Abstract
Streptococcus suis is a porcine pathogen, causing severe invasive infections. S. suis serotype 9 is increasingly causing disease in Dutch and Chinese pig herds, but it is unknown whether all serotype 9 isolates are equally virulent and markers that can identify virulent strains are not available. Therefore, discrimination between virulent isolates and carriage isolates typically not associated with disease, is currently not possible. We collected tonsillar S. suis isolates from 6 herds not previously diagnosed with S. suis infections, and clinical S. suis isolates of previously diseased pigs. We confirmed the virulence of a virulent type strain and one representative clinical isolate, and the lack of virulence of two carriage isolates, in a pig infection model. Phylogenetic analysis of whole genome sequences of 124 isolates resulted in 10 groups, of which two were almost uniquely populated by clinical isolates. The population structure of S. suis serotype 9 appears highly diverse. However, analysis of the capsule loci sequences showed variation in a single region which fully correlated with a virulent genotype. Transmission electron microscopy suggested differences in capsule thickness between carriage and clinical genotypes. In conclusion, we found that that the S. suis serotype 9 population in the Netherlands is diverse. A distinct virulence-associated lineage was identified and could be discriminated based on the capsule locus sequence. Whilst the difference in virulence cannot be directly attributed to the DNA sequence, the correlation of capsule locus sequence with virulence could be used in the development of diagnostic tests to identify potential virulent S. suis serotype 9 in pigs.
Subject terms: Bacterial genes, Bacteriology, Infectious-disease diagnostics
Introduction
Streptococcus suis is an important pathogen associated with a wide range of diseases in pigs including meningitis, arthritis and sepsis1,2, leading to major economic losses in the pig industry worldwide. Healthy pigs can carry S. suis asymptomatically on the mucosal surfaces of the tonsil, nasal cavities as well as the gastrointestinal tract1,3,4. Although serotype 2 is most often associated with clinical disease in pigs worldwide5 as well as with zoonotic disease in humans6–8, the proportion of serotype 9 among isolates from diseased pigs has increased substantially in the last two decades9. Nowadays, serotype 2 and 9 isolates are the most prevalent serotypes of invasive pig isolates in the Netherlands10,11. Zoonotic serotype 2 isolates generally belong to multi-locus sequence type (MLST) 1 and 710,12,13, whereas zoonotic serotype 2 isolates belonging to ST20 diverged from serotype 9 isolates belonging to ST1611. Although serotype 9 isolates are considered non-zoonotic, a single case of serotype 9 infection in humans was reported in Thailand14.
Among serotype 2 isolates, virulent as well as less virulent isolates can be discriminated15. Virulent isolates of serotype 2 can be recognized by expression of muramidase-released protein (MRP) and extracellular factor (EF)9,16 and virulence is associated with a considerable number of putative virulence factors in S. suis serotype 217. Moreover, several antigens were described as putative vaccine candidates17,18, although vaccines based on these antigens were not developed. The majority of the invasive serotype 9 isolates express a larger variant of MRP, but do not express EF19, whilst the virulence factor suilysin was shown to be expressed20. So far it is unknown whether virulent and less virulent isolates can be discriminated among serotype 9 isolates. In addition, antigens which generate a protective immune response against serotype 9 challenge, and which could be used for vaccine development, have not been described.
Given the large contribution of serotype 9 to S. suis related disease and outbreaks in pig husbandry, as well as its potential evolution into a zoonotic pathogen, a better understanding of the population structure of S. suis serotype 9 is urgently needed to improve diagnostics and control.
We studied S. suis serotype 9 isolates obtained from the tonsils of healthy pigs from farms without overt S. suis specific disease for over 1 year (carrier isolates) and isolates obtained from clinically diseased pigs (clinical isolates). We showed that a clinical serotype 9 isolate was capable to induce severe disease in pigs after experimental infection, thus reconfirming its virulence, whereas carrier isolates did not cause severe disease. This indicates the presence of different pathotypes among S. suis serotype 9 isolates, which we further investigated using whole genome sequencing. We observed a diverse population structure among carriage serotype 9 isolates, which was previously unknown, whilst clinical isolates belonged to a single clonal expansion. Finally, we describe differences in the capsule loci between carriage and clinical isolates and propose the cpsK gene as a diagnostic target for the detection of virulent serotype 9 isolates.
Results
Sample collection
Thirty-two clinical isolates, cultured from the brains of diseased pigs from 32 different unrelated pig herds in the Netherlands, as part of standard veterinary diagnostic procedures at the laboratory of the GD Deventer, were included. An additional 16 clinical serotype 9 isolates from the collections of consortium partners were included into the study, as well as 28 clinical serotype 9 isolates from our previous study11 and isolate 8067, which is a well-studied virulent isolate9. Carrier isolates were obtained from 6 unrelated farms, with multiple isolates per farm and a maximum of one isolate per animal (Supplementary Table 1). To collect carriage isolates, tonsil swabs of 50 sows and 223 piglets from 6 different farms in the Netherlands which did not experience S. suis disease during the previous 12 months, were analyzed by using a S. suis serotype 9 specific PCR. Pig breeds included different crossbreeds of Dutch Landrace with Yorkshire (Farms B_2, B_4, B_5, B_7), Finnish Landrace with Yorkshire (Farm B_3) and English Landrace with Large White (Farm B_6) which are all commercial breeds commonly used in the Netherlands. The number of sows in these farms varied between 196 and 800, which are common pig herd sizes in the Netherlands. A total of 36 out of the 50 sows (72%) and 106 out 223 (48%) piglets tested were positive in a serotype 9 specific PCR, with varying fractions of positive tonsil swabs between the farms (Supplementary Table 2). These data clearly indicate that high percentages of the piglets and sows carried S. suis serotype 9 isolates on their tonsils on farms without overt S. suis disease during the 12 months prior to sampling. Forty-six carriage isolates were selected for this study. One additional carriage isolate was provided by a consortium member from France. Taken together, 47 carriage isolates and 77 invasive isolates were used in this study (Supplementary Table 1).
Virulence of serotype 9 isolates
Isolates were defined as virulent if grown from sterile sites (blood, cerebrospinal fluid, brain) or in pure culture from pigs with clinical disease suspect of S. suis infection. Thus, clinical isolates included in this study are considered virulent. The virulence of serotype 9 isolates was confirmed in an infection experiment in piglets using two randomly selected carrier isolates obtained from the tonsils of healthy pigs of two different herds (21853 and 21900) and one clinical isolate (21970). Serotype 9 strain 8067, a clinical isolate previously used in experimental infections in piglets20 was used as a positive control. Caesarean derived, colostrum deprived (CDCD) piglets at 6 weeks of age were inoculated intravenously with a dose of 5·107 CFU and monitored for 8 days.
All 14 animals inoculated with the clinical isolates were euthanized within two days after inoculation because of serious signs of a S. suis infection, mainly arthritis and meningitis (Fig. 1). In contrast, 3 out of 16 piglets inoculated with the carrier strains were killed 4 and 7 days after inoculation with clinical signs of arthritis. The mean number of days until death of diseased pigs was 1.6 and 1.3 for animals inoculated with the clinical strains 8067 and 21970 and 7.6 and 7.5 days for the carrier strains 21853 and 21900, respectively (Table 1). Significant differences were observed in the number of specific clinical signs of disease between piglets inoculated with the clinical isolates and with the carrier isolates obtained from healthy piglets (Supplementary Fig. 1). S. suis was isolated at significantly higher frequency from the joints and meninges of piglets inoculated with the clinical isolates than after inoculation with isolates from healthy pigs (Fig. 2). Gross pathology findings revealed an acute arthritis due to inflammation of the synovial membrane or peri-articular soft tissues in one or more joints in all pigs (100%) of both groups inoculated with clinical isolates and 3 pigs (18%) of the groups inoculated with the carrier isolates. The histological findings in the synovia were consistent with S. suis typical inflammation ranging from diffuse mild inflammation to severe fibrinopurulent inflammation of the synovial membrane (Fig. 3). No inflammation of the serosae of the body cavities was seen in the pigs infected with carrier isolates, but this was found in 10 of 14 pigs (70%) infected with clinical strains. Typical histological changes in the brains consisted of inflammation of cerebral and cerebellum meninges varying from diffuse interstitial edema and congestion to fibrinopurulent meningitis and incidentally meningoencephalitis, which was observed in 29% of all pigs of both groups infected with clinical isolates, but in none of the pigs infected with the carrier isolates. In addition, only in the groups inoculated with clinical isolates an acute interstitial pneumonia with diffuse hyper cellularity of alveolar septae was observed in more than 70% of the pigs (Supplementary Fig. 1B).
Figure 1.
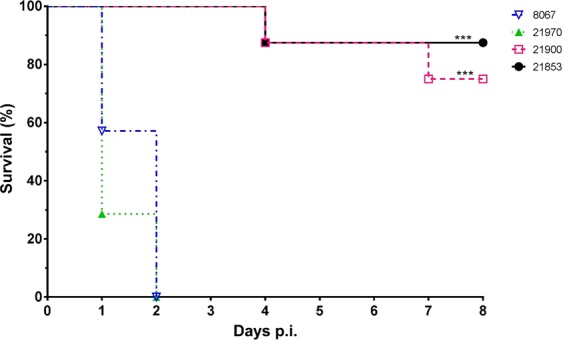
Kaplan-Meier survival curve of experimental infection of 30 pigs with two carriage isolates (21853 and 21900) and two clinical isolates (21970 and 8067). Pigs were randomly allocated to one of four groups and intravenously inoculated with 1 ml of PBS containing 5∙107 CFU. Groups infected with clinical isolates consisted of 7 pigs, whilst groups infected with carriage isolates consisted of 8 pigs. All pigs in both groups inoculated with the clinical isolates died or were euthanized after two days. In contrast, only 1 or 2 pigs inoculated with the carriage isolates died before the end of the experiment, but after at least 4 days after inoculation. ***P < 0.001 (Mantel-Cox Log-rank test).
Table 1.
| S. suis isolate | No. of pigs | Mortality (%) | Mean No. of days until death | Specific symptoms No. obs/total No. obs |
|---|---|---|---|---|
| 21853 | 8 | 13 | 7.6 | 4/128 |
| 21900 | 8 | 25 | 7.5 | 11/128 |
| 21970 | 7 | 100 | 1.3 | 20/22 |
| 8067 | 7 | 100 | 1.6 | 25/36 |
Figure 2.
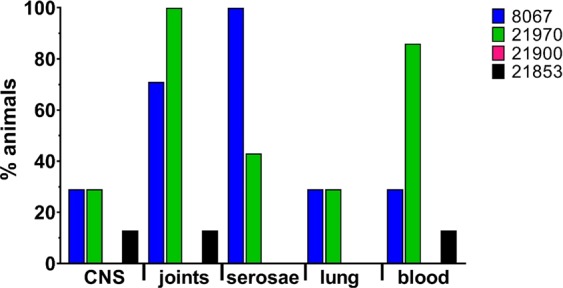
Percentage of animals from which S. suis could be isolated from the indicated organs postmortem. S. suis was isolated from CNS (meninges), one or several joints, serosae (peritoneal, pleural or pericardial), lungs and blood at a higher percentage of pigs inoculated with the clinical isolates 8067 (n = 7) and 21970 (n = 7) than from piglets inoculated with the carriage isolates 21900 (n = 8) and 21853 (n = 8). Each bar represents a group of pigs inoculated with one of the experimental S. suis isolates.
Figure 3.
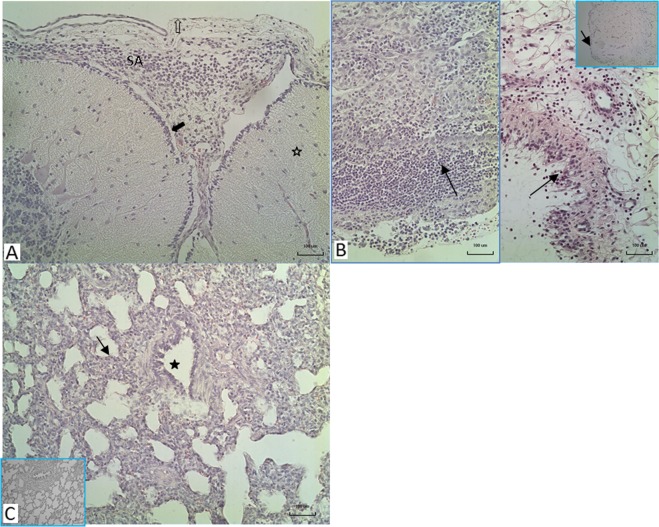
Histopathological findings in the brains, joints and lungs of pigs infected with clinical isolates of S. suis type 9. (A) Meningitis of the brain, here cerebellum (asterisk) consisting of infiltration of leucocytes and fibrin in the subarachnoidal space (SA), pia (solid arrow), arachnoid membrane (open arrow). (B) Various degrees of inflammation of the synovial membrane after infection ranging from severe fibrinous-suppurative inflammation (left part) to a mild to moderate mixed leucocyte infiltration in the intima (arrows) and subintima of the synovium, (inset: normal appearance of the synovium). (C) Acute interstitial pneumonia with lymphocyte and macrophage infiltration in the alveolar septae (arrow), no changes in the bronchioles (asterisk), (inset: normal histological appearance of lung); H&E staining ×10 objective magnification.
Bacterial population structure
Forty-seven S. suis serotype 9 isolates collected from farms without serotype 9 infections were sequenced together with 77S. suis serotype 9 isolates obtained from diseased pigs to determine the population structure and to investigate the genetic differences between carriage and clinical isolates. Draft assemblies of the isolates were annotated using the Prokka21. The clinical isolates demonstrated significant larger genome sizes compared to the carriage isolates (Supplementary Fig. 2). The annotation files were further used as input for Roary22, which created homology groups based on 95% identity at the amino acid level. The resulting core genome consisted of 1149 genes and the pangenome consisted of 6787 genes. A core genome alignment was used as input for a Bayesian Analysis of Population Structure (BAPS) to cluster the isolates into population groups. BAPS identified 10 clusters with a set maximum of 20 clusters (Supplementary Fig. 3). Out of 77 clinical isolates, 73 (95%) isolates clustered into BAPS group 2. Of the remaining 4 isolates, 3 isolates clustered into BAPS group 4 and 1 isolate clustered into BAPS group 8. BAPS group 8 is a residual group consisting of 3 isolates that are not similar to each other, but could also not be clustered in any other BAPS group. We therefore designated BAPS groups 2 and 4 as the invasive population groups. The 47 carriage isolates were more diverse as they clustered into 9 BAPS groups, and only BAPS group 4 did not contain carriage isolates (Supplementary Fig. 4). MLST Sequence types (STs) were extracted from the sequencing reads, which resulted in 14 novel STs of which 10 STs belonged to carriage isolates (Supplementary Table 3). The diversity of STs among the carriage isolates was consistent with the diversity of BAPS groups.
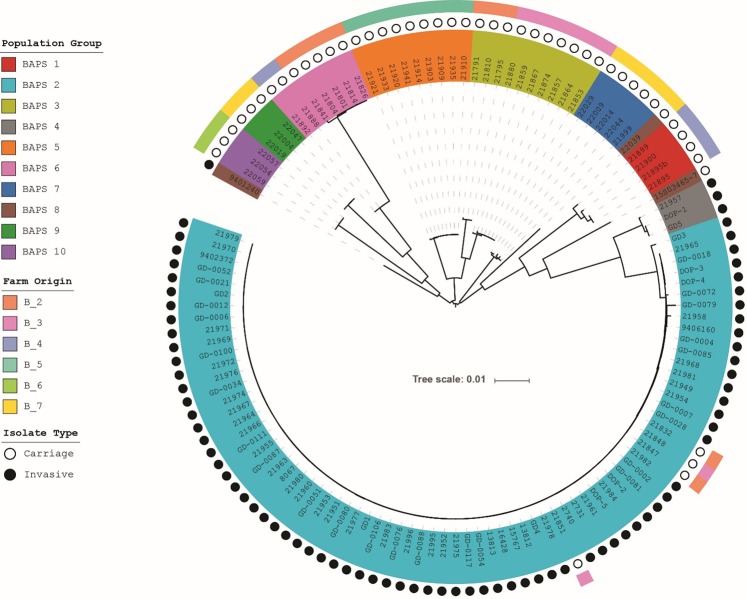
A phylogenetic tree of the core genome was created using RAxML23 to understand the relationships between the population groups (Supplementary Fig. 5). The invasive BAPS population groups 2 and 4 clustered near each other in the tree, whilst other population groups branched away from the invasive isolates revealing the population structure of serotype 9 isolates included in this study. BAPS group 8 consisted of 3 isolates, which did not group together in the tree indicating that these isolates are outliers in the serotype 9 population structure. The population tree was overlaid with the origin of the farms from which the carriage isolates were collected (Fig. 4). A correlation between BAPS groups and the farm origin was observed. Each carriage BAPS group was found to be associated with one or two farms only. Additionally, farms B_2 and B_3, without disease in the past 12 months, each had two isolates present belonging to invasive BAPS group 2. The virulent isolates used in the infection experiment were confirmed as representatives of the main clonal cluster of BAPS group 2 in a phylogenetic analysis of this group (Supplementary Fig. 6)
Figure 4.
Phylogenetic relationship between invasive and carriage isolates. The BAPS group colors are highlighted on the isolate’s name. The second ring of dots indicates the phenotype of the isolates as invasive (black) or carriage (white). The colors on the outer ring represent the farms from which the carriage isolates have been collected.
Capsule locus analysis
To investigate the relation between the single clonal expansion of invasive isolates and genomic virulence determinants we extracted the regions corresponding to the capsule genes from the various BAPS groups. The capsule locus ranged from cpsA (wzg; SSUD12_1370 in S. suis D12) until the glf gene (UDP-galactopyranose mutase; SSUD12_1356 in S. suis D1224). The genes were concatenated to form a nucleotide alignment, which was used to create a phylogenetic tree (Fig. 5). The structure of the tree differs from the structure of the tree generated from the core genome alignment (Supplementary Fig. 5). BAPS groups 1, 3, 5, 6 and 7 containing carriage isolates clustered together and opposite to the virulent BAPS groups 2 and 4. BAPS groups 8, 9 and 10 containing carriage isolates as well, are intermediates. Again, isolates of BAPS group 8 did not cluster together, and are outliers in their capsule locus as well as in their core genome.
Figure 5.
Phylogenetic tree based on the concatenated nucleotide alignment of capsule genes cpsA-cpsN. For clarity, names of the isolates are omitted. Colored circles indicate the BAPS population groups to which an isolate belongs. BAPS groups 1, 3 and 5–7 are highly similar and are located under the red circle of BAPS group 1. A filled or empty triangle respectively represents the presence or absence of a deletion between the cpsN and glf genes.
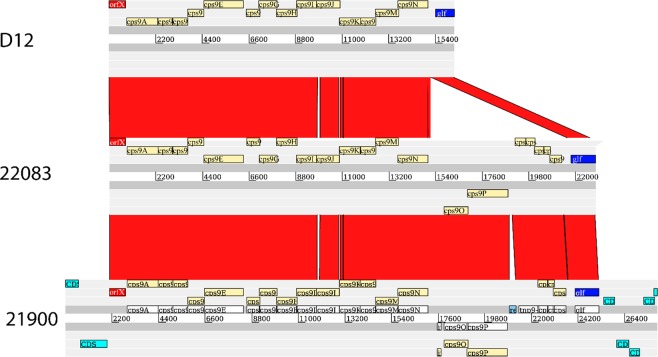
However, an analysis of the capsule locus did reveal that only two different capsule loci were present across BAPS population groups (indicated by filled and empty triangles in the phylogenetic tree in Fig. 5). More detailed investigation of these two capsule loci showed high similarity in gene content except for a deletion in the capsule locus of the invasive isolates before the glf gene and after cpsN (wzx or a putative flippase; SSUD12_1357 in S. suis D12) (Fig. 6). This deleted region encodes an integrase fragment, transposases, hypothetical proteins, and a Type I specificity (S) subunit and a Type I modification (M) subunit. The deletion is significantly associated with the invasive BAPS groups 2 and 4 (80/80) compared to the non-invasive BAPS groups 1, 3, 5, 6, 7, 9 and 10 (8/41); p-value < 0.001 (Pearson’s Chi-squared test).
Figure 6.
Comparison of the serotype 9 capsule loci of isolates D12, 22083 and 21900. Both D12 and 22083 were annotated by Prokka, whilst 21900 was annotated previously24. The capsule loci are similar except for the deletion after cpsN. The deleted area contains cps9O and cps9P, which may form a methyltransferase.
The proteins translated from the capsule locus of the various isolates were also individually compared to determine if certain genes differed more than others from the average difference. The genes for comparisons were chosen from isolates GD2, as a representative of virulent ST16 clonal complex isolates, and 21853, as representative of carriage isolates. The genes were compared at the protein level and the amino acid (aa) identities were plotted for each protein (Supplementary Fig. 7). The CpsA and CpsK proteins had less than 95% identity, and are therefore most discriminative between the two loci. Blast identified CpsA as a LytR-CpsA-PSR (LCP) protein containing two domains: DNA_PPF or DNA polymerase processivity factor (pfam02916) and LCP (pfam03816). The domain with the lowest amino acid identities between the two proteins was the DNA_PFF domain. CpsK was identified as a capsular polysaccharide synthesis protein with domain pfam05704. The largest differences in amino acids could be found between aa 200 and 250 (Supplementary Fig. 8). This deviant region is present in all isolates in BAPS groups 2 and 4.
A BLAST search was performed to investigate the association of the deviant region in the CpsK protein with clinical isolates. The search was done by aligning the invasive isolate GD2 CpsK complete protein with the NCBI database, which identified a few proteins with over 80% identity (Table 2). The majority of sequences with a high identity originate from invasive serotype 9 isolates contributed from our previous study11 and were included in this study and therefore excluded from Table 2. The foreign isolates with 99% amino acid identity had invasive as well as carriage phenotypes attributed to them and included isolates from China and the UK, as well as 3 isolates (LSS92, LSS70 and SS981) with unknown phenotypes.
Table 2.
List of foreign serotype 9 isolates not included in this study with their amino acid identities with CpsK of isolate GD2.
| S. suis isolate | Identity (%) | Phenotype | Country | Reference |
|---|---|---|---|---|
| LS2N | 99 | non-clinical | United Kingdom | Weinert et al.26 |
| LSS92 | 99 | NA | NA | NA |
| D12 | 99 | pneumonia | China | Zhang et al.26 |
| LS0P | 99 | non-clinical | United Kingdom | Weinert et al.26 |
| S91K | 99 | systemic (brain) | United Kingdom | Weinert et al.26 |
| DN13 | 99 | NA | China | NA |
| GZ0565 | 99 | meningitis | China | Wu et al. (2007) |
| LSS70 | 99 | NA | NA | NA |
| SS981 | 99 | NA | NA | NA |
| 22083 | 94 | NA | NA | Okura et al.24 |
| LS5Y | 94 | non-clinical | United Kingdom | Weinert et al.26 |
| LS6K | 94 | NA | United Kingdom | Weinert et al.26 |
| LSS55 | 94 | NA | NA | NA |
| LSS66 | 94 | NA | NA | NA |
For 6 matches, the amino acid identity was 94%. These matches included strain 21853 and serotype 9 reference strain 2208325. For these 6 isolates, the phenotype was unknown for 4 isolates and non-clinical for one isolate. Finally, 9401240 is a continuous outlier with its invasive phenotype in this study as well as the previous11. The known phenotypes of previously submitted serotype 9 strains are limited, but when present these metadata corroborate our phenotypical observations on virulence and carriage.
The differences observed in the capsule locus between the invasive and carriage isolates prompted us to look for differences in the capsule itself by means of Transmission Electron Microscopy (TEM). Carriage isolates 21900 and 21853 and invasive isolates 8067 and GD2 were selected for TEM (Supplementary Fig. 9). These isolates were also used in the virulence in vivo experiment except for isolate GD2, which was chosen instead of 21970 to include an ST16 isolate. The cps thickness of carriage isolates when fixated in McDowell’s fixative was 20.5 ± 7.1 nm and the thickness of invasive isolates 20.9 ± 6.4 nm. This difference was not significant (p = 0.81). However, when lysine-acetate was added to the fixative, it was impossible to measure the cps thickness of carriage isolates 21853 and 21900 due to absence of the layer, while the thickness of virulent isolates was 32.9 ± 15.4. No differences were observed between the two fixatives for the positive control S. suis S10 and negative control non-encapsulated S. suis J28.
Discussion
We performed a systematic analysis of carriage isolates from the tonsils of healthy pigs and clinical S. suis serotype 9 isolates in the Netherlands. The clinical isolates obtained from individual diseased pigs clustered mainly together in one phylogenetic group (BAPS group 2), whereas the carriage isolates, which originated from 6 different herds, were distributed across 8 different groups. The clinical isolates had higher gene counts, which is in contrast to previous findings, where invasive isolates of multiple serotypes had smaller genomes than carriage isolates26. These data could reflect the difference in virulence between the isolates belonging to the two different clusters, which was confirmed with representative isolates in an infection experiment. The results of this experiment indicated that the virulence of the tested clinical isolates and tonsillar isolates from healthy pigs differed significantly.
Although S. suis serotype 9 isolates are nowadays very frequently isolated from diseased pigs in the field, serotype 9 isolates were under experimental conditions shown to be less pathogenic compared to serotype 2 isolates20,27. Still, intravenous infection with high dose of S. suis serotype 9 (ST16) induced mortality and specific clinical symptoms28. In this study, we used highly susceptible CDCD piglets to compare the pathogenicity of the clinical and tonsillar serotype 9 isolates. By using an infection dose of 5·107 CFU, clear signs of S. suis disease were induced in the pigs after inoculation with virulent isolates, whereas only minor signs of S. suis disease were induced by the carrier isolates. The clinical course after infection with the virulent isolates, but not with carrier isolates, and the pathological findings in the meninges, the synovial membranes and the serosae, are in line with those observed under field conditions and experimental conditions. Whether S. suis is an important cause of pneumonia in pigs remains under debate and so far, clear experimental evidence in support is still lacking. Interestingly, in this study, in 70% of diseased pigs an acute, interstitial pneumonia with increased numbers of macrophages and lymphocytes in the alveolar wall, but no signs of an exudative broncho-pneumonia, was observed. These findings may be related to the occurring bacteremia, which may facilitate subsequent localized infections or secondary infection with other bacterial pathogens.
Here we analyzed a large collection of serotype 9 isolates from the Netherlands and revealed that these isolates belonged to a total of 21 different STs. Ten STs were associated with the clinical phenotype and 11 STs with the carriage phenotype. All together 14 new STs were determined amongst carriage isolates. Of all clinical isolates, 95% belonged to ST16 or a SLV of ST16. These results clearly indicate that isolates belonging to ST16 are mainly responsible for the disease caused by serotype 9 infections among pigs in the Netherlands. Thus far, clinical invasive serotype 9 isolates collected in the Netherlands were associated with ST1610,11, whereas in Asia serotype 9 isolates were very diverse and associated with a range of singleton STs29. Our data demonstrates that clinical isolates from The Netherlands have a different population structure than serotype 9 isolates from China. Interestingly, 2 out of 7 farms (B_2 and B_6) which were sampled to collect carriage isolates, had disease outbreaks caused by serotype 9 isolates after finishing the collection of these samples (personal communication from farmers). Interestingly, we showed that piglets sampled at farms B_2 and B_3 did carry ST16 isolates of the virulence associated BAPS group 2 (isolates 21832, 21848, 21847, and 21851) at the time of sampling. Unfortunately, we do not know whether the reported infections in farm B_2 were caused by (a SLV of) a ST16 isolate. Therefore, we cannot conclude whether these four isolates are asymptomatically carried virulent isolates or non-virulent isolates. The only non-novel ST among the carriage isolates, ST48, was found on farm B_6. According to the MLST database, ST48 was isolated in 1998 from the brain of a pig with meningitis in the UK (Y02316). This suggests that under certain conditions, ST48 isolates can be virulent. Unfortunately, it is not known which ST caused problems on farm B_6 after tonsil sampling.
Recently, Dong and co-workers compared 30S. suis serotype 9 isolates mainly obtained from China and Vietnam by MLST29. The isolates were designated to two different clusters based on the presence/absence of 23 different virulence-related genes. However, using a mice model, no difference in virulence was observed between isolates belonging to the two different clusters, further demonstrating the potential weakness of mouse models to assess virulence of a porcine pathogen.
Whole genome sequencing of invasive and carriage isolates from this study revealed that the average genome size of the virulent isolates was larger compared to the genome size of carriage isolates. This finding is in contrast to previous findings, where more virulent isolates had smaller genomes26. The genomes of the serotype 9 isolates contained two different capsular loci, which were correlated with virulent or carriage isolates respectively (Figs 5 and 6). Interestingly, the carriage isolates showed much more similarity with each other when comparing them on the capsule locus, than compared to the core genome, which suggests a higher conservation of the capsular locus compared to the core genome. The two different capsular loci were previously described for serotype 9 isolates 22083 and D1224. Isolate 22083 is the serotype 9 reference isolate with an unknown clinical phenotype, whereas D12 was an invasive isolate causing pneumonia in a pig from China. Compared to isolate 22083, the capsular locus of isolate D12 contains a deletion24. We here show that a carriage-associated capsule locus was identified in isolate 22083, whereas an virulence-associated capsule locus was identified in S. suis D12. Compared to most of the genes in the capsule locus, the fragment absent in the virulence-associated capsule locus contained genes transcribed in the reverse direction, encoding a Type I S and Type I M subunit. This indicates that a methyltransferase could be missing in the virulent genomes. Methylation patterns of the genome can be important for overall transcription levels30, but regulatory capabilities due to rapid switching as typical for type I restriction-modification systems are unlikely to explain the phenotype due to the lack of nearby additional Methylase_S domains31,32.
The most dissimilar genes in the capsule locus are cpsA and cpsK (Supplementary Fig. 7). CpsK is a capsular synthesis protein. The structure of the serotype 9 capsular polysaccharide was recently determined and the proposed function of CpsK is to transfer the last rhamnose to the glucitol33. We postulate that the cpsK gene is an interesting diagnostic target as the region between 600 and 750 nucleotides is highly different between the virulent BAPS 2 isolates and the carriage BAPS groups. The cpsK gene is also highly specific for serotype 9S. suis isolates as was shown by blast search. Protein blast hits with an identity over 80% included isolates from the UK that were previously classified as non-typable26. However, their genome sequences contained a serotype 9 capsule locus, which was consistent with the recently released S. suis_serotyping pipeline34. The function of CpsK in S. suis serotypes 1, 1/2, 2, and 14, was recently elucidated, and found to be crucial in the determination of the serotype. A single amino acid substitution resulted in a serotype switch from serotype 1 to 14 or 2 to 1/235.
Both the thickness and composition of the polysaccharide capsule are associated with virulence36. A visualization of the capsule layer by TEM did not reveal any differences in capsule thickness when McDowell’s fixative was used. However, after addition of lysine acetate, a clear difference was observed in the thickness of the capsule layers. The observed differences upon lysine acetate addition to the fixative could indicate a difference in capsule composition. The diamine lysine acetate aids in the fixation of hydrated structures, such as cps37. The influence of lysine acetate on the fixation of S. suis cps differs per serotype38. These results indicate a difference in capsule layer, but to what extend and how difference in the thickness of the capsule layer mediates virulence remains to be investigated.
With this study, we significantly broadened the knowledge on the important, but highly understudied S. suis serotype 9. We demonstrated a previously unseen depth in the population structure among carriage isolates of S. suis serotype 9 in The Netherlands. Two pathotypes were discriminated in an experimental infection model and studied by whole genome sequencing. Our analyses identified genomic signatures in the capsule locus that could differentiate between virulent and carriage S. suis isolates. This would potentially allow the development of a discriminatory diagnostic test which can aid in reducing the burden of S. suis serotype 9 related infections on pig farms. Future vaccine development strategies maybe therefore be focused on the virulent isolates exclusively. Such improved diagnostics and preventive vaccination can help in combatting S. suis infections in the field.
Methods
Sample collection
(1)S. suis serotype 9 strains from clinically diseased piglets. The GD Animal Health (Deventer, The Netherlands) kindly provided S. suis serotype 9 isolates obtained from the brains of diseased piglets. The 32 clinical S. suis isolates used in this study were obtained from 32 different pig herds in the Netherlands. An additional 16 clinical serotype 9 isolates from the collections of the partners were included. Moreover, 28 clinical serotype 9 isolates from our previous study11, as well as virulent isolate 80679 were included into the study (Supplementary Table 1).
(2)S. suis serotype 9 strains from tonsil samples of healthy pigs. Seven pig herds in the Netherlands, which did not use S. suis specific autovaccines nor used feed medication and where pigs did not show signs of meningitis, arthritis, or sudden death for over 1 year were selected. Tonsil swabs from sows and piglets (9–10 weeks of age) were collected as described before39. Swabs were transferred to the laboratory at 4 °C in 2 ml Todd–Hewitt broth with 0.25% Streptococcus Selective Supplement (Oxoid) and 0.2 μg/ml cristalviolet. To elute the bacteria swabs were subsequently sonicated in a water bath (Ultrasonic Cleaner, VWR symphony) at room temperature for 90 min. The material was then stored at −70 °C in the presence of 15% glycerol.
A serotype 9 specific PCR was used to identify tonsil samples positive for S. suis serotype 9 isolates40,41. Therefore 200 µl of the tonsil material was incubated overnight at 37 °C in 1.5 ml Todd Hewitt Broth containing 0.25% Streptococcus Selective Supplement (Oxoid) and 0.2 μg/ml cristalviolet. Fifty µl of the samples was subsequently used for DNA isolation41.
Tonsil samples positive in the PCR were used for isolation of S. suis serotype 9 isolates. Therefore, tonsil samples were plated on Columbia blood agar plates supplemented with 6% horse blood, 0.25% Streptococcus Selective Supplement (Oxoid) and 0.2 μg/ml cristalviolet. Plates were incubated overnight at 37 °C. Colonies were lifted onto sterile GeneScreen Plus membranes (New-England Nuclear Corp., Boston, USA) and hybridized to a 32P-labeled serotype 9 specific probe. Hybridizing colonies were picked from the original plates and used for further characterization and typing.
Experimental infection in pigs
Thirty caesarean-derived and colostrum deprived (CDCD) piglets (breed: Topigs 20) were obtained from 6 sows of a herd with a high health status and free of relevant pig pathogens. Piglets were produced and raised by WBVR housed at the animal facilities of WBVR with ad libitum access to water and feed. Three days before the inoculation the pigs were transferred to the final animal rooms. Animals were randomized by sex and weight and allocated to two groups of 7 and two groups of 8 piglets. At the age of 6 weeks the animals were injected intravenously with 1 ml of PBS containing 5 * 107 colony forming units (CFU) of S. suis serotype 9 isolates. To monitor the health status of piglets, body temperatures and clinical scores were systematically recorded twice a day, starting at 1 day before inoculation to record baseline levels. Piglets were followed clinically twice daily with special regard to signs of meningitis and arthritis. Blood was collected from the jugular vein at the day before inoculation and days 4, 6 and 8 after inoculation to monitor white blood cell counts (WBC) and bacteremia. WBCs were counted using an automated cell counter (Sysmex, pocH-100iV-diff).
Pathology
Pigs were sacrificed at 8 days post-infection or when pigs reached predefined humane end points and gross pathology performed with special emphasis on changes in joints of all limbs, meninges of cerebrum and cerebellum, serous surfaces of abdomen and thorax cavity, heart lung, spleen, liver and kidneys. From all of these organs/sites samples were taken by disposable inoculation loops for bacteriological examination. Tissue specimens from several CNS sites, heart, lung, joint capsules, liver, kidneys and spleen were formalin fixed for histological examination. Formalin fixed organ material was embedded in paraffin, sectioned at 3–5 µm, and subsequently stained with haematoxylin and eosin. Sections were microscopically screened for pathological changes.
Whole genome sequencing
The selection of isolates for sequencing consisted of 46 carriage isolates and 32 invasive isolates from the sample collection phase of this study. Companies in our consortium contributed an additional 16 invasive serotype 9 isolates and one carriage isolate from France. Finally, the 28 invasive serotype 9 isolates from our previous study11 were also included, as well as invasive isolate 80679 (Supplementary Table 1).
Bacterial isolates were sequenced by paired-end HiSeq sequencing with 2 * 125 basepairs. Adapters were trimmed from the reads using cutadapt (https://cutadapt.readthedocs.io/en/stable/) and the reads were further trimmed for quality by sickle (https://github.com/najoshi/sickle) with a cutoff Phred score of 20. Assemblies were constructed using SPAdes 3.9.042 using the long Illumina paired reads (2 * 150 basepairs) settings including the–careful setting. Contigs with <10 coverage and/or <500 basepairs were excluded from further analyses. Assembly statistics have been added in (Supplementary Table 4) and sequencing reads and assemblies have been submitted to the European Nucleotide Archive.
Genomic analysis
Serotype and sequence type of the isolates were obtained using SRST2 0.2.043 and the Ssuis_serotypingPipeline34. Isolates not belonging to the S. suis species either due to lack of a RecN gene or isolates that expressed serotypes other than serotype 9 were excluded from further analyses. Novel alleles and novel sequence types were submitted to the S. suis MLST database (http://pubmlst.org/ssuis/). Prokka 1.9 (https://github.com/tseemann/prokka) was run on the draft assemblies and the GFF output files were fed into Roary 3.6.822 with default settings. Roary clustered the genes into homology groups creating a core genome and a pangenome. The core genome alignment from Roary was used by BAPS 6.044 (in particular hierBAPS) to cluster the isolates into population groups with a single level of hierarchy and a prior upper boundary of 20 clusters (Supplementary Fig. 3). All phylogenetic trees were created using RAxML 8.1.623 with the GTRGAMMA substitution matrix and with as many bootstraps until the bootstopping criterion was reached. Best trees were visualized using iTOL v345. Annotated genomes were viewed using Artemis 16.0.046 and genomic comparisons (e.g. capsule loci) were performed using ACT 13.0.047.
Transmission electron microscopy
Bacteria were grown overnight in Todd Hewitt broth with 5% Yeast extract. The next morning, a growth curve was started and the bacteria were collected in log phase (OD = ~0.7). Bacteria were immediately prefixed in 1% glutaraldehyde, 4% paraformaldehyde in 0.1 M sodium cacodylate (Merck) with or without 1.5% (w/v) lysine acetate. After fixation, the samples were washed in 0.1 M phosphate buffer followed by a washing step in distilled water, osmicated for 60 minutes in 1% OsO4 in water and washed again in distilled water. Washed bacteria were subsequently dehydrated through a series of ethanol’s and embedded in resin LX-112 (Ladd). The resin blocks were polymerized for 48 hours at 60 °C. Ultrathin sections of 80 nm were cut on a Reichert EM UC6 with a diamond knife, collected on formvar coated grids and stained with uranyl acetate and lead citrate. Photographs were taken on a Tecnai electron microscope with a Veleta camera. Image analysis and measurements were performed with ImageJ (Version 1.52e). Images were obtained from 2 independent replicates of each isolate. A random selection of 8 bacteria of each replicate was measured, resulting in 16 measurements per isolate and 32 for each group of carriage and invasive isolates. A t-test was performed to determine significance of the differences. For bacteria fixed in McDowell’s with lysine acetate, 10 bacteria of each isolate were analyzed for the presence of a capsule layer.
Ethics statement
The established principles of laboratory animal use and the EU and Dutch laws related to animal experiments were adhered to in this study. The Dutch Commission for Animal Studies approved the project”Vaccine development to combat Streptococcus suis infections” under number AVD401002015140. The animal experiment was approved by the Ethical Committee of Wageningen Bioveterinary Research (The Netherlands), in accordance with the Dutch law on animal experiments (permit number 2016054).
Data Access
Sequencing data for this study have been deposited in the European Nucleotide Archive (ENA) in study accession: http://www.ebi.ac.uk/ena/data/view/PRJEB20548. Assemblies for isolates can be found under accession numbers as indicated in Supplementary Table 4. Additional metadata of the sequenced isolates can be found in Supplementary Table 4.
Supplementary information
Acknowledgements
The authors thank Frits Bouwkamp (G.D. Animal Health, Deventer, The Netherlands), Rudolf Raymakers (Veterinair Centrum Someren, Someren, The Netherlands), Hetty Schreurs (Wageningen Bioveterinary Research, Lelystad, The Netherlands), Bas Swildens (Faculty of Veterinary Medicine, University Utrecht, Utrecht, The Netherlands), Yvonne Verbeek (Dopharma Research B.V.) and Marc Martens (Topigs Norsvinn) for providing clinical S. suis serotype 9 isolates and/or for their help in identifying farms and collecting carrier isolates and Arie van der Ende (National Reference Laboratory of Bacterial Meningitis, Amsterdam UMC, University of Amsterdam, Amsterdam, the Netherlands) for his advice throughout the project. The authors thank the former Product Board for Livestock & Meat and the Ministry of Agriculture, Nature and Food Quality for financing strain collection. This work was part of public private partnership that was funded by the ministry of Economic Affairs of The Netherlands (AF15227).
Author contributions
C.S. and H.E.S. conceived the study. N.S.-Z. performed pathological analysis, A.G. conducted the animal experiment, C.S.-S. isolated S. suis carrier isolates, H.W. coordinated the collection of S. suis isolates, N.W. performed all genomic analyses. D.P. performed imaging experiments, K.A. contributed to analysis of data. N.W. drafted the manuscript, and K.A., A.G., N.S.-Z., C.S. and H.E.S. contributed to the writing of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-51576-0.
References
- 1.Staats JJ, Feder I, Okwumabua O, Chengappa MM. Streptococcus suis: past and present. Vet Res Commun. 1997;21:381–407. doi: 10.1023/A:1005870317757. [DOI] [PubMed] [Google Scholar]
- 2.Wertheim HF, Nghia HD, Taylor W, Schultsz C. Streptococcus suis: an emerging human pathogen. Clin Infect Dis. 2009;48:617–625. doi: 10.1086/596763. [DOI] [PubMed] [Google Scholar]
- 3.Gottschalk M, Xu J, Calzas C, Segura M. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future microbiology. 2010;5:371–391. doi: 10.2217/fmb.10.2. [DOI] [PubMed] [Google Scholar]
- 4.Su Y, Yao W, Perez-Gutierrez ON, Smidt H, Zhu WY. Changes in abundance of Lactobacillus spp. and Streptococcus suis in the stomach, jejunum and ileum of piglets after weaning. FEMS microbiology ecology. 2008;66:546–555. doi: 10.1111/j.1574-6941.2008.00529.x. [DOI] [PubMed] [Google Scholar]
- 5.Goyette-Desjardins G, Auger JP, Xu J, Segura M, Gottschalk M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect. 2014;3:e45. doi: 10.1038/emi.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang J, et al. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS medicine. 2006;3:e151. doi: 10.1371/journal.pmed.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takamatsu D, et al. Streptococcus suis in humans, Thailand. Emerging infectious diseases. 2008;14:181–183. doi: 10.3201/eid1401.070568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wertheim HF, et al. Streptococcus suis, an important cause of adult bacterial meningitis in northern Vietnam. PloS one. 2009;4:e5973. doi: 10.1371/journal.pone.0005973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wisselink HJ, Smith HE, Stockhofe-Zurwieden N, Peperkamp K, Vecht U. Distribution of capsular types and production of muramidase-released protein (MRP) and extracellular factor (EF) of Streptococcus suis strains isolated from diseased pigs in seven European countries. Vet Microbiol. 2000;74:237–248. doi: 10.1016/S0378-1135(00)00188-7. [DOI] [PubMed] [Google Scholar]
- 10.Schultsz C, et al. Differences in the population structure of invasive Streptococcus suis strains isolated from pigs and from humans in The Netherlands. PloS one. 2012;7:e33854. doi: 10.1371/journal.pone.0033854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willemse N, et al. An emerging zoonotic clone in the Netherlands provides clues to virulence and zoonotic potential of Streptococcus suis. Sci Rep. 2016;6:28984. doi: 10.1038/srep28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou, G. et al. Effects of Environmental and Management-Associated Factors on Prevalence and Diversity of Streptococcus suis in Clinically Healthy Pig Herds in China and the United Kingdom. Applied and environmental microbiology84, 10.1128/aem.02590-17 (2018). [DOI] [PMC free article] [PubMed]
- 13.King SJ, et al. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. Journal of clinical microbiology. 2002;40:3671–3680. doi: 10.1128/JCM.40.10.3671-3680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerdsin, A. et al. Emergence of Streptococcus suis serotype 9 infection in humans. J Microbiol Immunol Infect, 10.1016/j.jmii.2015.06.011 (2015). [DOI] [PubMed]
- 15.Vecht U, Wisselink HJ, van Dijk JE, Smith HE. Virulence of Streptococcus suis type 2 strains in newborn germfree pigs depends on phenotype. Infection and immunity. 1992;60:550–556. doi: 10.1128/iai.60.2.550-556.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vecht U, Wisselink HJ, Jellema ML, Smith HE. Identification of two proteins associated with virulence of Streptococcus suis type 2. Infection and immunity. 1991;59:3156–3162. doi: 10.1128/iai.59.9.3156-3162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baums CG, Valentin-Weigand P. Surface-associated and secreted factors of Streptococcus suis in epidemiology, pathogenesis and vaccine development. Anim Health Res Rev. 2009;10:65–83. doi: 10.1017/S146625230999003X. [DOI] [PubMed] [Google Scholar]
- 18.Goyette-Desjardins G, et al. Protection against Streptococcus suis Serotype 2 Infection Using a Capsular Polysaccharide Glycoconjugate Vaccine. Infection and immunity. 2016;84:2059–2075. doi: 10.1128/IAI.00139-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva LM, et al. Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet Microbiol. 2006;115:117–127. doi: 10.1016/j.vetmic.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 20.de Greeff A, et al. Genetic diversity of Streptococcus suis isolates as determined by comparative genome hybridization. BMC microbiology. 2011;11:161. doi: 10.1186/1471-2180-11-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics (Oxford, England) 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [Google Scholar]
- 22.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics (Oxford, England) 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okura M, et al. Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes of Streptococcus suis: potential mechanisms for generation of capsular variation. Applied and environmental microbiology. 2013;79:2796–2806. doi: 10.1128/aem.03742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brousseau R, et al. Streptococcus suis serotypes characterized by analysis of chaperonin 60 gene sequences. Applied and environmental microbiology. 2001;67:4828–4833. doi: 10.1128/AEM.67.10.4828-4833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinert LA, et al. Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis. Nat Commun. 2015;6:6740. doi: 10.1038/ncomms7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beineke A, et al. Comparative evaluation of virulence and pathology of Streptococcus suis serotypes 2 and 9 in experimentally infected growers. Vet Microbiol. 2008;128:423–430. doi: 10.1016/j.vetmic.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 28.Buttner N, et al. Streptococcus suis serotype 9 bacterin immunogenicity and protective efficacy. Veterinary immunology and immunopathology. 2012;146:191–200. doi: 10.1016/j.vetimm.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 29.Dong, W. et al. Multilocus sequence typing and virulence genotyping of Streptococcus suis serotype 9 isolates revealed high genetic and virulence diversity. FEMS microbiology letters364, 10.1093/femsle/fnx192 (2017). [DOI] [PubMed]
- 30.Casadesus J, Low D. Epigenetic gene regulation in the bacterial world. Microbiology and molecular biology reviews: MMBR. 2006;70:830–856. doi: 10.1128/mmbr.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willemse, N. & Schultsz, C. Distribution of Type I Restriction-Modification Systems in Streptococcus suis: An Outlook. Pathogens5, 10.3390/pathogens5040062 (2016). [DOI] [PMC free article] [PubMed]
- 32.Manso AS, et al. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun. 2014;5:5055. doi: 10.1038/ncomms6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vinogradov E, et al. Structure determination of Streptococcus suis serotype 9 capsular polysaccharide and assignment of functions of the cps locus genes involved in its biosynthesis. Carbohydr Res. 2016;433:25–30. doi: 10.1016/j.carres.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 34.Athey TB, et al. Determining Streptococcus suis serotype from short-read whole-genome sequencing data. BMC microbiology. 2016;16:162. doi: 10.1186/s12866-016-0782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy D, et al. A single amino acid polymorphism in the glycosyltransferase CpsK defines four Streptococcus suis serotypes. Sci Rep. 2017;7:4066. doi: 10.1038/s41598-017-04403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segura M, Fittipaldi N, Calzas C, Gottschalk M. Critical Streptococcus suis Virulence Factors: Are They All Really Critical? Trends Microbiol. 2017;25:585–599. doi: 10.1016/j.tim.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Jacques M, Graham L. Improved preservation of bacterial capsule for electron microscopy. J Electron Microsc Tech. 1989;11:167–169. doi: 10.1002/jemt.1060110212. [DOI] [PubMed] [Google Scholar]
- 38.Jacques M, Gottschalk M, Foiry B, Higgins R. Ultrastructural study of surface components of Streptococcus suis. J Bacteriol. 1990;172:2833–2838. doi: 10.1128/jb.172.6.2833-2838.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swildens B, et al. Detection of extracellular factor-positive Streptococcus suis serotype 2 strains in tonsillar swabs of live sows by PCR. Vet Microbiol. 2005;109:223–228. doi: 10.1016/j.vetmic.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 40.Smith HE, et al. The cps genes of Streptococcus suis serotypes 1, 2, and 9: development of rapid serotype-specific PCR assays. Journal of clinical microbiology. 1999;37:3146–3152. doi: 10.1128/jcm.37.10.3146-3152.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wisselink HJ, et al. Detection of virulent strains of Streptococcus suis type 2 and highly virulent strains of Streptococcus suis type 1 in tonsillar specimens of pigs by PCR. Vet Microbiol. 1999;67:143–157. doi: 10.1016/S0378-1135(99)00036-X. [DOI] [PubMed] [Google Scholar]
- 42.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inouye M, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng L, Connor TR, Siren J, Aanensen DM, Corander J. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol. 2013;30:1224–1228. doi: 10.1093/molbev/mst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics (Oxford, England) 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carver TJ, et al. ACT: the Artemis Comparison Tool. Bioinformatics (Oxford, England) 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.