Abstract
Background
Inflammatory processes contribute to the pathophysiology of multiple chronic conditions. Genetic factors play a crucial role in modulating the inflammatory load, but the exact mechanisms are incompletely understood.
Objective
To assess genetic determinants of 16 circulating cytokines and cell adhesion molecules (inflammatory phenotypes) in Finns.
Methods
Genome-wide associations of the inflammatory phenotypes were studied in Northern Finland Birth Cohort 1966 (N=5284). A subsequent meta-analysis was completed for 10 phenotypes available in a previous genome-wide association study, adding up to 13 577 individuals in the study. Complementary association tests were performed to study the effect of the ABO blood types on soluble adhesion molecule levels.
Results
We identified seven novel and six previously reported genetic associations (p<3.1×10−9). Three loci were associated with soluble vascular cell adhesion molecule-1 (sVCAM-1) level, one of which was the ABO locus that has been previously associated with soluble E-selectin (sE-selectin) and intercellular adhesion molecule-1 (sICAM-1) levels. Our findings suggest that the blood type B associates primarily with sVCAM-1 level, while the A1 subtype shows a robust effect on sE-selectin and sICAM-1 levels. The genotypes in the ABO locus associating with higher soluble adhesion molecule levels tend to associate with lower circulating cholesterol levels and lower cardiovascular disease risk.
Conclusion
The present results extend the knowledge about genetic factors contributing to the inflammatory load. Our findings suggest that two distinct mechanisms contribute to the soluble adhesion molecule levels in the ABO locus and that elevated soluble adhesion molecule levels per se may not increase risk for cardiovascular disease.
Keywords: genome-wide association, inflammatory load, svcam-1, abo blood type
Introduction
It is currently established that inflammatory load may play a role in the aetiology of autoimmune and infectious diseases, but also in a broad range of other diseases, such as chronic cardiometabolic disorders,1 neurodegenerative diseases2 and cancer.3 The risk for these diseases increases with age,4 and due to the world’s ageing population5 their prevalence is likely to expand. Moreover, these diseases often co-occur, which is likely due to shared inflammation-related pathophysiology.6
Inflammation is the body’s physiological response to harmful stimuli involving multiple molecular and cellular interactions attempting to restore disturbances in tissue or systemic homeostasis. Circulating cytokines, growth factors, chemokines and cell adhesion molecules (CAMs) (hereafter inflammatory phenotypes) are fundamental mediators of inflammatory responses. Genes encoding these molecules and their receptors play a crucial role in mediating the related functions. Previous studies have identified loci associating with levels of inflammatory phenotypes,7–9 but the understanding of the exact regulatory mechanisms is still incomplete.
To add insights into the genetic mechanisms contributing to the inflammatory load, we performed a genome-wide association study (GWAS) of 16 circulating inflammatory phenotypes in 5284 individuals from Northern Finland Birth Cohort 1966 (NFBC1966) and a subsequent meta-analysis of 10 phenotypes in three other Finnish population cohorts,7 adding up to a total of 13 577 individuals in the study. We report identification of seven novel and replication of six loci associating with levels of the circulating inflammatory markers.
Methods
Study populations, genotyping and inflammatory phenotype quantification
Northern Finland Birth Cohort 1966
The NFBC1966 comprises 96% of all births during 1966 in the two northernmost provinces in Finland; altogether 12 058 children were live-born into the cohort, and the follow-ups occurred at the ages of 1, 14, 31 and 46 years.10 The data analysed in the present study are from the 31 years’ follow-up when clinical examinations and blood sampling were completed for altogether 6033 individuals, 5284 of whom had body mass index (BMI), inflammatory phenotypes and genotype data available (a maximum number of individuals per inflammatory marker of 5100). Genotyping of the samples was completed using 370 k Illumina HumanHap arrays (Illumina, California, USA), and subsequent imputation was performed based on the 1000 Genomes reference panel. A total of 16 inflammatory phenotypes were quantified from overnight fasting plasma samples using Bio-Rad’s Bio-Plex 200 system (Bio-Rad Laboratories, California, USA) with Milliplex Human Chemokine/Cytokine and CVD/Cytokine kits (Cat# HCYTOMAG-60K-12 and Cat# SPR349; Millipore, St Charles, Missouri, USA) and Bio-Plex Manager Software V.4.3 as previously described.11 The 16 inflammatory phenotypes studied in the NFBC1966 were interleukin (IL) 1-alpha, IL1-beta (IL1β), IL4, IL6, IL8, IL17, IL1 receptor antagonist (IL1ra), interferon gamma-induced protein 10 (IP10), monocyte chemoattractant protein 1 (MCP1), tumour necrosis factor alpha (TNFα), vascular endothelial growth factor (VEGF), plasminogen activator inhibitor 1, soluble CD40 ligand, soluble E-selectin (sE-selectin), soluble intercellular adhesion molecule-1 (sICAM-1) and soluble vascular cell adhesion molecule 1 (sVCAM-1).
GWAS summary statistics from three Finnish population cohorts
Meta-analyses were conducted for 10 phenotypes available in a previous GWAS.7 The study included up to 8293 Finnish individuals from the Cardiovascular Risk in Young Finns Study (YFS)12 and FINRISK (www.thl.fi/finriski),13 adding up to 13 577 individuals studied in the present meta-analyses. Shortly, YFS is a population-based follow-up study started in 1980 comprising randomly chosen individuals from Finnish cities Helsinki, Kuopio, Tampere, Oulu and Turku. The YFS data included in the previous GWAS are from 2019 individuals who participated in the follow-up in 2007 and who had both inflammatory phenotype and genotype data available. FINRISK is a Finnish population survey conducted every 5 years to monitor chronic diseases and their risk factors. The surveys use independent, random and representative samples from different geographical areas of Finland. The data included in the present meta-analyses were from participants of the 1997 and 2002 surveys. Genotypes were obtained using 670 k Illumina HumanHap arrays (Illumina) and imputed based on 1000 Genomes reference panel. Inflammatory markers were quantified using Bio-Rad’s premixed Bio-Plex Pro Human Cytokine 27-plex Assay and 21-plex Assay, and Bio-Plex 200 reader with Bio-Plex V.6.0 software (Bio-Rad Laboratories) as previously described.14 Samples were serum in YFS, EDTA plasma in FINRISK1997 and heparin plasma in FINRISK2002.
Statistical analyses
GWAS and meta-analysis
To allow meta-analysis between the present results and the previous GWAS, the data processing and analysis model were done according to Ahola-Olli et al.7 First, rank-based inverse transformation was applied to normalise the phenotypes. Preceding the analyses, linear regression models were fitted to adjust the transformed inflammatory phenotypes for age, sex, BMI and the 10 first genetic principal components to control for population stratification. The resulting residuals were again normalised with inverse transformation, and the adjusted and transformed residuals were used as phenotypes in the analyses.
Genome-wide association tests were performed using snptest V.2.5.1 software.15 Allele effects were estimated using an additive model (-frequentist 1), and the option to centre and scale the phenotypes was disabled (-use_raw_phenotypes). The GWAS results were filtered by including markers with model fit info >0.8 and minor allele count >10. Filtered data were used to perform meta-analyses by METAL software (V.2011-03-25)16 for the 10 phenotypes (IL1β, IL1ra, IL4, IL6, IL8, IL17, IP10, MCP1, TNFα and VEGF) available in the previous GWAS.7 Genomic control correction was enabled (GENOMICCONTROL ON) to account for population stratification and cryptic relatedness. To estimate the heterogeneity of effect sizes between NFBC1966 and the previous GWAS, calculation of heterogeneity statistics based on Cochrane’s Q-test was enabled (ANALYZE HETEROGENEITY).
Supplemental genome-wide tests in NFBC1966
Individuals showing symptoms of an acute infection were omitted from the supplemental genome-wide tests performed in the NFBC1966 population. Here, individuals reported having fever at the time of the blood sampling and individuals having C-reactive protein (CRP) level >10 mg/L were excluded. Otherwise the analysis models were as above.
Conditional analyses and variance explained
To assess whether the identified loci harbour multiple independent association signals, we conducted conditional analyses by further adjusting the models with the locus-specific lead variants. The association tests were repeated within a 2 Mb window around the lead SNP for the phenotypes studied in the NFBC1966 population only. For the meta-analysed phenotypes, we applied a method proposed by Yang et al 17 that enables conditional analyses of GWAS summary statistics. NFBC1966 was used as a reference sample to estimate linkage disequilibrium (LD) corrections in these analyses. The proportion of variance explained was calculated using all independent variants using the following formula:
Here β is the variant’s effect estimate on the inflammatory phenotype and MAF denotes minor allele frequency.
Complementary association tests on soluble adhesion molecule levels
Complementary association tests were conducted to better evaluate the molecular mechanism explaining the two potentially independent association signals with soluble CAM levels in the ABO locus. Here, linear models were repeated within a 2 Mb window and further adjusted for the ABO blood type or rs507666 genotype tagging the A1 subtype.18 In addition, we determined the effect estimates of ABO blood types and ABO blood types stratified by rs507666 genotype on sE-selectin, sICAM-1 and sVCAM-1 levels: the adjusted and transformed CAM concentrations were as outcomes in the linear models and ABO blood types as categorical variables (individuals with blood type A vs non-A, and so on). Corresponding models were fitted for the rs507666-stratified blood types (individuals with blood type A and rs507666 G/G vs others, and so on).
Shared genetic influences on inflammatory and cardiovascular phenotypes
As previous evidence suggests that elevated concentrations of circulating markers of inflammation increase the risk of cardiovascular diseases (CVD),19 20 we further evaluated how variants in the loci associating with inflammatory phenotypes may relate to other cardiovascular traits. We used the gwas-pw method developed by Pickrell et al 21 that estimates whether a locus harbours a genetic variant influencing one of the two phenotypes compared (models 1 and 2), if the same variant influences both phenotypes (model 3), or if separate variants within a locus influence the two phenotypes (model 4). Using the gwas-pw and open-access data provided by CARDIoGRAM,22 MEGASTROKE consortium23 and Global Lipids Genetics Consortium,24 we evaluated the shared genetic determinants of circulating levels of low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), total cholesterol (TotC) and total triglycerides (TG), as well as risk of coronary artery disease (CAD), ischaemic stroke and the inflammatory phenotypes showing significant genetic associations in the present study. The genome breakpoint data set for individuals with European ancestry provided at https://bitbucket.org/nygcresearch/ldetect-data was used in the gwas-pw analyses to split the genome into approximately independent blocks.25
Results
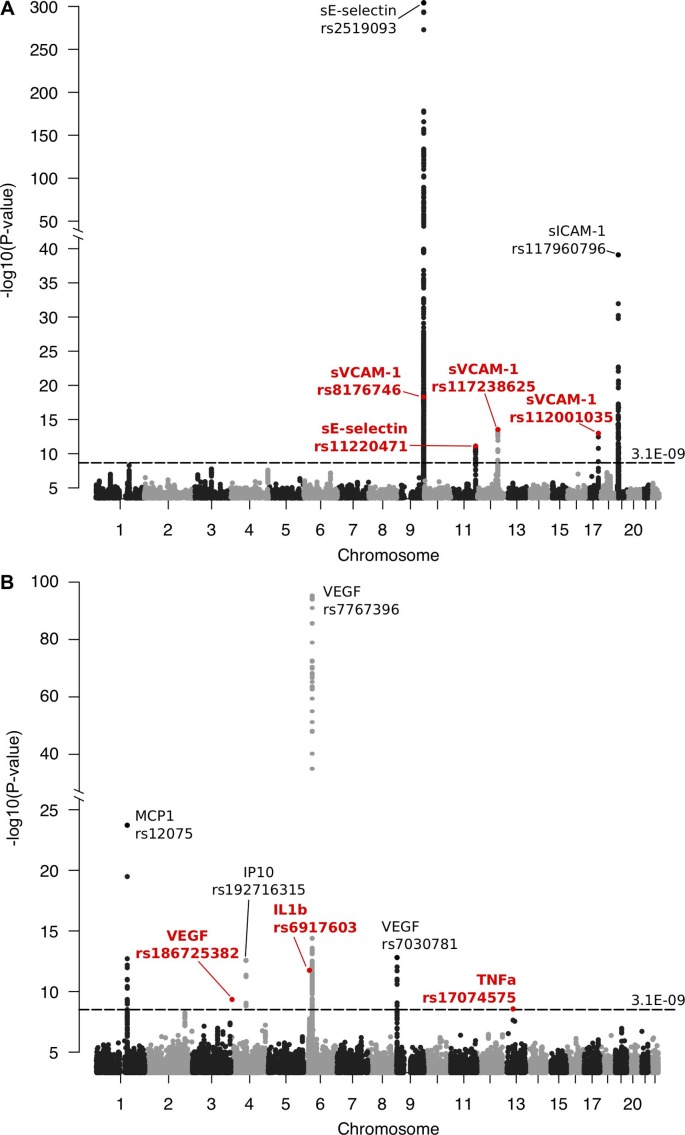
The basic characteristics of the NFBC1966 study population are provided in table 1. Inflammatory phenotype distributions are tabulated in online supplementary table S1, and their correlation structure is shown in online supplementary figure S1. Using a threshold of p<3.1×10−9 for statistical significance (standard genome-wide significance level p<5×10−8 corrected for 16 phenotypes tested), we identified seven novel and six previously reported loci associating with one or more of the inflammatory phenotypes. The results are summarised in table 2 and combined Manhattan plots are shown in figure 1. Manhattan plots and Q-Q plots for each inflammatory phenotype are provided in online supplementary figure S2 A–Z. Genomic inflation factor values range between 0.99 and 1.02, suggesting no inflation in the test statistics (online supplementary table S2). Online supplementary table S3 lists the traits associated previously with the loci showing novel associations with inflammatory phenotypes in the present study.
Table 1.
Basic characteristics of the Northern Finland Birth Cohort 1966 study population
| Characteristics | |
| Total number of individuals | 5284 |
| Number of men (%) | 2543 (48.1) |
| Age, years | 31.1±0.4 |
| Body mass index, kg/m2 | 24.4±4.0 |
| Glucose, mmol/L | 5.1±0.7 |
| Low-density lipoprotein-cholesterol, mmol/L | 3.0±0.9 |
| High-density lipoprotein-cholesterol, mmol/L | 1.6±0.4 |
| Systolic blood pressure, mm Hg | 124.2±13.6 |
| Diastolic blood pressure, mm Hg | 76.8±11.7 |
Values are mean±SD.
Table 2.
Significant loci associating with the circulating inflammatory phenotypes
| Study | Marker | Locus | Chr:Position | Candidate gene | Nearest gene(s) | Annotation | dbSNP reference | INFO | EA | EAF | Beta | P value | HetPVal | Variance explained | Total variance explained |
| NFBC1966 | sE-selectin | 9q34.2 | 9:136 141 870 | ABO | ABO | Intronic | rs2519093 | 0.994 | T | 0.188 | −0.903 | 4.48e-305 | NA | 0.249 | 0.258 |
| 11q24.2 | 11:126 266 665 | ST3GAL4 | ST3GAL4 | Intronic | rs11220471 | 0.967 | G | 0.212 | −0.162 | 7.72e-12 | NA | 0.009 | |||
| sICAM-1 | 9q34.2 | 9:136 141 870 | ABO | ABO | Intronic | rs2519093 | 0.994 | T | 0.188 | −0.352 | 7.43e-48 | NA | 0.038 | 0.118 | |
| 19p13.2 | 19:10 383 403 | ICAM1 | ICAM1 | Intronic | rs117960796 | 0.802 | A | 0.012 | −1.669 | 8.03e-40 | NA | 0.066 | |||
| 19p13.2 | 19:10 497 360 | ICAM1 | CDC37 | Intergenic | rs74428614 | 0.992 | A | 0.163 | 0.226 | 1.14e-16* | NA | 0.014 | |||
| sVCAM-1 | 9q34.2 | 9:136 131 322 | ABO | ABO | Missense | rs8176746 | 1.000 | T | 0.129 | 0.256 | 5.06e-19 | NA | 0.015 | 0.038 | |
| 12q23.3 | 12:104 448 391 | HSP90B1 | GLT8D2 | Intronic | rs117238625 | 0.981 | A | 0.023 | 0.510 | 2.90e-14 | NA | 0.012 | |||
| 17q24.2 | 17:66 823 805 | ABCA8 | ABCA8 | Intergenic | rs112001035 | 0.883 | A | 0.060 | −0.324 | 1.04e-13 | NA | 0.012 | |||
| Meta-analyses | IL1β | 6p22.1 | 6:30 017 071 | HLA locus | Intronic | rs6917603 | 1.000 | C | 0.251 | −0.163 | 1.76e-12 | 1.00 | 0.010 | 0.015 | |
| 6p22.1 | 6:30 013 887 | HLA locus | Intronic | rs9261224 | 1.000 | T | 0.035 | 0.261 | 1.31e-09 * | 1.00 | 0.005 | ||||
| IP10 | 4q21.1 | 4:76 899 176 | CXCL10 | SAD1 | Intronic | rs192716315 | 0.851 | C | 0.003 | 1.513 | 2.71e-13 | 1.00 | 0.014 | 0.014 | |
| MCP1 | 1q23.2 | 1:159 175 354 | ACKR1 | ACKR1 | Missense | rs12075 | 1.000 | A | 0.469 | 0.148 | 1.43e-33 | 1.51e-13 | 0.011 | 0.011 | |
| TNFα | 13q14.3 | 13:51 141 997 | DLEU1 | DLEU1 | Intronic | rs17074575 | 0.803 | G | 0.002 | 2.131 | 2.71e-09 | 1.00 | 0.018 | 0.018 | |
| VEGF | 4p16.2 | 4:5 636 073 | STK32B | EVC2 | Intronic | rs186725382 | 0.875 | A | 0.001 | −2.380 | 4.53e-10 | 1.00 | 0.011 | 0.052 | |
| 6p21.1 | 6:43 927 050 | VEGFA | C6orf223 | Intergenic | rs7767396 | 1.000 | A | 0.523 | 0.284 | 8.35e-105 | 1.22e-69 | 0.040 | 0.056 | ||
| 9p24.2 | 9:2 686 273 | VLDLR | VLDLR, KCNV2 | Intergenic | rs7030781 | 0.959 | T | 0.373 | -0.099 | 1.57e-13 | 5.34e-04 | 0.005 |
Statistical significance is considered at p<3.1×10−9. Novel findings are highlighted with bold font. All positions correspond to human genome build 37.
*Indicates associations that are significant after conditioning the analyses on the locus-specific lead variant on the preceding row.
EA, effect allele; EAF, effect allele frequency; HLA, human leukocyte antigen; HetPVal, p value of heterogeneity as estimated by Cochrane’s Q-test; IL1β, interleukin 1-beta; INFO, imputation score in NFBC1966; IP10, interferon gamma-induced protein 10; MCP1, monocyte chemoattractant protein 1; NA, not available; NA, not available; NFBC1966, Northern Finland Birth Cohort 1966; TNFα, tumour necrosis factor alpha; VEGF, vascular endothelial growth factor; dbSNP, single nucleotide polymorphism database; sE-selectin, soluble E-selectin; sICAM-1, soluble intercellular adhesion molecule -1; sVCAM-1, soluble vascular cell adhesion molecule-1.
Figure 1.
The combined Manhattan plots for significant associations with inflammatory markers studied in (A) Northern Finland Birth Cohort 1966 and in (B) meta-analyses with three other Finnish population cohorts. Significance threshold p<3.1×10−9 derives from the standard p value limit for genome-wide significance p<5×10−8 corrected for 16 markers examined in the present study. Novel association signals are highlighted with red font and replicated loci are marked with black font. sE-selectin, soluble E-selectin; IL1b, interleukin 1-beta; IP10, interferon gamma-induced protein 10; MCP1, monocyte chemoattractant protein 1; sICAM-1, soluble intercellular adhesion molecule-1; sVCAM-1, soluble vascular cell adhesion molecule-1; TNFa, tumour necrosis factor alpha; VEGF, vascular endothelial growth factor.
jmedgenet-2018-105965supp001.pdf (42.2MB, pdf)
Cell adhesion molecules
The ABO locus shows large effects on sE-selectin, sICAM-1 and sVCAM-1 levels
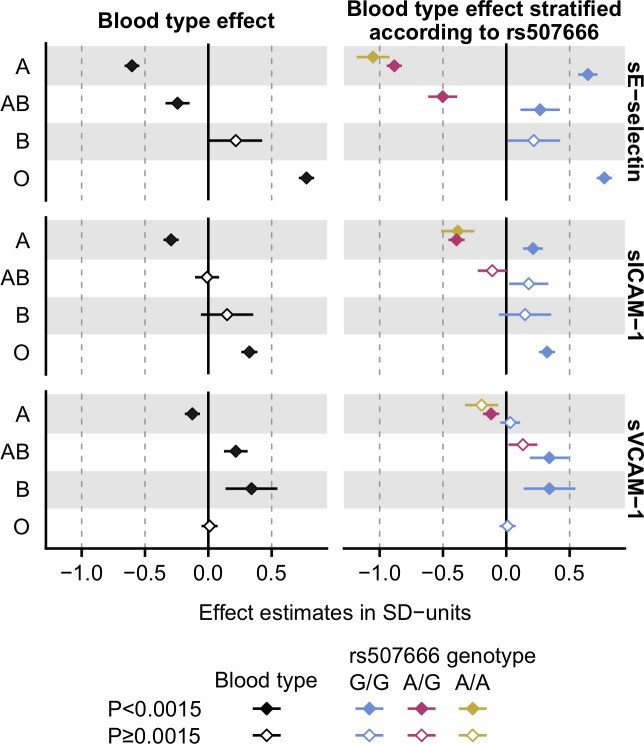
We observed a novel effect on sVCAM-1 concentration in 9q34.2 near ABO (ABO, alpha 1–3 n-acetylgalactosaminyltransferase and alpha 1–3-galactosyltransferase) in the NFBC1966 population. This locus showed a robust association also with sE-selectin and sICAM-1 concentrations as previously reported.18 26 27 Noteworthy, the lead variant for sE-selectin and sICAM-1 associations (rs2519093) was different from the lead variant for sVCAM-1 association (rs8176746). The former variant is in LD (r2=1 in NFBC1966) with rs507666 tagging the ABO blood type A subtype A1, whereas the latter variant tags the blood type B.18
As the GWAS results suggested two potentially separate association signals with the soluble CAM levels in the ABO locus, we conducted complementary association tests to better evaluate the molecular mechanisms explaining the associations. The association of the rs8176746 with sVCAM-1 concentration was significant when adjusted for the rs507666 indicative of the A1 subtype (p=4.98×10−15). On the contrary, the associations of the rs2519093 with concentrations of sE-selectin and sVCAM-1 were highly significant when adjusted for the ABO blood type (p=3.40×10−123 and p=3.43×10−17, respectively). Overall, these results suggest that the association of the rs8176746 with sVCAM-1 level is independent of the A1 subtype, while the association of the rs2519093 with sE-selectin and sICAM-1 levels is independent of the ABO blood type. Statistical significances were abolished when the rs8176746 association with sVCAM-1 was adjusted for ABO blood type and rs2519093 association with sE-selectin or sICAM-1 was adjusted for rs507666.
To further evaluate the related molecular mechanisms, we determined the effect estimates of the ABO blood types and ABO blood types stratified by rs507666 genotype on soluble CAM levels. The blood type A showed negative associations with the levels of all the three CAMs and the effect was the most robust on the sE-selectin level (figure 2, left panel). However, major discrepancies in the effect directions were seen when the analyses were stratified by the rs507666 genotype (figure 2, right panel). Congruent with previous reports,18 26 the present results suggest that the A1 subtype/rs507666 influences sE-selectin or sICAM-1 levels. In contrast, the blood type B seems to attribute predominantly to sVCAM-1 level, while the A1 subtype/rs507666 shows only a modest effect on sVCAM-1.
Figure 2.
The effects of the ABO blood types and the A1 subtype on soluble adhesion molecule levels. The effects of the ABO blood types on sE-selectin, sICAM-1 and sVCAM-1 levels were evaluated in linear models, where adjusted (sex, age, body mass index and the 10 first genetic principal components) and transformed soluble adhesion molecule concentrations were used as outcomes and the ABO blood type served as categorical variable (A vs non-A, and so on). Corresponding models were fitted for the ABO blood types stratified by the rs507666-A allele count (0, 1 or 2), where the A allele tags the ABO subtype A1 having enhanced glycosyltransferase activity.18 No individuals were found to have B or O blood type and one or more copies of the rs507666-A allele, and thus it was not possible to perform stratification within these blood types. sE-selectin, soluble E-selectin; sICAM-1, soluble intercellular adhesion molecule-1; sVCAM-1, soluble vascular cell adhesion molecule-1.
HSP90B1 and ABCA8 loci associate with sVCAM-1 levels
We identified two other novel loci for sVCAM-1 (12q23.3 and 17q24.2) in the NFBC1966 population. In chr12 the lead variant rs117238625 is in LD (r2=1 in NFBC1966) with rs117468318 that locates in the 5’ untranslated region (UTR) region of HSP90B1 (heat shock protein 90 kDa beta member 1) and, according to RegulomeDB,28 is likely to affect transcription factor binding. The association signal in chr17 locates near ABCA8 (ATP binding cassette subfamily A member 8) encoding one of the ATP binding cassette transporters.
Variations in sialyltransferase encoding genes show an effect on sE-selectin level
For sE-selectin level, we identified a novel association in 11q24.2 in the region of ST3GAL4 (ST3 beta-galactoside alpha-2,3-sialyltransferase 4). We identified a suggestive signal with sE-selectin level also in 3q12.1 near ST3GAL6 (ST3 beta-galactoside alpha-2,3-sialyltransferase 6), but the association was not significant after multiple correction (p=1.75×10−08). Both of the sialyltransferase genes have been implicated in the production of functional E-selectin, P-selectin and L-selectin ligands in mice.29
Two independent association signals on sICAM-1 level near ICAM1
We replicated the previously reported association for sICAM-1 level in 19p13.2 near ICAM1 (intracellular adhesion molecule 1).18 30 When the primary association test was conditioned for the lead variant rs117960796, another significant association was detected (rs74428614, p=1.14×10−16) indicative of more than one independent variant contributing to sICAM-1 level in this locus.
Vascular endothelial growth factor
In the meta-analyses, we identified a novel locus 4p16.2 with a large effect on VEGF (β=−2.38 SD). This locus harbours genes EVC (EvC ciliary complex subunit 1), EVC2 (EvC ciliary complex subunit 2) and STK32B (serine/threonine kinase 32B). In addition, we replicated two previously reported loci associating with VEGF levels in 6p21.1 near VEGFA (vascular endothelial growth factor A) and in 9p24.2 near VLDLR (very-low-density lipoprotein receptor).7
Proinflammatory cytokines
Locus near DLEU1 shows a large effect on TNFa
We identified a novel variant with a large effect on TNFα levels (β=2.13 SD) in 13q14.3 near DLEU1 and DLEU7 (deleted in lymphocytic leukaemia 1 and 7) in the meta-analyses.
The human leukocyte antigen (HLA) locus shows a small effect on IL1β
A novel variant at 6p22.1 in the human leucocyte antigen locus associating with IL1β level was identified in the meta-analyses. In the conditional analyses, we observed two independent association signals at this locus (table 1, online supplementary figure S2J). The same locus and the same lead variant rs6917603 showed also a suggestive effect on IL4 level (online supplementary figure S2L), but the meta-analysed result was not significant after multiple correction (p=5.56×10−09).
Chemokines
We replicated previously reported loci near CXCL10 (C-X-C motif chemokine ligand 10) and ACKR1 (atypical chemokine receptor 1) associating with IP10 levels and with MCP1 levels, respectively.7
Supplemental genome-wide tests in NFBC1966
Altogether 236 individuals having fever or CRP >10 mg/L were excluded from the supplemental genome-wide tests performed in the NFBC1966 population. The results of the supplementary analyses were congruent with the original findings (online supplementary table S4).
Comparisons of genetic effects on inflammatory phenotypes versus other traits
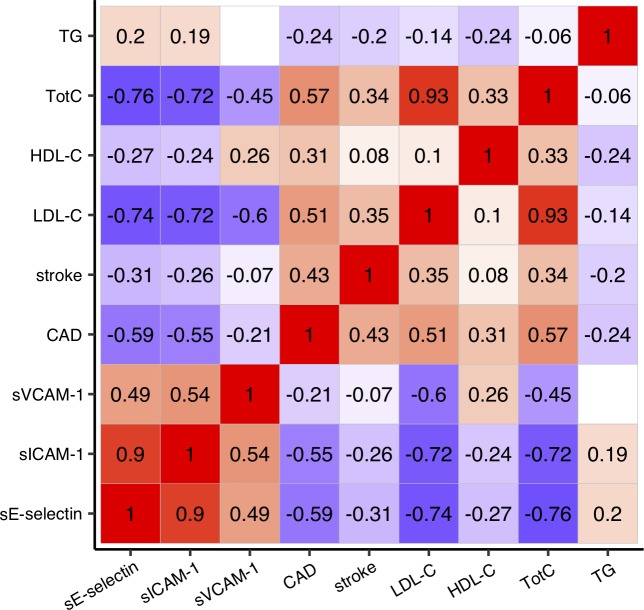
Elevated circulating concentrations of inflammatory markers increase the risk for CVD.19 20 We used the gwas-pw method21 to evaluate the presence of shared genetic determinants between inflammatory phenotypes showing significant genetic association in the present study and other cardiovascular health-related traits (LDL-C, HDL-C, TotC, TG, CAD risk, ischaemic stroke risk) obtained from open-access sources.22–24 Altogether 56 genomic regions showed robust statistical evidence for containing a genetic variant influencing one or more of the inflammatory phenotypes and at the same time one or more of the other traits studied (model 3 posterior probability greater than 0.99; online supplementary figure S3). The ABO locus was one of the loci harbouring variants influencing multiple traits. In this locus, we observed negative linear relationships between the SNP effects on sE-selectin and sICAM-1 levels and CAD risk, stroke risk, as well as LDL-C, HDL-C and TotC levels (figure 3). The results in the other loci are provided in online supplementary figure S3.
Figure 3.
SNP effects on soluble adhesion molecule levels versus other cardiovascular health-related traits in the ABO locus. The Pearson’s r of the genetic effects (Z-scores) were estimated using a set of SNPs that located in the ABO locus (defined as the LD block25 containing ABO gene in the gwas-pw21 analyses) and that were available in both the present study and open-access data sets.22–24 Positive correlations are indicated with red color, negative correlations are indicated with blue color, and correlations with p≥0.05 are left blank. The scatter plot representations as well as correlations in the other loci are shown in online supplementary figure S3. CAD, coronary artery disease; HDL-C, high-density lipoprotein cholesterol; LD, linkage disequilibrium; LDL-C, low-density lipoprotein cholesterol; sE-selectin; soluble E-selectin; sICAM-1, soluble intercellular adhesion molecule-1; sVCAM-1, soluble vascular cell adhesion molecule-1; TG, total triglycerides; TotC, total cholesterol.
Discussion
The present study examines the genetic determinants of 16 circulating inflammatory phenotypes in 5284 individuals from Northern Finland with a subsequent meta-analysis of 10 phenotypes in 3 other Finnish populations, adding up to a total of 13 577 participants. We report seven novel and replication of six previously published genetic associations.
We identified a novel association for sVCAM-1 concentration at the ABO locus. This locus was also associated with sE-selectin and sICAM-1 levels as observed previously.18 26 The present GWAS suggested two distinct association signals in the ABO locus for the sE-selectin and sICAM-1 levels versus sVCAM-1 level, and the supplementary tests provided further support for at least two mechanisms contributing to circulating concentrations of CAMs in this locus. The two mechanisms include the blood type A subtype A1, which has a robust lowering effect on sE-selectin and sICAM-1 levels,18 26 and the blood type B which seems to have an increasing effect on sVCAM-1 level. The lowering effect of the A1 subtype on sE-selectin and sICAM-1 could arise from increased glycosyltransferase activity that possibly modifies the shedding of the CAMs from the endothelium and/or their clearance rate from circulation.18 26 The underlying mechanism explaining the association between the blood type B and higher sVCAM-1 concentration remains unknown and warrants research. VCAM-1-mediated adhesion involves interaction with galectin-3, a protein that has a specificity for galactosides.31 As the B antigen holds an additional galactose monomer compared with the A and O antigens, and galectins are known to recognise blood type antigens,32 it raises the speculation that the amount of unbound sVCAM-1 in the circulation could be influenced by a possible competitive binding of galectin-3 with sVCAM-1 and the B antigen.
To evaluate the shared genetic mechanisms, we compared the correspondence of genetic effects on inflammatory phenotypes versus cardiovascular health-related traits. We observed a negative relationship between the genetic effects on CAM levels and the genetic effects on LDL-C and TotC levels, as well as lower risk for CAD and ischaemic stroke, in the ABO locus. This denotes that the genotypes in the ABO locus associating with higher levels of soluble CAMs tended to associate with lower circulating cholesterol levels as well as lower risk of cardiovascular outcomes. This was unexpected since previous evidence suggests that increased soluble CAM levels are linked with atherosclerosis progression and vascular outcomes.20 33 Possible explanations unravelling the negative correlation advocate that soluble CAMs may compete with leucocyte adhesion to the endothelial molecules or that enhanced ectodomain shedding may contribute to the reduced recruitment of leucocytes to the subendothelial space, thereby promoting cardioprotective effects.34 Our results suggesting a negative relationship between the genetic effects on soluble CAM and circulating cholesterol levels advocate that altered cholesterol metabolism could contribute to the CAD risk associated with the ABO locus; the genetic effects of the same SNPs on LDL-C or TotC show positive correlation with CAD risk. Nevertheless, further studies are warranted to understand the exact mechanisms.
Another novel association with sVCAM-1 level was detected in chr12. The lead SNP of this locus is in LD with rs117468318 (r2=1 in NFBC1966) that locates in the 5’UTR of HSP90B1 encoding heat shock protein gp96 and, according to RegulomeDB,28 is likely to affect transcription factor binding, suggesting a possible regulatory mechanism for the detected association. HSP90B1/gp96 is a chaperone that is essential for assembly of 14 of 17 integrin pairs in the haematopoietic system.35 Integrin α4β1 is an important ligand of VCAM-1; if altered transcription of HSP90B1 had a downstream effect on integrin α4β1 level, this could further modify the level of unbound sVCAM-1 in circulation.
The third novel locus showing association with sVCAM-1 level was identified in chr17 near ABCA8. The lead SNP rs112001035 is an expression quantitative trait locus (eQTL) for ABCA8 in multiple tissue types.36 If ABCA8 is involved in the regulation of HDL level via interaction with ABCA137 and if plasma HDL levels contribute to VCAM-1 expression,38 then altered expression of the ABCA8 could influence circulating levels of sVCAM-1 by modulating HDL particle concentration. However, this hypothesis is not supported by the fact that the effect of the lead SNP on HDL particle concentration is negligible in a metabolomics GWAS (β=−0.043 SD, p=0.049).39 There is evidence suggesting that ABCA8 may be involved in sphingolipid metabolism,40 and it has been hypothesised that ABCA8 may be involved in the formation of specific membrane domains during ApoA-I lipidation.37 Thus, the association between the ABCA8 and sVCAM-1 level could be related to altered HDL composition possibly contributing to endothelial homeostasis rather than absolute particle concentration. However, more evidence is needed to draw conclusions.
We detected a novel effect of rs11220471 in chr11 near ST3GAL4 on sE-selectin levels in the NFBC1966 population. ST3GAL4 encodes a member of the glycosyltransferase 29 family of enzymes involved in protein glycosylation. In mice, St3Gal4 is needed for synthesis of functional selectin ligands.29 The altered levels or structure of selectin ligands due to variation in ST3GAL4 could contribute to the levels of unbound sE-selectin in circulation, providing a biologically rational mechanism for the detected association.
In the meta-analyses, we detected a novel large-effect locus for VEGF in chr4 (β=−2.38 SD) near STK32B. Mutations in this locus have been associated previously with coeliac disease,41 CAD42 and Ellis-van Creveld syndrome.43 STK32B may play a role in the hedgehog signalling pathway, which has been implicated in metastasis and angiogenesis in cancer44 and downregulated in coeliac disease.45 The hedgehog signalling has shown to be involved in the regulation of VEGF expression during developmental angiogenesis in avian embryo.46 Thus, previous literature and our results advocate that STK32B may be involved in the regulation of VEGF levels possibly via hedgehog signalling-related mechanism.
The other novel findings obtained in meta-analysis include a large-effect locus on TNFα level in chr13 (β=2.13 SD). The locus in 13q14.3 associating with TNFα locates near DLEU1 and DLEU7. This region is recurrently deleted in tumours and haematopoietic malignancies.47 DLEU1 is a part of a transcriptionally coregulated gene cluster that modulates the activity of the nuclear factor kappa B (NF-kB) pathway,48 which is also modulated by TNFα.49 It is largely unknown how the DLEU1 and related DLEU2 regulate NF-kB activity50; our result suggests that TNFα signalling might be involved in this mechanism.
At last, we identified a small-effect locus in chr6 harbouring two independent association signals on IL1β and showing suggestive association also on IL4 level. This association signal is in the region coding the human leucocyte antigen proteins, and further experimental evidence would be needed to identify the exact mechanism how the locus contributes to IL levels.
The strengths and limitations of our study should be considered. The sample size of the present study should provide adequate power for detecting genetic associations with circulating markers of systemic inflammation.8 The use of genetically isolated populations, such as inhabitants of Northern Finland, should further enhance the power for locus identification in GWAS settings. We were able to perform meta-analyses only for 10 out of the total of 16 inflammatory phenotypes, and the novel findings are largely based on NFBC1966 population only. Thus, replication of the present findings in other populations would be helpful. In particular, the associations of the novel rare, large-effect variants need to be interpreted with caution until the associations are validated in other populations. The interassay coefficient of variability measures for sE-selectin and VEGF in particular are notably larger than 15%, which is considered to be the limit for acceptable values (online supplementary table S1). However, to our consideration, all the findings identified in the present study locate on genome regions with biologically relevant genes. Furthermore, the replications of the previously reported loci speak for the data adequacy and add confidence to the novel associations. Finally, as we have not included functional experiments in this work, we are limited to previous literature when explaining the potential biological mechanism behind the identified associations.
The present results provide novel information on genetic mechanisms influencing levels of inflammatory phenotypes in circulation. The evident role of the ABO locus in the regulation of the soluble CAM levels likely encompasses at least two distinct mechanisms influencing sE-selectin, sICAM-1 and sVCAM-1 levels. Our findings provide evidence that increased soluble CAM concentrations per se may not be a risk factor for cardiovascular outcomes. In particular, genetic variation associating with increased sE-selectin or sICAM-1 levels at the ABO locus seems to contribute to lower cardiovascular risk. Furthermore, genetic effects at the ICAM1 locus providing a direct molecular link to sICAM-1 concentration do not correlate with the genetic effects on CAD risk nor stroke risk. Overall, the present study extends the knowledge about the molecular pathways involved in inflammatory load.
Acknowledgments
We gratefully acknowledge the contributions of the participants in the Northern Finland Birth Cohort 1966 study. We also thank all the field workers and laboratory personnel for their efforts. Data on coronary artery disease/myocardial infarction have been contributed by CARDIoGRAMplusC4D investigators and have been downloaded from www.CARDIOGRAMPLUSC4D.ORG.
Footnotes
SS and JK contributed equally.
Correction notice: This article has been corrected since it was published Online First. Values in table 2 have been amended.
Contributors: ES, SS and JK conceptualised the study. ES, MK and AA-O performed the statistical analyses. TK, K-HH, MS, KS and SJ performed the quantifications of the inflammatory markers. OR, MP, VS, TL, HV, MS, SJ, JJ, SK-K, MM, K-HH, M-RJ, SS and JK provided funding or other resources to conduct the study. SS and JK supervised the study. ES, SS and JK wrote the original manuscript. All authors contributed to revising the content and approved the final version.
Funding: This work was supported by the University of Oulu Graduate School (ES, MK), the Finnish Foundation for Cardiovascular Research (VS), Biocenter Oulu (SS), European Commission (DynaHEALTH – H2020 – 633595; SS), Academy of Finland (297338 and 307247; JK) and Novo Nordisk Foundation (NNF17OC0026062; JK). NFBC1966 received financial support from University of Oulu (Grant No 65354), Oulu University Hospital (Grant No 2/97 and 8/97), Ministry of Health and Social Affairs (Grant No 23/251/97, 160/97 and 190/97), National Institute for Health and Welfare, Helsinki (Grant No 54121), and Regional Institute of Occupational Health, Oulu, Finland (Grant No 50621 and 54231). The Young Finns Study has been financially supported by the Academy of Finland (grants 286284, 134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi) and 41071 (Skidi)); the Social Insurance Institution of Finland; Competitive State Research Financing of the Expert Responsibility area of Kuopio, Tampere and Turku University Hospitals (grant X51001); Juho Vainio Foundation; Paavo Nurmi Foundation; Finnish Foundation for Cardiovascular Research; Finnish Cultural Foundation; Sigrid Juselius Foundation; Tampere Tuberculosis Foundation; Emil Aaltonen Foundation; Yrjö Jahnsson Foundation; Signe and Ane Gyllenberg Foundation; Diabetes Research Foundation of Finnish Diabetes Association; EU Horizon 2020 (grant 755320 for TAXINOMISIS); European Research Council (grant 742927 for MULTIEPIGEN project); and Tampere University Hospital Supporting Foundation.
Competing interests: VS has participated in a conference trip sponsored by Novo Nordisk and received a honorarium from the same source for participating in and advisory board meeting. He also has ongoing research collaboration with Bayer.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Donath MY. Targeting inflammation in the treatment of type 2 diabetes: time to start. Nat Rev Drug Discov 2014;13:465–76. [DOI] [PubMed] [Google Scholar]
- 2. Heneka MT, Kummer MP, Latz E. Innate immune activation in neurodegenerative disease. Nat Rev Immunol 2014;14:463–77. [DOI] [PubMed] [Google Scholar]
- 3. Coussens LM, Werb Z. Inflammation and cancer. Nature 2002;420:860–7. 10.1038/nature01322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goldberg EL, Dixit VD. Drivers of age-related inflammation and strategies for healthspan extension. Immunol Rev 2015;265:63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization World report on ageing and health 2015.
- 6. de Craen AJM, Posthuma D, Remarque EJ, van den Biggelaar AHJ, Westendorp RGJ, Boomsma DI. Heritability estimates of innate immunity: an extended twin study. Genes Immun 2005;6:167–70. [DOI] [PubMed] [Google Scholar]
- 7. Ahola-Olli AV, Würtz P, Havulinna AS, Aalto K, Pitkänen N, Lehtimäki T, Kähönen M, Lyytikäinen L-P, Raitoharju E, Seppälä I, Sarin A-P, Ripatti S, Palotie A, Perola M, Viikari JS, Jalkanen S, Maksimow M, Salomaa V, Salmi M, Kettunen J, Raitakari OT. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet 2017;100:40–50. 10.1016/j.ajhg.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, Oliver-Williams C, Kamat MA, Prins BP, Wilcox SK, Zimmerman ES, Chi A, Bansal N, Spain SL, Wood AM, Morrell NW, Bradley JR, Janjic N, Roberts DJ, Ouwehand WH, Todd JA, Soranzo N, Suhre K, Paul DS, Fox CS, Plenge RM, Danesh J, Runz H, Butterworth AS. Genomic atlas of the human plasma proteome. Nature 2018;558:73–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ligthart S, Vaez A, Võsa U, Stathopoulou MG, de Vries PS, Prins BP, Van der Most PJ, Tanaka T, Naderi E, Rose LM, Wu Y, Karlsson R, Barbalic M, Lin H, Pool R, Zhu G, Macé A, Sidore C, Trompet S, Mangino M, Sabater-Lleal M, Kemp JP, Abbasi A, Kacprowski T, Verweij N, Smith AV, Huang T, Marzi C, Feitosa MF, Lohman KK, Kleber ME, Milaneschi Y, Mueller C, Huq M, Vlachopoulou E, Lyytikäinen L-P, Oldmeadow C, Deelen J, Perola M, Zhao JH, Feenstra B, Amini M, Lahti J, Schraut KE, Fornage M, Suktitipat B, Chen W-M, Li X, Nutile T, Malerba G, Luan Jian'an, Bak T, Schork N, Del Greco M F, Thiering E, Mahajan A, Marioni RE, Mihailov E, Eriksson J, Ozel AB, Zhang W, Nethander M, Cheng Y-C, Aslibekyan S, Ang W, Gandin I, Yengo L, Portas L, Kooperberg C, Hofer E, Rajan KB, Schurmann C, den Hollander W, Ahluwalia TS, Zhao J, Draisma HHM, Ford I, Timpson N, Teumer A, Huang H, Wahl S, Liu Y, Huang J, Uh H-W, Geller F, Joshi PK, Yanek LR, Trabetti E, Lehne B, Vozzi D, Verbanck M, Biino G, Saba Y, Meulenbelt I, O'Connell JR, Laakso M, Giulianini F, Magnusson PKE, Ballantyne CM, Hottenga JJ, Montgomery GW, Rivadineira F, Rueedi R, Steri M, Herzig K-H, Stott DJ, Menni C, Frånberg M, St Pourcain B, Felix SB, Pers TH, Bakker SJL, Kraft P, Peters A, Vaidya D, Delgado G, Smit JH, Großmann V, Sinisalo J, Seppälä I, Williams SR, Holliday EG, Moed M, Langenberg C, Räikkönen K, Ding J, Campbell H, Sale MM, Chen Y-DI, James AL, Ruggiero D, Soranzo N, Hartman CA, Smith EN, Berenson GS, Fuchsberger C, Hernandez D, Tiesler CMT, Giedraitis V, Liewald D, Fischer K, Mellström D, Larsson A, Wang Y, Scott WR, Lorentzon M, Beilby J, Ryan KA, Pennell CE, Vuckovic D, Balkau B, Concas MP, Schmidt R, Mendes de Leon CF, Bottinger EP, Kloppenburg M, Paternoster L, Boehnke M, Musk AW, Willemsen G, Evans DM, Madden PAF, Kähönen M, Kutalik Z, Zoledziewska M, Karhunen V, Kritchevsky SB, Sattar N, Lachance G, Clarke R, Harris TB, Raitakari OT, Attia JR, van Heemst D, Kajantie E, Sorice R, Gambaro G, Scott RA, Hicks AA, Ferrucci L, Standl M, Lindgren CM, Starr JM, Karlsson M, Lind L, Li JZ, Chambers JC, Mori TA, de Geus EJCN, Heath AC, Martin NG, Auvinen J, Buckley BM, de Craen AJM, Waldenberger M, Strauch K, Meitinger T, Scott RJ, McEvoy M, Beekman M, Bombieri C, Ridker PM, Mohlke KL, Pedersen NL, Morrison AC, Boomsma DI, Whitfield JB, Strachan DP, Hofman A, Vollenweider P, Cucca F, Jarvelin M-R, Jukema JW, Spector TD, Hamsten A, Zeller T, Uitterlinden AG, Nauck M, Gudnason V, Qi L, Grallert H, Borecki IB, Rotter JI, März W, Wild PS, Lokki M-L, Boyle M, Salomaa V, Melbye M, Eriksson JG, Wilson JF, Penninx BWJH, Becker DM, Worrall BB, Gibson G, Krauss RM, Ciullo M, Zaza G, Wareham NJ, Oldehinkel AJ, Palmer LJ, Murray SS, Pramstaller PP, Bandinelli S, Heinrich J, Ingelsson E, Deary IJ, Mägi R, Vandenput L, van der Harst P, Desch KC, Kooner JS, Ohlsson C, Hayward C, Lehtimäki T, Shuldiner AR, Arnett DK, Beilin LJ, Robino A, Froguel P, Pirastu M, Jess T, Koenig W, Loos RJF, Evans DA, Schmidt H, Smith GD, Slagboom PE, Eiriksdottir G, Morris AP, Psaty BM, Tracy RP, Nolte IM, Boerwinkle E, Visvikis-Siest S, Reiner AP, Gross M, Bis JC, Franke L, Franco OH, Benjamin EJ, Chasman DI, Dupuis J, Snieder H, Dehghan A, Alizadeh BZ, LifeLines Cohort Study, CHARGE Inflammation Working Group . Genome Analyses of >200,000 Individuals Identify 58 Loci for Chronic Inflammation and Highlight Pathways that Link Inflammation and Complex Disorders. Am J Hum Genet 2018;103:691–706. 10.1016/j.ajhg.2018.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Järvelin MR, Sovio U, King V, Lauren L, Xu B, McCarthy MI, Hartikainen AL, Laitinen J, Zitting P, Rantakallio P, Elliott P. Early life factors and blood pressure at age 31 years in the 1966 northern Finland birth cohort. Hypertension 2004;44:838–46. [DOI] [PubMed] [Google Scholar]
- 11. Saukkonen T, Mutt SJ, Jokelainen J, Saukkonen AM, Raza GS, Karhu T, Härkönen P, Eckel J, Herzig KH, Rajala U, Keinänen-Kiukaanniemi S. Adipokines and inflammatory markers in elderly subjects with high risk of type 2 diabetes and cardiovascular disease. Sci Rep 2018;8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raitakari OT, Juonala M, Rönnemaa T, Keltikangas-Järvinen L, Räsänen L, Pietikäinen M, Hutri-Kähönen N, Taittonen L, Jokinen E, Marniemi J, Jula A, Telama R, Kähönen M, Lehtimäki T, Åkerblom HK, Viikari JSA. Cohort profile: the cardiovascular risk in young finns study. Int J Epidemiol 2008;37:1220–6. [DOI] [PubMed] [Google Scholar]
- 13. Borodulin K, Tolonen H, Jousilahti P, Jula A, Juolevi A, Koskinen S, Kuulasmaa K, Laatikainen T, Männistö S, Peltonen M, Perola M, Puska P, Salomaa V, Sundvall J, Virtanen SM, Vartiainen E. Cohort profile: the National FINRISK study. Int J Epidemiol 2017;47:696–696i. [DOI] [PubMed] [Google Scholar]
- 14. Santalahti K, Maksimow M, Airola A, Pahikkala T, Hutri-Kähönen N, Jalkanen S, Raitakari OT, Salmi M. Circulating cytokines predict the development of insulin resistance in a prospective Finnish population cohort. J Clin Endocrinol Metab 2016;101:3361–9. [DOI] [PubMed] [Google Scholar]
- 15. Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906–13. [DOI] [PubMed] [Google Scholar]
- 16. Willer CJ, Li Y, Abecasis GR. Metal: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang J, Ferreira T, Morris AP, Medland SE, Madden PAF, Heath AC, Martin NG, Montgomery GW, Weedon MN, Loos RJ, Frayling TM, McCarthy MI, Hirschhorn JN, Goddard ME, Visscher PM. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 2012;44:369–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paré G, Chasman DI, Kellogg M, RYL Z, Rifai N, Badola S, Miletich JP, Ridker PM. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet 2008;4:e1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ridker H, Buring R. C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. Ital Heart J Suppl 2000;1:1066–7. [PubMed] [Google Scholar]
- 20. Hwang SJ, Ballantyne CM, Sharrett AR, Smith LC, Davis CE, Gotto AM, Boerwinkle E. Circulating adhesion molecules VCAM-1, ICAM-1, and E-selectin in carotid atherosclerosis and incident coronary heart disease cases: the Atherosclerosis Risk in Communities (ARIC) study. Circulation 1997;96:4219–25. [DOI] [PubMed] [Google Scholar]
- 21. Pickrell JK, Berisa T, Liu JZ, Segurel L, Tung JY, Hinds D. Detection and interpretation of shared genetic influences on 42 human traits 2017;48:709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nikpay M, Goel A, Won H-H, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, Webb TR, Zeng L, Dehghan A, Alver M, Armasu SM, Auro K, Bjonnes A, Chasman DI, Chen S, Ford I, Franceschini N, Gieger C, Grace C, Gustafsson S, Huang J, Hwang S-J, Kim YK, Kleber ME, Lau KW, Lu X, Lu Y, Lyytikäinen L-P, Mihailov E, Morrison AC, Pervjakova N, Qu L, Rose LM, Salfati E, Saxena R, Scholz M, Smith AV, Tikkanen E, Uitterlinden A, Yang X, Zhang W, Zhao W, de Andrade M, de Vries PS, van Zuydam NR, Anand SS, Bertram L, Beutner F, Dedoussis G, Frossard P, Gauguier D, Goodall AH, Gottesman O, Haber M, Han B-G, Huang J, Jalilzadeh S, Kessler T, König IR, Lannfelt L, Lieb W, Lind L, Lindgren CM, Lokki M-L, Magnusson PK, Mallick NH, Mehra N, Meitinger T, Memon F-U-R, Morris AP, Nieminen MS, Pedersen NL, Peters A, Rallidis LS, Rasheed A, Samuel M, Shah SH, Sinisalo J, Stirrups KE, Trompet S, Wang L, Zaman KS, Ardissino D, Boerwinkle E, Borecki IB, Bottinger EP, Buring JE, Chambers JC, Collins R, Cupples LA, Danesh J, Demuth I, Elosua R, Epstein SE, Esko T, Feitosa MF, Franco OH, Franzosi MG, Granger CB, Gu D, Gudnason V, Hall AS, Hamsten A, Harris TB, Hazen SL, Hengstenberg C, Hofman A, Ingelsson E, Iribarren C, Jukema JW, Karhunen PJ, Kim B-J, Kooner JS, Kullo IJ, Lehtimäki T, Loos RJF, Melander O, Metspalu A, März W, Palmer CN, Perola M, Quertermous T, Rader DJ, Ridker PM, Ripatti S, Roberts R, Salomaa V, Sanghera DK, Schwartz SM, Seedorf U, Stewart AF, Stott DJ, Thiery J, Zalloua PA, O'Donnell CJ, Reilly MP, Assimes TL, Thompson JR, Erdmann J, Clarke R, Watkins H, Kathiresan S, McPherson R, Deloukas P, Schunkert H, Samani NJ. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121–30. 10.1038/ng.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese AK, Van Der Laan SW, Gretarsdottir S, Anderson CD, Chong M, Adams HHH, Ago T, Almgren P, Amouyel P, Ay H, Bartz TM, Benavente OR, Bevan S, Boncoraglio GB, Brown RD, Butterworth AS, Carrera C, Carty CL, Chasman DI, Chen WM, Cole JW, Correa A, Cotlarciuc I, Cruchaga C, Danesh J, De Bakker PIW, Destefano AL, Den Hoed M, Duan Q, Engelter ST, Falcone GJ, Gottesman RF, Grewal RP, Gudnason V, Gustafsson S, Haessler J, Harris TB, Hassan A, Havulinna AS, Heckbert SR, Holliday EG, Howard G, Hsu FC, Hyacinth HI, Ikram MA, Ingelsson E, Irvin MR, Jian X, Jiménez-Conde J, Johnson JA, Jukema JW, Kanai M, Keene KL, Kissela BM, Kleindorfer DO, Kooperberg C, Kubo M, Lange LA, Langefeld CD, Langenberg C, Launer LJ, Lee JM, Lemmens R, Leys D, Lewis CM, Lin WY, Lindgren AG, Lorentzen E, Magnusson PK, Maguire J, Manichaikul A, McArdle PF, Meschia JF, Mitchell BD, Mosley TH, Nalls MA, Ninomiya T, O’Donnell MJ, Psaty BM, Pulit SL, Rannikmäe K, Reiner AP, Rexrode KM, Rice K, Rich SS, Ridker PM, Rost NS, Rothwell PM, Rotter JI, Rundek T, Sacco RL, Sakaue S, Sale MM, Salomaa V, Sapkota BR, Schmidt R, Schmidt CO, Schminke U, Sharma P, Slowik A, Sudlow CLM, Tanislav C, Tatlisumak T, Taylor KD, Thijs VNS, Thorleifsson G, Thorsteinsdottir U, Tiedt S, Trompet S, Tzourio C, Van Duijn CM, Walters M, Wareham NJ, Wassertheil-Smoller S, Wilson JG, Wiggins KL, Yang Q, Yusuf S, Bis JC, Pastinen T, Ruusalepp A, Schadt EE, Koplev S, Björkegren JLM, Codoni V, Civelek M, Smith NL, Trégouët DA, Christophersen IE, Roselli C, Lubitz SA, Ellinor PT, Tai ES, Kooner JS, Kato N, He J, Van Der Harst P, Elliott P, Chambers JC, Takeuchi F, Johnson AD, Sanghera DK, Melander O, Jern C, Strbian D, Fernandez-Cadenas I, Longstreth WT, Rolfs A, Hata J, Woo D, Rosand J, Pare G, Hopewell JC, Saleheen D, Stefansson K, Worrall BB, Kittner SJ, Seshadri S, Fornage M, Markus HS, Howson JMM, Kamatani Y, Debette S, Dichgans M, Amin N, Aparicio HS, Arnett DK, Attia J, Beiser AS, Berr C, Buring JE, Bustamante M, Caso V, Cheng YC, Choi SH, Chowhan A, Cullell N, Dartigues JF, Delavaran H, Delgado P, Dörr M, Engström G, Ford I, Gurpreet WS, Hamsten A, Heitsch L, Hozawa A, Ibanez L, Ilinca A, Ingelsson M, Iwasaki M, Jackson RD, Jood K, Jousilahti P, Kaffashian S, Kalra L, Kamouchi M, Kitazono T, Kjartansson O, Kloss M, Koudstaal PJ, Krupinski J, Labovitz DL, Laurie CC, Levi CR, Li L, Lind L, Lindgren CM, Lioutas V, Liu YM, Lopez OL, Makoto H, Martinez-Majander N, Matsuda K, Minegishi N, Montaner J, Morris AP, Muiño E, Müller-Nurasyid M, Norrving B, Ogishima S, Parati EA, Peddareddygari LR, Pedersen NL, Pera J, Perola M, Pezzini A, Pileggi S, Rabionet R, Riba-Llena I, Ribasés M, Romero JR, Roquer J, Rudd AG, Sarin AP, Sarju R, Sarnowski C, Sasaki M, Satizabal CL, Satoh M, Sattar N, Sawada N, Sibolt G, Á S, Smith A, Sobue K, Soriano-Tárraga C, Stanne T, Stine OC, Stott DJ, Strauch K, Takai T, Tanaka H, Tanno K, Teumer A, Tomppo L, Torres-Aguila NP, Touze E, Tsugane S, Uitterlinden AG, Valdimarsson EM, Van Der Lee SJ, Völzke H, Wakai K, Weir D, Williams SR, Wolfe CDA, Wong Q, Xu H, Yamaji T. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet 2018;50:524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang H-Y, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkilä K, Hyppönen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikäinen L-P, Magnusson PKE, Mangino M, Mihailov E, Montasser ME, Müller-Nurasyid M, Nolte IM, O’Connell JR, Palmer CD, Perola M, Petersen A-K, Sanna S, Saxena R, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney ASF, Döring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen A-L, Hayward C, Hernandez D, Hicks AA, Holm H, Hung Y-J, Illig T, Jones MR, Kaleebu P, Kastelein JJP, Khaw K-T, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimäki T, Lin S-Y, Lindström J, Loos RJF, Mach F, McArdle WL, Meisinger C, Mitchell BD, Müller G, Nagaraja R, Narisu N, Nieminen TVM, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancáková A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen Y-DI, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrières J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Järvelin M-R, Jula A, Kähönen M, Kaprio J, Kesäniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, März W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njølstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PEH, Sheu WH-H, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BHR, Ordovas JM, Boerwinkle E, Palmer CNA, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR, Global Lipids Genetics Consortium,, Service SK, . Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berisa T, Pickrell JK. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics 2015;32:283–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karakas M, Baumert J, Kleber ME, Thorand B, Dallmeier D, Silbernagel G, Grammer TB, Rottbauer W, Meisinger C, Illig T, März W. Koenig W. a variant in the ABO gene explains the variation in soluble E-selectin Levels-Results from dense genotyping in two independent populations. PLoS One 2012;7:e51441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paterson AD, Lopes-Virella MF, Waggott D, Boright AP, Hosseini SM, Carter RE, Shen E, Mirea L, Bharaj B, Sun L, Bull SB. Genome-wide association identifies the ABO blood group as a major locus associated with serum levels of soluble E-selectin. Arterioscler Thromb Vasc Biol 2009;29:1958–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012;22:1790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang WH, Nussbaum C, Grewal PK, Marth JD, Sperandio M. Coordinated roles of ST3Gal-VI and ST3Gal-IV sialyltransferases in the synthesis of selectin ligands. Blood 2012;120:1015–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suhre K, Arnold M, Bhagwat AM, Cotton RJ, Engelke R, Raffler J, Sarwath H, Thareja G, Wahl A, Delisle RK, Gold L, Pezer M, Lauc G, Selim M, Mook-Kanamori DO, Al-Dous EK, Mohamoud YA, Malek J, Strauch K, Grallert H, Peters A, Kastenmüller G, Gieger C, Graumann J. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rao SP, Wang Z, Zuberi RI, Sikora L, Bahaie NS, Zuraw BL, Liu F-T, Sriramarao P. Galectin-3 functions as an adhesion molecule to support eosinophil rolling and adhesion under conditions of flow. J Immunol 2007;179:7800–7. [DOI] [PubMed] [Google Scholar]
- 32. Stowell SR, Arthur CM, Dias-Baruffi M, Rodrigues LC, Gourdine JP, Heimburg-Molinaro J, Ju T, Molinaro RJ, Rivera-Marrero C, Xia B, Smith DF, Cummings RD. Innate immune lectins kill bacteria expressing blood group antigen. Nat Med 2010;16:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pradhan AD, Rifai N, Ridker PM. Soluble intercellular adhesion molecule-1, soluble vascular adhesion molecule-1, and the development of symptomatic peripheral arterial disease in men. Circulation 2002;106:820–5. [DOI] [PubMed] [Google Scholar]
- 34. Kiechl S, Pare G, Barbalic M, Qi L, Dupuis J, Dehghan A, Bis JC, Laxton RC, Xiao Q, Bonora E, Willeit J, Xu Q, Witteman JCM, Chasman D, Tracy RP, Ballantyne CM, Ridker PM, Benjamin EJ, Ye S. Association of variation at the ABO locus with circulating levels of soluble intercellular adhesion molecule-1, soluble P-selectin, and soluble E-selectin: a meta-analysis. Circ Cardiovasc Genet 2011;4:681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Staron M, Yang Y, Liu B, Li J, Shen Y, Zuniga-Pflucker JC, Aguila HL, Goldschneider I, Li Z. Gp96, an endoplasmic reticulum master chaperone for integrins and Toll-like receptors, selectively regulates early T and B lymphopoiesis. Blood 2010;115:2380–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, Foster B, Moser M, Karasik E, Gillard B, Ramsey K, Sullivan S, Bridge J, Magazine H, Syron J, Fleming J, Siminoff L, Traino H, Mosavel M, Barker L, Jewell S, Rohrer D, Maxim D, Filkins D, Harbach P, Cortadillo E, Berghuis B, Turner L, Hudson E, Feenstra K, Sobin L, Robb J, Branton P, Korzeniewski G, Shive C, Tabor D, Qi L, Groch K, Nampally S, Buia S, Zimmerman A, Smith A, Burges R, Robinson K, Valentino K, Bradbury D, Cosentino M, Diaz-Mayoral N, Kennedy M, Engel T, Williams P, Erickson K, Ardlie K, Winckler W, Getz G, DeLuca D, MacArthur D, Kellis M, Thomson A, Young T, Gelfand E, Donovan M, Meng Y, Grant G, Mash D, Marcus Y, Basile M, Liu J, Zhu J, Tu Z, Cox NJ, Nicolae DL, Gamazon ER, HK I, Konkashbaev A, Pritchard J, Stevens M, Flutre T, Wen X, Dermitzakis ET, Lappalainen T, Guigo R, Monlong J, Sammeth M, Koller D, Battle A, Mostafavi S, McCarthy M, Rivas M, Maller J, Rusyn I, Nobel A, Wright F, Shabalin A, Feolo M, Sharopova N, Sturcke A, Paschal J, Anderson JM, Wilder EL, Derr LK, Green ED, Struewing JP, Temple G, Volpi S, Boyer JT, Thomson EJ, Guyer MS, Ng C, Abdallah A, Colantuoni D, Insel TR, Koester SE, Roger Little A, Bender PK, Lehner T, Yao Y, Compton CC, Vaught JB, Sawyer S, Lockhart NC, Demchok J, Moore HF. The Genotype-Tissue expression (GTEx) project. Nat Genet 2013;45:580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trigueros-Motos L, Van Capelleveen JC, Torta F, Castao D, Zhang LH, Chai EC, Kang M, Dimova LG, Schimmel AWM, Tietjen I, Radomski C, Tan LJ, Thiam CH, Narayanaswamy P, DH W, Dorninger F, Yakala GK, Barhdadi A, Angeli V, Dub MP, Berger J, Dallinga-Thie GM, Tietge UJF, Wenk MR, Hayden MR, Kees Hovingh G, Singaraja RR. ABCA8 regulates cholesterol efflux and high-density lipoprotein cholesterol levels. Arterioscler Thromb Vasc Biol 2017;37:2147–55. [DOI] [PubMed] [Google Scholar]
- 38. Dimayuga P, Zhu J, Oguchi S, Chyu KY, XOH X, Yano J, Shah PK, Nilsson J, Cercek B. Reconstituted HDL containing human apolipoprotein A-1 reduces VCAM-1 expression and neointima formation following periadventitial cuff-induced carotid injury in apoE null mice. Biochem Biophys Res Commun 1999;264:465–8. [DOI] [PubMed] [Google Scholar]
- 39. Kettunen J, Demirkan A, Würtz P, Draisma HHM, Haller T, Rawal R, Vaarhorst A, Kangas AJ, Lyytikäinen LP, Pirinen M, Pool R, Sarin AP, Soininen P, Tukiainen T, Wang Q, Tiainen M, Tynkkynen T, Amin N, Zeller T, Beekman M, Deelen J, Van Dijk KW, Esko T, Hottenga JJ, Van Leeuwen EM, Lehtimäki T, Mihailov E, Rose RJ, De Craen AJM, Gieger C, Kähönen M, Perola M, Blankenberg S, Savolainen MJ, Verhoeven A, Viikari J, Willemsen G, Boomsma DI, Van Duijn CM, Eriksson J, Jula A, Järvelin MR, Kaprio J, Metspalu A, Raitakari O, Salomaa V, Eline Slagboom P, Waldenberger M, Ripatti S, Ala-Korpela M. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim WS, Hsiao J-HT, Bhatia S, Glaros EN, Don AS, Tsuruoka S, Shannon Weickert C, Halliday GM. ABCA8 stimulates sphingomyelin production in oligodendrocytes. Biochem J 2013;452:401–10. [DOI] [PubMed] [Google Scholar]
- 41. Östensson M, Montén C, Bacelis J, Gudjonsdottir AH, Adamovic S, Ek J, Ascher H, Pollak E, Arnell H, Browaldh L, Agardh D, Wahlström J, Nilsson S, Å T-N. A possible mechanism behind autoimmune disorders discovered by genome-wide linkage and association analysis in celiac disease. PLoS One 2013;8:e70174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Slavin TP, Feng T, Schnell A, Zhu X, Elston RC. Two-marker association tests yield new disease associations for coronary artery disease and hypertension. Hum Genet 2011;130:725–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Temtamy SA, Aglan MS, Valencia M, Cocchi G, Pacheco M, Ashour AM, Amr KS, Helmy SMH, El-Gammal MA, Wright M, Lapunzina P, Goodship JA, Ruiz-Perez VL. Long interspersed nuclear element-1 (LINE1)-mediated deletion of EVC, EVC2, C4orf6, and STK32B in ellis-van Creveld syndrome with borderline intelligence. Hum Mutat 2008;29:931–8. [DOI] [PubMed] [Google Scholar]
- 44. Parris TZ, Aziz L, Kovács A, Hajizadeh S, Nemes S, Semaan M, Chen CY, Karlsson P, Helou K. Clinical relevance of breast cancer-related genes as potential biomarkers for oral squamous cell carcinoma. BMC Cancer 2014;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liang R, Hinds R, Abud HE, Cheng W. Hedgehog signalling is downregulated in celiac disease. Can J Gastroenterol 2013;27:e5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moran CM, Myers CT, Lewis CM, Krieg PA. Hedgehog regulates angiogenesis of intersegmental vessels through the VEGF signaling pathway. Dev Dyn 2012;241:1034–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wolf S, Mertens D, Schaffner C, Korz C, Dohner H, Stilgenbauer S. Lichter P. B-cell neoplasia associated gene with multiple splicing (BCMS): the candidate B-CLL gene on 13q14 comprises more than 560 kb covering all critical regions. Hum Mol Genet 2001;10:1275–85. [DOI] [PubMed] [Google Scholar]
- 48. Garding A, Bhattacharya N, Claus R, Ruppel M, Tschuch C, Filarsky K, Idler I, Zucknick M, Caudron-Herger M, Oakes C, Fleig V, Keklikoglou I, Allegra D, Serra L, Thakurela S, Tiwari V, Weichenhan D, Benner A, Radlwimmer B, Zentgraf H, Wiemann S, Rippe K, Plass C, Döhner H, Lichter P, Stilgenbauer S, Mertens D. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the in cis downregulation of a gene cluster that targets NF-kB. PLoS Genet 2013;9:e1003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schütze S, Wiegmann K, Machleidt T, Krönke M. TNF-induced activation of NF-kappa B. Immunobiology 1995;193:193–203. [DOI] [PubMed] [Google Scholar]
- 50. Mao X, Su Z, Mookhtiar AK. Long non-coding RNA: a versatile regulator of the nuclear factor-κB signalling circuit. Immunology 2017;150:379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jmedgenet-2018-105965supp001.pdf (42.2MB, pdf)