and 2E). This suggests that spliceosome inhibition triggers the generation of newly synthesized circHomer1.
Next, we asked whether the depletion of core spliceosome components would have a similar effect on circRNA generation as pharmacological inhibition of the spliceosome. Therefore, we knocked down SF3B1 and SF3A2, two components of the U2 snRNP (Will and Luhrmann, 2011), and investigated the expression of circHomer1. We observed a 52% reduction of SF3B1 and a 38% reduction of SF3A2 expression in knockdown cells compared with the scrambled control (Figures 2F and 2G). In agreement with the results obtained after Isoginkgetin treatment, depletion of SF3B1 or SF3A2 led to a significant increase in circHomer1 levels (Figure 2H).
Long Flanking Introns with Repeat Sequences Facilitate circRNA Formation under Spliceosome Inhibition
As spliceosome inhibition increased the generation of certain circRNAs but not others, we wondered which features of circRNA transcripts may have facilitated their expression. To address this, we investigated the characteristics of the introns flanking circularizing exons, as they have been shown to influence circRNA generation (Jeck et al., 2013, Westholm et al., 2014). First, we compared the length of all rat introns, introns flanking circularizing exons, and the remaining circRNA introns that are not flanking the back-splice junction (henceforth referred to as circRNA middle intron). We found that the introns flanking circularizing exons are significantly longer than all rat introns and circRNA middle introns (Figures 3A and S3A). No difference in intron length distribution was observed between all neuronal circRNAs and those that were unchanged or down-regulated after the Isoginkgetin treatment. Intriguingly, we found that circRNAs whose expression was facilitated by reduced spliceosome activity possessed the longest flanking introns (Figure 3A), with the up- and downstream flanking introns sharing a similar length (Figure S3B). Our results suggest that these long introns confer an increased probability of circRNA formation under conditions of reduced spliceosome activity.
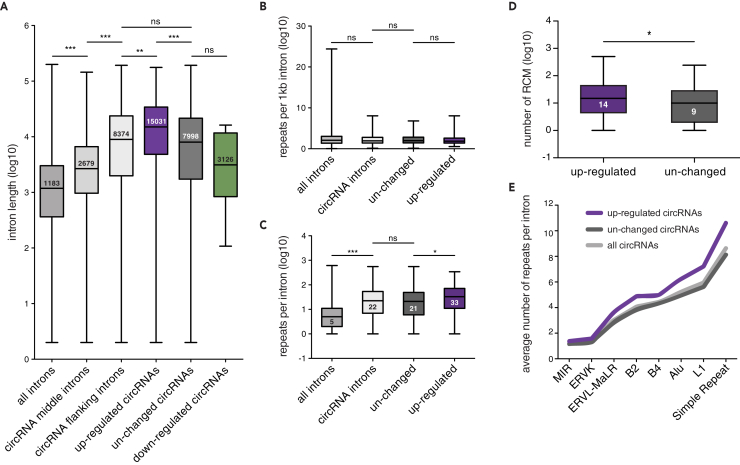
Figure 3.
Distinct Intronic Features Facilitate circRNA Formation under Conditions of Reduced Spliceosome Activity
(A) Length distribution of introns. Introns flanking back-spliced exons are significantly longer than the median rat intron and circRNA middle introns (***p < 0.0001, Kruskal-Wallis test followed by Dunn's multiple comparisons test). Flanking introns of up-regulated circRNAs are significantly longer than flanking introns of unchanged circRNAs and all circRNAs (***p < 0.0001, **p < 0.01, Kruskal-Wallis test followed by Dunn's multiple comparisons test).
(B) Frequency of repeat sequence occurrence per 1 kb. No difference is observed between all introns, all circRNA-flanking introns, flanking introns of unchanged circRNAs, and up-regulated circRNAs (ns > 0.05, Kruskal-Wallis test followed by Dunn's multiple comparisons test).
(C) Number of repeat sequences detected per flanking intron. Flanking introns of circRNAs harbor more repeats than the average rat intron. Up-regulated circRNAs contain significantly more repeat sequences than the flanking introns of unchanged circRNAs (***p < 0.0001, *p < 0.05, Kruskal-Wallis test followed by Dunn's multiple comparisons test).
(D) Number of Reverse complementary motifs (RCM) detected in flanking intron pairs. Flanking introns of up-regulated circRNAs possess significantly more RCMs compared with unchanged circRNAs (*p < 0.05, Mann Whitney U test).
(E) Comparison of the average number of repeats observed in the flanking intron. Distinct repeat families (x axis) are more abundant in flanking introns of up-regulated circRNAs compared with unchanged circRNAs, whereas the number of repeat families observed between all circRNAs and the unchanged population is similar. Box-whisker plots show minimum, first quartile, median, third quartile, and maximum of a set of data (A–D).
There are two possible explanations for why circRNAs with very long flanking introns become up-regulated. First, it could be that long introns are more effectively retained than short introns after Isoginkgetin treatment. Second, the long introns could contain certain features in their sequences that promote circularization. To test the first hypothesis, we determined the differential expression of each detected intron between Isoginkgetin and control condition (see Transparent Methods). We observed 9,667 up-regulated introns after spliceosome inhibition (Figure S3C), with the Isoginkgetin-enriched introns being slightly shifted toward the first intron (Figure S3D). However, we found that intron length does not influence the strength of intron retention (Figure S3E). The fact that long introns are not preferentially retained suggests that information contained in the intron sequences is responsible for enhanced circRNA formation.
We therefore investigated the features of the flanking introns of up-regulated circRNAs. Specifically, we examined the presence and frequency of repeat sequences, as well as their complementarity, which can bring splice sites in close proximity and thus facilitate circRNA formation (Jeck et al., 2013, Liang and Wilusz, 2014, Zhang et al., 2014). The frequency of repeat sequences observed per 1 kb of flanking intron was similar between all circRNAs, unchanged circRNAs, and up-regulated circRNAs (Figure 3B). However, flanking introns of up-regulated circRNAs possessed overall more repeat sequences per intron (Figure 3C), most likely due to their increased length, with the up- and downstream flanking introns harboring a similar number of repeats (Figure S3F). Moreover, intron pairs of up-regulated circRNAs carried a significantly higher number of reverse complementary motifs when compared with intron pairs of unchanged circRNAs (Figure 3D). Interestingly, we found several repeat families to be enriched in the flanking introns of up-regulated circRNAs compared with the unchanged circRNA population. Among them, the number of simple repeats, L1, and Alu repeats were increased the strongest (Figure 3E), with 63.8% of the Alu repeats found in intron pairs arranged in a complementing orientation (Figure S3G). When we looked into the structural properties of a subset of flanking introns (see Transparent Methods), introns of unchanged circRNAs showed a slightly lower minimum free energy normalized to intron length (norm.MFE) than up-regulated circRNAs that were tested (Figure S3H). This result indicated that unchanged circRNAs possessed a more stable secondary structure in their flanking introns than up-regulated circRNAs.
Our data suggest that reduced spliceosome activity prolongs the presence of introns, enabling long and repeat-rich flanking introns to interact with one another, thus facilitating the biogenesis of circRNAs. An increase in intron presence could be also achieved by increasing the number of transcripts produced. Therefore, we over-expressed human POLR2 in primary neurons and investigated the expression of circHomer1 (see Transparent Methods). Immunostaining against CTD phosphor Ser5 was used to measure the levels of active RNA-Pol2 in the nucleus of transfected and untransfected neurons. We observed a significant increase in CTD phospho Ser5 4 days after transfection (Figure S3I). In agreement with our hypothesis, we detected elevated levels of circHomer1 in cells that over-expressed RNA-Pol2 compared with untransfected controls (Figure S3J).
Taken together, we show that flanking introns play an important role in circRNA formation in rat neurons. Our data suggest that under conditions of reduced spliceosome activity the generation of circRNAs with longer flanking introns will be enhanced (Figure 4). This preference may be the result of the increased number and type of repeat sequences that these long introns harbor. Our data indicate that in rat neurons the spliceosome and intron features work in synchrony to generate circRNAs.
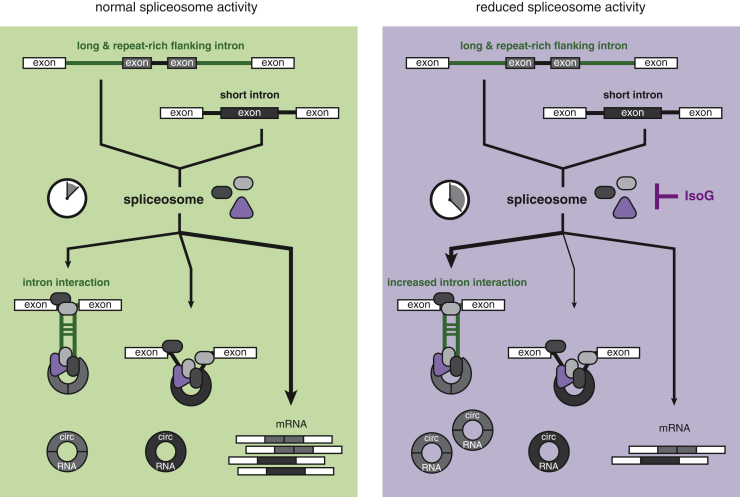
Figure 4.
Proposed Model for Neuronal circRNA Formation When Spliceosome Activity Is Reduced
The pre-mRNA will give rise to a mature mRNA or circRNA isoform. Circularizing exons are flanked by short (black) or long introns (green). Under normal spliceosome activity introns are timely removed, providing little opportunity for the flanking introns of circRNAs to interact with another. Generation of circRNAs with short and long flanking introns is balanced (light green box). When the spliceosome activity is reduced, such as by treatment with Isoginkgetin, introns are retained longer. These conditions are beneficial for long, repeat-rich flanking introns to interact with another and thus facilitate of the processing of their cognate circRNAs (light purple box).
Discussion
The enrichment of circRNAs in the brain and their dynamic expression during development and plasticity suggest that the biogenesis of neuronal circRNAs must be under tight regulation. Although some biogenesis factors of circRNAs have been previously described, owing to the lack of studies performed in primary neurons the mechanisms by which neuronal circRNAs are generated remain elusive. In this study, we pharmacologically inhibited the spliceosome and used RNA-seq to obtain a full view of the circRNA landscape in rat primary hippocampal neurons (Figure 1A). We found hundreds of robustly expressed circRNAs that were generated by a single exon or multiple exons (Figures 1D and 1F, Data S1). Intriguingly, a minimal sequence length seems to be required for successful circularization, as single-exon circRNAs were processed from one unusually long exon, whereas multi-exon circRNAs were generated by several shorter exons (Figure 1G). This is in line with observations in human circRNAs (Zhang et al., 2014). Previously, pharmacological inhibition or depletion of the spliceosome was shown to either decrease (Starke et al., 2015) or increase the expression of the circRNAs tested (Kramer et al., 2015, Liang et al., 2017, Stegeman et al., 2018). However, we found that reduced spliceosome activity has distinct effects on circRNA expression: whereas some circRNAs become up- or down-regulated, others remain unchanged in their expression level (Figure 2A). We successfully validated our results using several independent methods and further ruled out potential contribution of circRNA accumulation to the up-regulation during the Isoginkgetin treatment (Figures 2B, 2D, 2E, and 2H). It could be that the extent of spliceosome inhibition we achieved differed from those of previous studies or that reduced spliceosome activity affects circRNA biogenesis in distinct species differently. However, our RNA-seq approach offers two advantages over existing studies: first, we captured a wider view of the expression changes of the circRNA landscape after spliceosome inhibition. Second, the identification of hundreds of changed and unchanged circRNAs enabled us to investigate gene features that determines the directionality of their regulation.
To this end, we focused on the characterization of the introns flanking circularizing exons, as they can facilitate back-splicing (Ivanov et al., 2015, Jeck et al., 2013, Liang and Wilusz, 2014, Zhang et al., 2014, Zhang et al., 2016). Interestingly, flanking introns of up-regulated circRNAs were nearly double the length of flanking introns of all other circRNA populations and harbored 57% more repeat elements and reverse complementary motifs than the unchanged circRNA population (Figures 3A, 3C, and 3D). When we looked into the type of repeats, we found that certain repeat classes and families were enriched in up-regulated circRNAs (Figure 3E). While we detected inversely oriented Alu repeats (Figure S3G), which have been previously associated with facilitation of circRNA biogenesis (Jeck et al., 2013, Liang and Wilusz, 2014, Zhang et al., 2014), we also identified many new repeat elements that may benefit exon circularization, such as LINE (L1) and long terminal repeats (ERVL_MALR, ERVK). For the investigation of the structural properties of flanking introns, we had to restrict the analysis to the 66 shortest introns of up-regulated and an equivalent number of unchanged circRNAs. Here, we found that unchanged circRNAs were predicted with slightly lower norm.MFE than the tested up-regulated circRNAs (Figure S3H).
We propose that under conditions of reduced spliceosome activity, when introns become strongly retained (Figures S3C–S3E), long and repeat-rich flanking introns can interact easier with another and thus facilitate the processing of their cognate circRNAs. Following our hypothesis, a modulation of transcription rate would also affect intron retention and therefore circRNA formation. Indeed, we observed a significant increase in circHomer1 expression when RNA polymerase 2 was over-expressed in primary neurons (Figures S3I and S3J). Along the same line, increase of transcription speed in 293T cells has been previously shown to enhance the production of some circRNAs (Zhang et al., 2016).
We propose the following working model based on our observations (Figure 4): The pre-mRNA transcript is spliced into mRNA or circRNA isoforms. Under conditions of normal spliceosome activity, introns are removed in a timely fashion thus reducing the opportunity for back-splicing to occur. Under these conditions the production of circRNAs flanked by short and long introns is balanced. Isoginkgetin interferes with the spliceosome assembly (O'Brien et al., 2008). As a consequence, introns are retained for longer. circRNA flanked by long and repeat-rich introns will benefit from these conditions, as the opportunity for their flanking introns to interact with another increases, therefore enhancing the biogenesis of their cognate circRNAs. As the interaction with intron features can affect how the spliceosome processes the circRNA, our findings show that a delicate interplay between biogenesis factors determines the circRNA output of a given gene loci. Our model is in line with previous work in Drosophila (Liang et al., 2017) - suggesting that the regulation of circRNA biogensis is evolutionarily conserved across species. Many human diseases are closely linked to a disruption of the basal splicing machinery, such as retinitis pigmentosa and spinal muscular atrophy (Faustino and Cooper, 2003). It will be interesting to investigate the expression of circRNAs under these pathological conditions, as it may shed light on the role of circRNA abundance on disease states and their potential as targets for designing therapeutic approaches.
Limitations of the Study
In this study, we described the role of the spliceosome on circRNA formation in primary hippocampal neurons, specifically the fact that under conditions of reduced spliceosome activity the generation of neuronal circRNAs flanked by long and repeat-rich introns becomes favored. As our experiments were performed on cultured neurons, future studies would be needed to confirm similar regulation in vivo. Moreover, it is known that the expression of circRNAs varies across distinct brain regions (Rybak-Wolf et al., 2015, Westholm et al., 2014, You et al., 2015). Therefore, it would be interesting to investigate how reduced splicing activity affects circRNA formation in different brain areas.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank David Unzue and Georgi Tushev for discussion. We thank David Unzue for assistance with the bioinformatic analysis. We thank Ina Bartnik, Nicole Fürst, and Anja Staab for the preparation of cultured hippocampal neurons. Work in the laboratory of E.M.S. is supported by the Max Planck Society and The European Research Council under the European Union's Horizon 2020 research and innovation program (grant agreement No 743216). We also acknowledge the support of DFG CRC 902 (E.M.S and M.M.-M) and 1080 (E.M.S) and the DFG Cluster of Excellence for Macromolecular Complexes (E.M.S and M.M.-M).
Author Contributions
M.W., M.M.-M., and E.M.S designed experiments; M.W. conducted and analyzed experiments; and J.H. and W.C. performed RNA-seq experiments. M.W. and E.M.S wrote the manuscript, and all authors edited and revised the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: October 25, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.08.058.
Data and Code Availability
The accession number for the raw sequencing data reported in this paper is NCBI BioProject: PRJNA561523. Processed data used for analyses in this manuscript are included in Data S1. All scripts used are briefly described in methods, and will be available upon request.
Supplemental Information
References
- Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Barrett S.P., Wang P.L., Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell S.A., Snavely A., Landry H.M., Churchman L.S., Gray J.M., Springer M. Total RNA-seq to identify pharmacological effects on specific stages of mRNA synthesis. Nat. Chem. Biol. 2017;13:501–507. doi: 10.1038/nchembio.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakeman P.R., Lanahan A.A., O'Brien R., Roche K., Barnes C.A., Huganir R.L., Worley P.F. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- Cocquerelle C., Mascrez B., Hetuin D., Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R., Ma X.K., Chen L.L., Yang L. Increased complexity of circRNA expression during species evolution. RNA Biol. 2017;14:1064–1074. doi: 10.1080/15476286.2016.1269999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errichelli L., Dini Modigliani S., Laneve P., Colantoni A., Legnini I., Capauto D., Rosa A., De Santis R., Scarfo R., Peruzzi G. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Giovannone B., Tsiaras W.G., de la Monte S., Klysik J., Lautier C., Karashchuk G., Goldwurm S., Smith R.J. GIGYF2 gene disruption in mice results in neurodegeneration and altered insulin-like growth factor signaling. Hum. Mol. Genet. 2009;18:4629–4639. doi: 10.1093/hmg/ddp430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M.C., Liang D., Tatomer D.C., Gold B., March Z.M., Cherry S., Wilusz J.E. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen L.S., Hansen T.B., Veno M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu C.X., Xue W., Zhang Y., Jiang S., Yin Q.F., Wei J., Yao R.W., Yang L., Chen L.L. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell. 2017;67:214–227.e7. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- Liang D., Tatomer D.C., Luo Z., Wu H., Yang L., Chen L.L., Cherry S., Wilusz J.E. The output of protein-coding genes shifts to circular RNAs when the Pre-mRNA processing machinery is limiting. Mol. Cell. 2017;68:940–954.e3. doi: 10.1016/j.molcel.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- O'Brien K., Matlin A.J., Lowell A.M., Moore M.J. The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. J. Biol. Chem. 2008;283:33147–33154. doi: 10.1074/jbc.M805556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasman Z., Been M.D., Garcia-Blanco M.A. Exon circularization in mammalian nuclear extracts. RNA. 1996;2:603–610. [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L.H., Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Stegeman R., Hall H., Escobedo S.E., Chang H.C., Weake V.M. Proper splicing contributes to visual function in the aging Drosophila eye. Aging cell. 2018;17:e12817. doi: 10.1111/acel.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veno M.T., Hansen T.B., Veno S.T., Clausen B.H., Grebing M., Finsen B., Holm I.E., Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.L., Bao Y., Yee M.C., Barrett S.P., Hogan G.J., Olsen M.N., Dinneny J.R., Brown P.O., Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm J.O., Miura P., Olson S., Shenker S., Joseph B., Sanfilippo P., Celniker S.E., Graveley B.R., Lai E.C. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L., Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:1–23. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., Akbalik G., Wang M., Glock C., Quedenau C. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.L., Yang L., Chen L.L. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the raw sequencing data reported in this paper is NCBI BioProject: PRJNA561523. Processed data used for analyses in this manuscript are included in Data S1. All scripts used are briefly described in methods, and will be available upon request.