Abstract
Age-related macular degeneration (AMD) is the leading cause of blindness in developed countries, and is characterized by slow retinal degeneration linked to chronic reactive oxygen species (ROS) in the retinal pigmented epithelium (RPE). The molecular mechanisms leading to RPE dysfunction in response to ROS are unclear. Here, human stem cell-derived RPE samples were stressed with ROS for 1 or 3 weeks, and both intracellular and secreted proteomes were quantified by mass spectrometry. ROS increased glycolytic proteins but decreased mitochondrial complex I subunits, as well as membrane proteins required for endocytosis. RPE secreted over 1,000 proteins, many of which changed significantly due to ROS. Notably, secreted APOE is decreased 4-fold, and urotensin-II, the strongest known vasoconstrictor, doubled. Furthermore, secreted TGF-beta is increased, and its cognate signaler BMP1 decreased in the secretome. Together, our results paint a detailed molecular picture of the retinal stress response in space and time.
Subject terms: Proteomic analysis, Stem-cell biotechnology, Ageing
Introduction
AMD is the leading cause of blindness in people over age 50, and represents an area of significant unmet clinical need. AMD is characterized by retinal degeneration in the center of the retina, the macula. Three tissues comprise a minimally functional unit of the retina, RPE is the epithelial layer between the light-sensitive photoreceptors (PRs) and vasculature (choroid). RPE is especially important among this triplet because it forms the outer blood-retinal barrier due to the tight-junctions between the cells. RPE is also the main support layer for the PRs. Some RPE functions that support photoreceptor survival and function include: (i) receipt of nutrients from vasculature and transport of nutrients to PRs1, (ii) phagocytosis of shed photoreceptor outer segments, and (iii) secretion of signals including growth factors and cytokines2. The functional disruption and atrophy of the RPE is a key factor in the progression of degenerative conditions in the retina, leading to the death of other cell types in the retina, including the light-sensitive rod and cone PRs, resulting in significant vision loss. Progression of AMD is associated with chronic ROS especially in the RPE layer3. ROS in the retina is produced mostly due to high rates of metabolism in RPE4 and the resulting electron leakage from mitochondria5. Notably, a large number of oxidative molecules may result from phagocytosis of PR outer segments6. Overall, the main site of oxidative injury appears to be the mitochondria, and pathological studies suggest that RPE damage is an early event in AMD7, which justifies the need to understand the oxidative stress response mechanisms in RPE. Chronic oxidative damage in RPE was shown to increase the expression of transcripts for proteins that are found in drusen8. Stem cell-derived RPE was previously used as a model system with chronic, low-level oxidative stress from paraquat (PQ) to study the NRF2-mediated transcriptional responses9. PQ is well tolerated by RPE10 and is a suitable mimic for pathological oxidative stress that occurs in AMD, which is thought to be mostly generated in mitochondria11. Although the connection between oxidative stress and RPE dysfunction is clear, the exact molecular changes that mediate dysfunction remain poorly defined.
Altered protein secretion and the related changes in extracellular matrix (ECM) organization are also known to be involved in AMD, especially loss of barrier function and vascular invasion12. Several reports describe RPE secretion of various proteins, including ARMS213, which interacts with other proteins that are mutated in AMD, such as fibulin-6, although the explicit function of secreted ARMS2 is unknown. Other studies using RPE cells have looked at specific protein secretions, such as apolipoprotein E (APOE)14,15. The dry form of AMD is associated with deposits rich in lipids and proteins called drusen. The protein components of drusen were found to include several complement proteins and APOE16. One recent study also reported use of induced pluripotent stem cell (iPSC)-derived RPE from control and AMD patients to study altered transcription and protein secretion17. The study carried out RNA-seq analysis and targeted secretomic analysis in non-stressed RPE cells using ELISA which focused on complement pathway components and amyloid β. This study found that RPE from patients with AMD upregulated mRNA transcripts for complement system components, that some of those components were increased in media by ELISA, and that nicotinamide could partially alleviate that effect in both AMD and control lines. What is still not well understood is how the RPE secretome changes over time during chronic oxidative stress to communicate its state to adjacent PR and vasculature, which is a focus of our study.
Although much is known about the pathology of AMD, we still do not understand the exact molecular mechanisms that lead from chronic stress to tissue dysfunction and the eventual pathology described above. In such cases, unbiased, system-wide measurements of molecular remodeling in response to stress can help provide new ideas for mechanistic follow-up experiments. Mass spectrometry-based proteomics is one such method18. Recent advancements, particularly using data-independent acquisition (DIA)19 and faster hardware, have enabled quantification of over 5,000 proteins per hour20–23. This strategy provides more complete data across replicate samples with excellent quantitative accuracy, thereby enabling more comprehensive discovery of biological pathways.
Studies of the human retina often use immortal cell lines such as ARPE1924, but these lines can differ significantly in their behavior relative to primary RPE cells. Primary human RPE cells are the ideal source of tissue for studies of AMD, but their variability across donors and limited universal availability detract from their utility. Another promising way to study AMD is with RPE generated from human pluripotent stem cells generated using protocols that mimic developmental cues25,26. These cells have been characterized to have features similar to in vivo RPE and are currently being tested in clinical trials for RPE replacement27,28. Here we used stem cell-derived RPE tissue as a model system and performed unbiased DIA mass spectrometry-based proteomics to understand the intracellular mechanisms that mediate RPE ROS tolerance, and how those changes are communicated to surrounding cells through secreted proteins (the secretome). We collected data from the intracellular proteome and the protein secretome after one or three weeks of PQ treatment to simulate short-term and long-term stresses. Further more, to understand how RPE adapts from short-term and long-term ROS, we compare the proteome and secretome after three weeks of stress with that after one week. Our results show that low-concentration PQ treatment of RPE for one or three weeks causes significant alterations of metabolic proteins, suggestive of a shift in RPE from lipid to sugar metabolism. We also find a number of changes in abundance of proteins that function in structural organization of cells and protein translation. From the secretome data, we find that ROS significantly alters proteins in the pathways of ECM-receptor interaction, focal adhesion, and complement and coagulation cascades. Remarkably, we also found significant decreases in APOE, amyloid-precursor-like protein, and the strongest known vasoconstrictor urotensin-2. Together, our results paint a picture of the human RPE’s proteomic landscape in response to short- and long-term ROS stress over space and time that point to possible mechanisms of cellular damage in AMD.
Results
Workflow and general overview
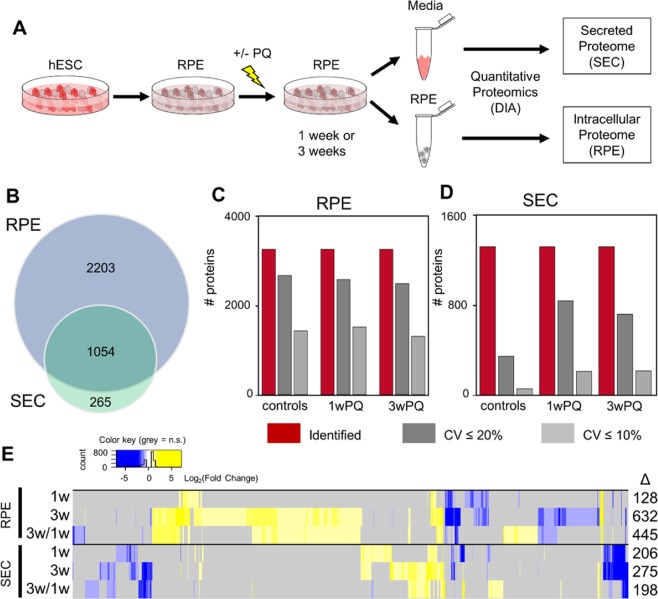
An overview of the experimental strategy is shown in Fig. 1A. RPE derived from hESC were treated with PQ for one or three weeks, and the conditioned media containing secreted proteins was collected to quantify changes in the composition of secreted proteins (secretome, SEC). Parallel experiments were carried out where RPE cells were harvested in order to quantify the intracellular proteome changes. We collected DIA data from each sample and identified and quantified proteins with Spectronaut29 and the pan-human spectral library21. Using this strategy, we identified a total of 3,257 proteins from the intracellular proteome samples, and a total of 1,319 proteins from the secretome samples. Among those proteins, 1,054 proteins were identified in both groups (Fig. 1B). We assessed the quality of our proteome quantification by comparing the coefficients of variation for all identified protein groups within each treatment group (Fig. 1C,D). Based on the proportions of each group that had CV <20% or <10%, the quality of the data from the intracellular proteome quantification was better than the quality of the secretome. This difference in variability between the RPE and SEC experiments is reflected in the number of protein changes we were able to detect from each experiment (Fig. 1E, Supplementary Table 1). For both the RPE and SEC experiments, one-week or three-week PQ treatments were compared to controls. In addition, three-week treatment results were also compared to the one-week treatment data in order to determine which proteins might be involved in the transition from short term to long term ROS adaptation.
Figure 1.
Proteomics study workflow and results overview. (A) Human embryonic stem cells (hESC) were differentiated into retinal pigment epithelium (RPE) cells, allowed to mature, and then treated with paraquat (PQ) or vehicle control for one week or three weeks. After treatment, serum-free media was collected to measure the protein secretome (SEC), and cells were harvested to measure the intracellular proteome (RPE). (B) Venn diagram showing the number and overlap of proteins identified in the two datasets. (C) Number of protein groups that meet certain criteria from each condition. Total identified protein groups are in red, the subset of protein groups that were quantified with <20% coefficient of variation (CV) in dark grey, and the subset of protein groups that were quantified with <10% CV are in light grey. (D) The same as (C) for SEC data. (E) Heatmap showing overview of statistically significant changes (q-value <0.01, fold-change >50%) found in each of the comparisons. For the intracellular proteomics experiments, three biological replicates from each treatment timepoint were compared with four control biological replicates (two controls from each timepoint). For the secreted proteome experiments, four biological replicates from each timepoint were compared with eight control biological replicates (four from each timepoint).
Intracellular proteome changes
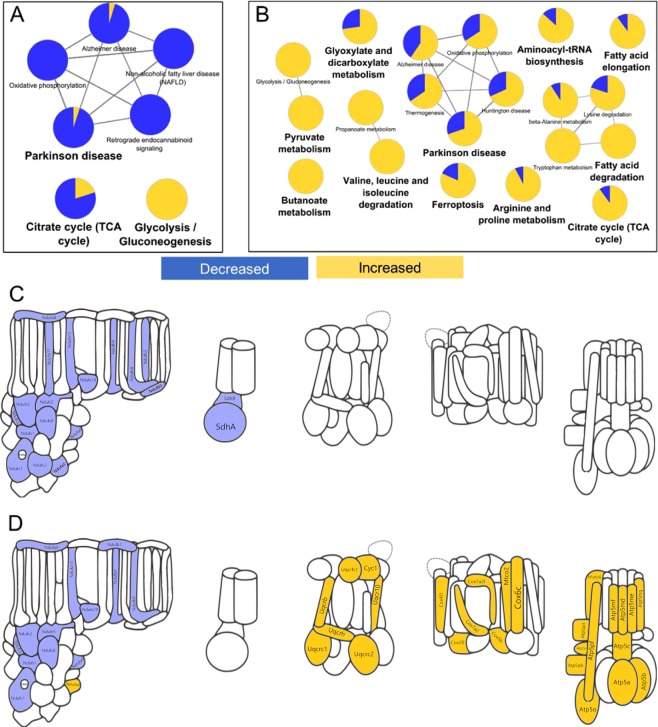
Intracellular proteome changes were examined to understand how RPE responds to short term or long term ROS. After one week of PQ treatment, only 128 proteins were altered in abundance. KEGG pathway enrichment analysis of those proteins revealed only a handful of pathways: the citric acid cycle, glycolysis, and a cluster of pathways related to oxidative phosphorylation (OxPhos, Fig. 2A). All of these pathways are related to cellular energy production. Notably, the direction of protein changes in these pathways clearly suggests a decrease in mitochondrial metabolism and an increase in glycolysis. After 3 weeks of ROS stress in RPE, the changes were much more pronounced. Over 600 proteins changed and could be mapped to 20 KEGG pathways (Fig. 2B). Again, most of these pathways were associated with energy production, except for the protein synthesis-related pathway aminoacyl-tRNA biosynthesis, members of which mostly increased. Many pathways related to amino acid metabolism, fatty acid metabolism, and glycolysis were found. After the three-week PQ treatment, changes to the cluster of OxPhos pathways were more pronounced potentially due to activation of cellular compensatory mechanisms to chronic oxidative stress. To better understand how short-term and long-term ROS from PQ treatment were influencing the mitochondrial electron transport chain (ETC), we generated a cartoon representation of ETC complex structures and colored their subunits according to whether they were increased or decreased (Fig. 2C,D). Remarkably, short term ROS caused a concerted decrease in subunits of ETC complex I, as well as a decrease of complex II subunits SDHA and SDHB (Fig. 2C). This result supports reports that complex I is the major site of ROS production by paraquat30. Chronic ROS also caused a nearly concerted decrease in ETC complex I subunits; however, concerted increases in subunits of complexes III, IV, and V were now also apparent (Fig. 2D). This data suggests that cells compensate for the reduction in complex I activity by increased translation of complex III-V subunits.
Figure 2.
RPE intracellular proteome changes resulting from acute and chronic ROS. KEGG pathway analysis showing enriched pathways (p-value < 0.001) of the intracellular RPE proteome changes after (A) one week or (B) three weeks of treatment with PQ. Subunits of the electron transport chain showing statistically significant changes after (C) one week or (D) three weeks of treatment with PQ. Throughout this figure, yellow indicates an increase, and blue indicates a decrease.
The most- and least-changed intracellular proteins after three weeks of ROS stress do not appear in the pathway enrichment analysis. The most increased proteins were GDF15, PLIN2, and SQSTM1 (Fig. S1A). Known NRF2-regulated proteins NQO1, SRXN1, and HMOX1 are also found in the six most-increased proteins9,31. The most-decreased proteins were COL6A1, LRP2, PCBP4, CD74, GLUL, and the complement cascade inhibitor, SERPING1 (Fig. S1B).
Intracellular adaptation from short-term and long-term ROS
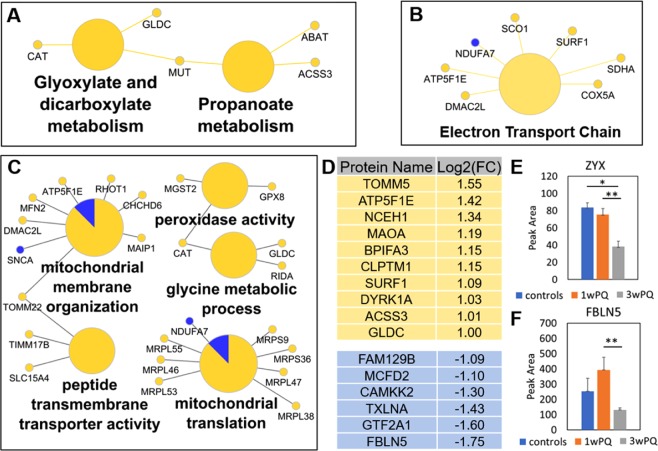
Because proteomic data was collected from both one-week and three-week ROS treatments, the evolution of proteome response to ROS exposure from short-term to long-term can be computed by directly comparing these two time points (3w/1w). In order to understand only the adaptation of the ROS response from short term to long term, any proteins that changed in the 1w/controls or 3w/controls comparisons were excluded, leaving 92 protein changes. KEGG pathway enrichment analysis using this subset of altered proteins found Glyoxylate and dicarboxylate metabolism and Propanoate metabolism pathways (enrichment p-values < 5e-4, Fig. 3A). Only 5 proteins (CAT, GLDC, MUT, ABAT ACSS3) were found in these two pathways, all of which were increased. Wikipathway term enrichment analysis (Fig. 3B) revealed changes to the ETC pathway (enrichment p-value = 4e-7), including SCO1 and SURF1 which assemble cytochrome C oxidase. Finally, the set of altered proteins were checked for enrichment of gene ontology (GO) molecular functions and biological processes (Fig. 3C). Notably, several proteins involved in mitochondrial membrane organization and protein import, as well as several mitochondrial ribosomal proteins translation were increased. Among these proteins, alpha-synuclein and NDUFA7 were the only downregulated members. Proteins with peroxidase activity (Catalase, MGST2 and GPX8) were also upregulated.
Figure 3.
Temporal dynamics of RPE intracellular proteome from one week to three weeks of chronic ROS. Proteins that changed only in the three-weeks/one-week comparison found enriched in (A) KEGG pathways, (B) Wikipathways, or (C) Gene Ontology Molecular Function and Biological Processes. All pathway enrichment analysis used a minimum p-value of 0.001. (D) List of proteins that increased or decreased at least 2-fold. (E) Quantification of Zyxin protein from the average of the following peptides: SPGAPGPLTLK2+, QNVAVNELCGR2+, and FGPVVAPK2+. (F) Quantification of Fibulin-5 protein from the peptide DQPFTILYR2+. Error bars show the standard error. Significant changes defined as at least 1.5-fold and q-value < 0.05 (*), q-value < 0.01 (**), and q-value < 0.001 (***).
In the list of altered proteins there were many interesting changes that were not part of these concerted pathway changes. Of these, two categories jump out: calcium signaling and structural protein changes. Among the significantly decreased proteins were structural proteins Zyxin (Zyx, UniProt: Q15942, Fig. 3E) and fibulin 5 (FBLN5, UniProt: Q9UBX5, Fig. 3F). The latter was the most downregulated protein. FBLN5 is especially interesting in the context of AMD because it localizes to Bruch’s membrane in the eye32 and mutations in FBLN5 are known to be associated with AMD33,34. Three of the most decreased proteins are involved in calcium signaling, Multiple coagulation factor deficiency protein 2 (binds to calcium ions)35, alpha-taxilin (may play a role in calcium-dependent exocytosis)36, and calcium/calmodulin-dependent protein kinase kinase 2. Related to calcium signaling, a member of the calcium-activated chloride channels, Anoctamin-1037 (UniProt: Q9NW15), was increased almost 2-fold (Supplementary Table 1). Finally, in the lists of the most increased and decreased proteins (at least 2-fold change from three-weeks /one-week), several proteins have yet unknown function in the retina, such as CLPTM1, BPIFA3, MAOA, and FAM129B (Fig. 3D).
Secretome remodeling
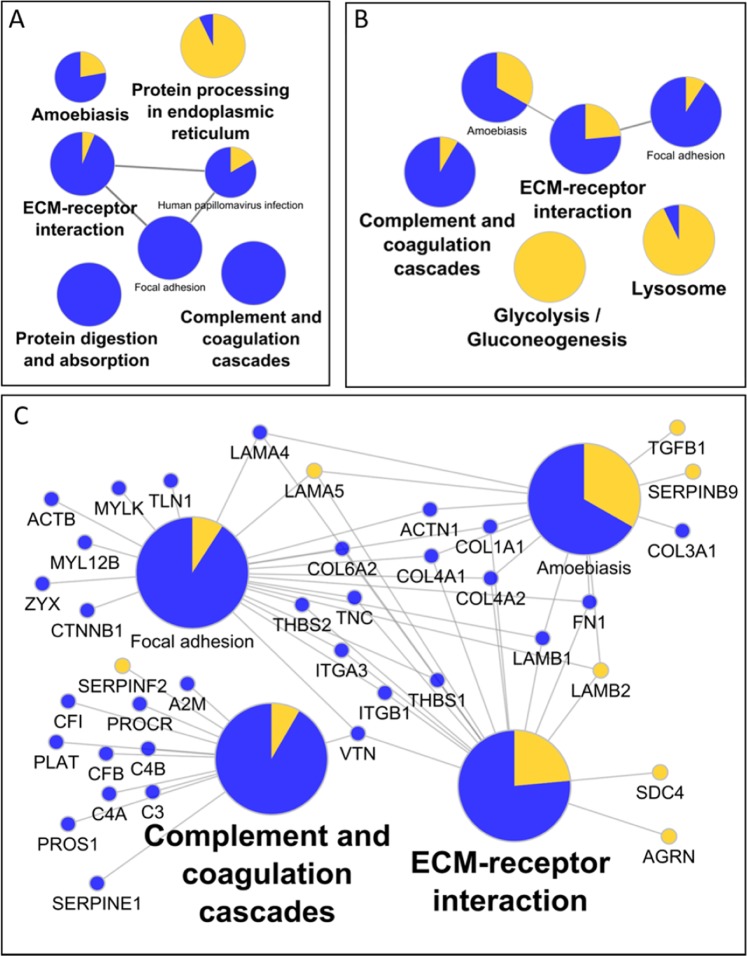
Remodeling of the protein secretome in response to short-term and long-term ROS was examined. Protein changes representative of several KEGG pathways changed after one week of PQ treatment, including “complement and coagulation cascades”, “protein digestion and absorption”, and a cluster related to ‘ECM-receptor interaction” and “Focal adhesion” (Fig. 4A). After three weeks of PQ treatment, proteins in the glycolysis and lysosome pathways were also found to increase in the extracellular medium (Fig. 4B). In common between the one-week and three-week changes were the cell and ECM structure-related pathways of “ECM-receptor interaction”, “focal adhesion”, “amoebiasis”, as well as the “complement and coagulation cascades”. Figure 4C shows these common pathways plotted with the protein changes that make up and connect these pathways. Related to the structural protein pathways, we see decreases in several intracellular and extracellular organization proteins, including actin, actinin, thrombospondins 1 and 2, fibronectin, and collagens. The Laminin changes were mixed, with subunits alpha-5 and beta-2 increased, but alpha-4 and beta-1 were decreased. Within the complement cascade several factors were decreased, including: CFI, CFB, C3, C4A, and C4B. Overall, the data indicates significant deregulation of the protein machinery for the complement, coagulation, and organizational systems results from ROS stress.
Figure 4.
Functional characteristics of secreted protein changes. KEGG pathways of proteins altered after (A) one week or (B) three weeks of PQ treatment. (C) Expansion of proteins related to ‘ECM-receptor interaction’ and ‘Complement and Coagulation Cascades’ that change after 3 weeks of PQ treatment.
There are several notable protein changes among the secretome quantification that did not appear in the pathway analysis. Since we noticed an increase in TGF-β1 (Figs 4C, S2Ai) we looked for related extracellular signaling proteins, and found that BMP1 decreased three- to four-fold with short-term and long-term treatment (Fig. S2Aii). TGF-β2 was decreased more subtly around 30% after one or three weeks of stress, but did not reach our arbitrary cutoff for significance (Fig. S2Aiii). Also altered was apolipoprotein E (APOE), which is linked to AMD wherein the APOE2 allele increases risk, while APOE4 is protective, in contrast to Alzheimer’s disease38,39. We observed that APOE was down over 2-fold after 1 week of stress, and nearly 4-fold after 3 weeks of stress (Fig. S2Bi). Another related protein strongly associated with high myopia, amyloid β-like protein 2 (APLP2), was significantly increased after three weeks of stress (Fig. S2Bii). Finally, given that development of AMD is associated with both loss and gain of vasculature, it is notable that urotensin-II (UTS2), the strongest known vasoconstrictor, was more than tripled after one or three weeks of PQ induced stress (Fig. S2Biii).
Given the apparent ECM remodeling we also checked for any changes in secreted proteases. We found a large number of proteases and protease-related proteins altered in the secretome data. We did identify two matrix metalloproteases (MMPs), MMP2 and MMP15, but only MMP2, a type IV collagenase, was significantly decreased (Fig. S2Ci). One of the most upregulated proteases was ADAM9, a known ‘sheddase’ protease that cleaves extracellular regions of membrane proteins40 and is known to regulate pathologic angiogenesis in the retina41 (Fig. S2Cii). Finally, a serine carboxypeptidase, Retinoid-inducible serine carboxypeptidase (SCPEP1), was increased over 50% after one or three weeks of PQ stress (Fig. S2Ciii).
Short term to long term adaptation of the Secretome
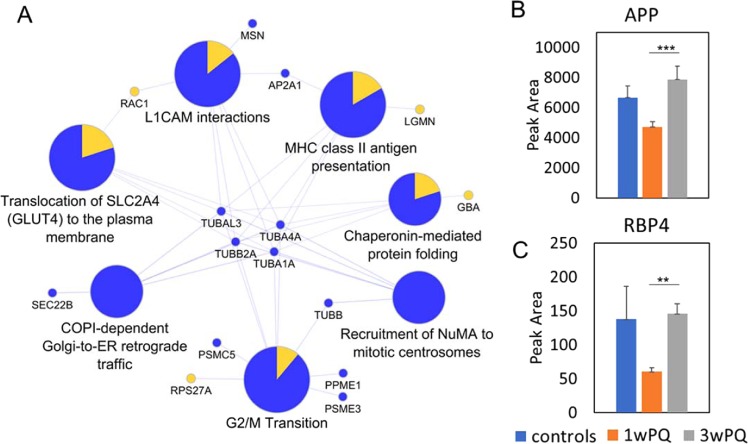
As described for the intracellular proteome data, we can understand the adaptive response in the secretome using the proteins that only change between one week and three weeks of stress. This filtering leaves only 29 increased and 34 decreased proteins. Reactome pathway analysis of these altered proteins returned many altered candidate pathways centered around the decreased levels of tubulins (Fig. 5A). Overall, there are many less concerted changes in this data subset than the others. However, there are more interesting individual changes. Amyloid precursor protein (APP) was increased between one and three weeks of stress (Fig. 5B). The most increased protein in the adaptation of the Secretome was tetranectin (almost 14-fold increase), which is a potential marker of several diseases and may play a role in exocytosis42–44. Retinol-binding protein 4 (RBP4) was also increased over 2-fold (Fig. 5C). RBP4 plays an important role in facilitating transportation and cellular uptake of retinol45.
Figure 5.
Short term to long term ROS stress adaptation in secreted protein profiles. (A) Subset of the Reactome pathways enriched in only the Secretome proteins that change from 1 week to 3 weeks of stress (pathway enrichment corrected p-values < 0.01). (B) Quantification of Amyloid precursor protein (APP). (C) Quantification of Retinol-binding protein 4 (RBP4). Error bars show the standard error. Significant changes defined as at least 1.5-fold and q-value < 0.05 (*), q-value < 0.01 (**), and q-value < 0.001 (***).
Discussion
The results presented here indicate that oxidative stress induces proteome remodeling inside RPE indicative of a metabolic shift from respiration toward glycolysis. A shift to glycolysis has been observed in other systems to reduce oxidative damage under high energy demand46. We also found intracellular proteome changes suggestive of ECM and cellular organization changes. Some intracellular protein changes have less-clear implications and require follow-up experiments. For example, CD74, a major histocompatibility class II antigen that acts as a transcription regulator linked to cell survival47 was decreased. GDF15, a distant member of the BMP subfamily and target of p5348, was also highly upregulated after chronic ROS, although its function in RPE has not been studied.
Short-term and long-term oxidative stress also induce significant remodeling of the secreted proteome. Notably, there were many altered proteins in the complement system which is known to be affected in AMD14,49,50. We also observe many changes in proteins related to ECM, focal adhesion, and TGFβ signaling51, which together may regulate neovascularization. Most interestingly, we find a number of Alzheimer’s-related proteins change in the secretome of oxidatively stressed RPE, including APP, APLP2, and APOE. The different isoforms of APOE have been associated with altered risk of AMD39,52,53, and APOE was previously demonstrated to be secreted from RPE and regulate lipid uptake15.
It is also interesting to consider the proteins we identified that did not change, such as HTRA1. Single-nucleotide polymorphisms (SNPs) near the ARMS2/HTRA1 genes are associated with altered risk of developing AMD54–56. We detected HTRA1 protein both inside RPE and in the secretome. Intracellularly, HTRA1 was increased between one and three weeks of stress but did not meet our initial statistical criteria for defining a protein change (39% increase, q-value = 0.016). Extracellularly, HTRA1 was increased 65% but the q-value was only 0.047. We also identified HTRA2 from the intracellular proteome data, and HTRA2 was significantly increased between the chronic stress and control groups (54% increase, q-value < 0.001).
Our study uniquely addresses both the temporal and spatial reorganization of the RPE proteome. We expect this data will serve as an important resource for future mechanistic and therapeutic studies. For example, the data suggest new proteins that may mediate the observed retinal disfunction in AMD in tissues adjacent to RPE, the photoreceptors and vasculature (Fig. 6). Increased secretion of TGFβ1 protein and decreased secretion of BMP1 protein may promote harm to adjacent photoreceptors and retina tissue; TGFβ1 protein was reported to play a role in proliferative vitreoretinal diseases57 and neovascular AMD58, and BMP-Smad1/5/8 signaling was reported to promote survival of retinal ganglion cells59. Signaling by TGFβ1 and BMP proteins are also known to modulate macrophage transitions to pro- or anti-inflammatory state60–63, and macrophage activity in the retina has been connected to AMD64. The decreased APOE secretion may decrease the RPE’s ability to uptake lipids and possibly the shed photoreceptor outer segments. Increased secretion of proteases ADAM9 and SCPEP1 might breakdown the basement membrane, and increased urotensin-II might cause choroidal vasculature to constrict and limit the oxygen delivery to the region. All of these ideas are opportunities for follow-up research uniquely enabled by this study. An important limitation to note about our study is that in order to quantify extracellular proteins without interference from proteins in serum used to prepare media, secretome experiments were carried out by switching cells serum-free media for 24 hours. This was not done for the intracellular proteomics experiments, and somewhat limits direct comparison of the two datasets. Regardless of this caveat, this intracellular and secretome remodeling of RPE cells shown by proteomics provides a highly granular view of the short-term and long-term oxidative stress responses.
Figure 6.
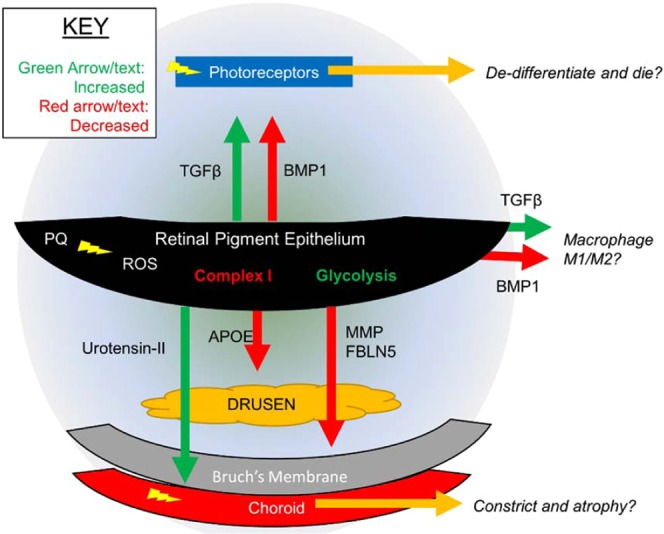
Proposed Spatial Model of RPE Response to Chronic ROS.
Methods
Chemicals and reagents
LC-MS grade acetonitrile and water were manufactured by Burdick and Jackson and purchased from Thermo-Fisher. Urea and the BCA assay kit (Pierce) were also purchased from Thermo-Fisher. LC-MS grade formic acid and 1 M triethanolamine bicarbonate (TEAB), pH 8.5, paraquat (PQ) and tris base were purchased from Sigma-Aldrich. Sequencing-grade modified trypsin was purchased from Promega.
Generation of RPE from human embryonic stem cells
Much of the methods for RPE generation from stem cells and the treatment with PQ was identical to a previous publication9, and therefore some of the following text matches those methods verbatim. All stem cell work was approved by the Buck Institute SCRO committee, and all methods were performed in accordance with federal and local stem cell research guidelines and regulations. Human ESCs (WA-01; National Institutes of Health registry #0043) were maintained in Essential 8 medium (Gibco, Grand Island, NY, USA) and 1% penicillin-streptomycin-amphotericin B solution (Lonza, Walkersville, MD, USA). Cells were grown on Matrigel (BD Biosciences, St. Paul, MN, USA)-coated plates and serially passaged using 0.5 mM EDTA solution. Genotyping revealed that cells did not have any key AMD single nucleotide polymorphism (SNP)65,66. Human ESCs were differentiated to retinal lineage using our previously published protocol26,67–69. The RPE regions were manually picked and expanded.
The RPE cells generated from hESCs were cultured in α-MEM medium (Life Technologies, Grand Island, NY, USA) containing 1% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA, USA), L-glutamine (VWR, Radnor, PA, USA), taurine (Sigma-Aldrich Corp., St. Louis, MO, USA), hydrocortisone (Sigma-Aldrich Corp.), and tri-iodo-thyronine (Sigma-Aldrich Corp.) on Matrigel-coated plates or filter membranes (VWR)70. Cells were subcultured using Accutase (Gibco) in the presence of thiazovivin (1µM), a Rho-associated protein kinase pathway inhibitor that allows passaging RPE cells for over eight passages71. The RPE cells were in culture for up to seven passages without any appreciable loss in their ability to mature into polarized RPE cells. The cells were allowed to grow to 100% confluence and used for experiments following maturation.
Cell treatments and collection
RPE cells were treated as described previously9. Briefly, RPE cells were seeded in 12-well dishes and allowed to mature for up to three weeks following final passage. Fully confluent plates of cells showing typical cobblestone morphology and presence of pigmentation were treated every other day with 160 μM Paraquat (PQ) diluted in RPE media described above for one week or three weeks. In parallel on the same multi-well dish, control RPE were grown with the same media lacking PQ. After treatment, the cells were washed once with PBS and then trypsinized. Media containing KSR was then added to quench the trypsin activity after 5 minutes, and cells were concentrated by centrifugation. The media was aspirated and the cells were washed once with PBS before being frozen at −80 °C until further processing.
Sample preparation for intracellular proteome measurement
RPE cell pellets were lysed and proteins were simultaneously precipitated according to the MPLEx protocol72. To each frozen cell pellet, 100 μL of PBS was added, and then 5 volumes of cold (−20 °C) chloroform-methanol (2:1 [vol/vol]) was added to each pellet and cells were vortexed until they thawed and dissolved in the solvent. Samples were incubated on ice for 5 minutes, vortexed in the cold room for 5 minutes, and then centrifuged at 15,700 relative centrifugal force (RCF) for 10 minutes at 4 °C, which helped clarify a biphasic solution with proteins at the interphase. The top aqueous layer was removed taking care to avoid the protein disc at the interphase, and then 500 μL of cold (−20 °C) methanol was added resulting in a monophasic solution. Samples were then vortexed briefly and centrifuged at 15,700 RCF for 10 minutes at 4 °C to pellet proteins. The supernatant was removed and the protein pellet was dissolved again with 1 mL of cold (−20 °C) methanol by vortexing to wash away any remaining lipids, and proteins were pelleted again by centrifugation at 15,700 RCF for 10 minutes in the cold room. The methanol wash was removed, and the protein pellet was dried briefly in a vacuum centrifuge. The dried protein pellet was then dissolved in 8 M Urea and 100 mM TEAB containing protease inhibitor cocktail, and protein concentration was quantified using the BCA assay. Protein disulfides were reduced by adding 4.5 mM DTT (final concentration) and heating to 37 degrees for 30 minutes, and then samples were cooled to room temperature before adding 10 mM iodoacetamide (final concentration) to alkylate free thiols. Samples were then diluted 4-fold with 100 mM TEAB to reduce the Urea concentration, and enzymatic protein hydrolysis was initiated by the addition of trypsin (Sequencing Grade Modified Trypsin, Frozen, Promega) at a ratio of 1:50, trypsin: substrate weight.
Sample preparation for secreted proteome measurement
RPE cells were stressed as described above for one week or three weeks, and then cells were washed three times with 1x PBS before adding 2 mL of serum-free RPE media. Although many cell lines are sensitive to serum-free conditions, RPE can be cultured serum-free after maturation73 and can even be passaged in serum-free media without affecting their viability74. Cells were cultured for 24 hours and then the media was collected and frozen at −80 °C. Media containing secreted proteome was then concentrated using a centrifugal ultrafiltration membrane (Amicon Ultra-15, MWCO 30 kDa) by centrifugation at 7,500 X gravity for 10 minutes at 4 °C. To denature proteins the buffer was exchanged three times by adding 1 mL of 2 M Urea in 50 mM Tris, pH 8.0 and centrifuging. The hold-up volume of the buffer-exchanged protein was then removed from the ultrafilter and protein was quantified using the BCA assay. Protein disulfides were then reduced by adding 4.5 mM DTT (final concentration) and heating to 37 degrees for 30 minutes, and then samples were cooled to room temperature before adding 10 mM iodoacetamide (final concentration) to alkylate free thiols. Enzymatic protein hydrolysis was initiated by the addition of trypsin (Sequencing Grade Modified Trypsin, Frozen, Promega) at a ratio of 1:50, trypsin: substrate weight.
Mass spectrometry data collection and analysis
Peptides were analyzed by nanoflow liquid chromatography – tandem mass spectrometry analysis (nanoLC-MS/MS) on a Sciex 5600 TripleTOF mass spectrometer using data-independent acquisition (DIA) using variable-width precursor isolation windows as described previously75. Peptide separation for DIA data collection was performed using a linear gradient of 5% mobile phase B (98% Acetonitrile, 0.05% formic acid, 1.95% water) to 40% B over 200 minutes with a flow rate of 300 nL/min.
Data analysis
Proteins were identified and quantified from DIA-MS data using Spectronaut29 and the pan-human spectral library21. Spectronaut settings were all defaults except that quantification filtering was set to Q-value percentile = 25% for intracellular proteome, and Q-value percentile = 20% for secreted proteome. Spectronaut performs local retention time-based signal normalization. Statistically-significant protein changes (defined as q < 0.01 and at least 50% change) were assessed for enriched pathways and visualized using the ClueGO plugin of Cytoscape76. Additional analysis was carried out using custom scripts in R77.
Sample groups and statistical comparisons
Data from the intracellular proteome was collected from three wells of treated cells and two wells of control cells per time point, resulting in a total of 10 samples for intracellular proteome. To ensure that we only detect changes that result from short term and long term ROS, we compared the treated samples in triplicate with the four control samples, two from each treatment timepoint. Data from the protein secretome was collected from four replicates of treated cells and four replicates of control cells at each timepoint. As described for the intracellular proteomics comparison, samples from each treatment timepoint were compared with a pool of all eight control samples.
Supplementary information
Acknowledgements
J.G.M. was supported by an NIH training grant (T32 AG000266). Work in Dr. Lamba’s lab is supported by NIH grant EY025779. The authors acknowledge support from the NIH shared instrumentation grant for the TripleTOF system at the Buck Institute (1 S10 OD016281). Additional support was provided by a core grant from the National Institutes of Health (EY02162) and by Research to Prevent Blindness unrestricted grants to the UCSF Department of Ophthalmology (to DAL).
Author contributions
Conceptualization, J.G.M., B.W.G. and D.A.L.; Methodology, J.G.M., T.Y.G., B.W.G. and D.A.L.; Software, J.G.M.; Validation, J.G.M. and T.Y.G.; Formal Analysis, J.G.M.; Investigation, J.G.M. and T.Y.G.; Resources, B.S., B.W.G. and D.A.L.; Data Curation, J.G.M.; Writing – Original Draft, J.G.M.; Writing – Reviewing and Editing, J.G.M., B.W.G. and D.A.L.; Visualization, J.G.M.; Supervision, J.G.M., B.S., B.W.G. and D.A.L.; Project Administration, J.G.M., B.S., B.W.G. and D.A.L.; Funding Acquisition, J.G.M., B.W.G. and D.A.L.
Data availability
All raw mass spectrometry data files are available from massive.ucsd.edu under dataset identifier MSV000083398 (password: intrpesec).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jesse G. Meyer, Email: jessegmeyer@gmail.com
Deepak A. Lamba, Email: Deepak.Lamba@ucsf.edu
Supplementary information
is available for this paper at 10.1038/s41598-019-51777-7.
References
- 1.Chao JR, et al. Human retinal pigment epithelial cells prefer proline as a nutrient and transport metabolic intermediates to the retinal side. Journal of Biological Chemistry. 2017;292:12895–12905. doi: 10.1074/jbc.M117.788422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strauss O. The Retinal Pigment Epithelium in Visual Function. Physiol Rev. 2005;85:845. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 3.Jarrett SG, Boulton ME. Consequences of oxidative stress in age-related macular degeneration. Molecular Aspects of Medicine. 2012;33:399–417. doi: 10.1016/j.mam.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu D-Y, Cringle SJ. Oxygen Distribution and Consumption within the Retina in Vascularised and Avascular Retinas and in Animal Models of Retinal Disease. Progress in Retinal and Eye Research. 2001;20:175–208. doi: 10.1016/S1350-9462(00)00027-6. [DOI] [PubMed] [Google Scholar]
- 5.Brand MD, et al. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radical Biology and Medicine. 2004;37:755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 6.Miceli MV, Liles MR, Newsome DA. Evaluation of Oxidative Processes in Human Pigment Epithelial Cells Associated with Retinal Outer Segment Phagocytosis. Experimental Cell Research. 1994;214:242–249. doi: 10.1006/excr.1994.1254. [DOI] [PubMed] [Google Scholar]
- 7.Cai J, Nelson KC, Wu M, Sternberg P, Jones DP. Oxidative damage and protection of the RPE. Progress in Retinal and Eye Research. 2000;19:205–221. doi: 10.1016/S1350-9462(99)00009-9. [DOI] [PubMed] [Google Scholar]
- 8.Rabin DM, Rabin RL, Blenkinsop TA, Temple S, Stern JH. Chronic oxidative stress upregulates Drusen-related protein expression in adult human RPE stem cell-derived RPE cells: A novel culture model for dry AMD. Aging (Albany NY) 2013;5:51–66. doi: 10.18632/aging.100516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia TY, Gutierrez M, Reynolds J, Lamba DA. Modeling the Dynamic AMD-Associated Chronic Oxidative Stress Changes in Human ESC and iPSC-Derived RPE Cells. Investigative Opthalmology & Visual Science. 2015;56:7480. doi: 10.1167/iovs.15-17251. [DOI] [PubMed] [Google Scholar]
- 10.Lu L, Hackett SF, Mincey A, Lai H, Campochiaro PA. Effects of different types of oxidative stress in RPE cells: MEASUREMENT OF OXIDATIVE DAMAGE. Journal of Cellular Physiology. 2006;206:119–125. doi: 10.1002/jcp.20439. [DOI] [PubMed] [Google Scholar]
- 11.Blanco-Ayala T, Andérica-Romero AC, Pedraza-Chaverri J. New insights into antioxidant strategies against paraquat toxicity. Free Radical Research. 2014;48:623–640. doi: 10.3109/10715762.2014.899694. [DOI] [PubMed] [Google Scholar]
- 12.Rizzolo, L. J. Development and Role of Tight Junctions in the Retinal Pigment Epithelium. in International Review of Cytology258, 195–234 (Academic Press, 2007). [DOI] [PubMed]
- 13.Kortvely E, et al. ARMS2 Is a Constituent of the Extracellular Matrix Providing a Link between Familial and Sporadic Age-Related Macular Degenerations. Investigative Opthalmology & Visual Science. 2010;51:79. doi: 10.1167/iovs.09-3850. [DOI] [PubMed] [Google Scholar]
- 14.Johnson LV, et al. Cell culture model that mimics drusen formation and triggers complement activation associated with age-related macular degeneration. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18277–18282. doi: 10.1073/pnas.1109703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishida BY, et al. Regulated expression of apolipoprotein E by human retinal pigment epithelial cells. Journal of Lipid Research. 2004;45:263–271. doi: 10.1194/jlr.M300306-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, et al. Abundant Lipid and Protein Components of Drusen. PLOS ONE. 2010;5:e10329. doi: 10.1371/journal.pone.0010329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saini JS, et al. Nicotinamide Ameliorates Disease Phenotypes in a Human iPSC Model of Age-Related Macular Degeneration. Cell Stem Cell. 2017;20:635–647.e7. doi: 10.1016/j.stem.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 19.Meyer JG, Schilling B. Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert Review of Proteomics. 2017;14:419–429. doi: 10.1080/14789450.2017.1322904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelstrup CD, et al. Performance Evaluation of the Q Exactive HF-X for Shotgun Proteomics. J. Proteome Res. 2018;17:727–738. doi: 10.1021/acs.jproteome.7b00602. [DOI] [PubMed] [Google Scholar]
- 21.Rosenberger G, et al. A repository of assays to quantify 10,000 human proteins by SWATH-MS. Scientific. Data. 2014;1:140031. doi: 10.1038/sdata.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. The Use of Variable Q1 Isolation Windows Improves Selectivity in LC–SWATH–MS Acquisition. Journal of Proteome Research. 2015;14:4359–4371. doi: 10.1021/acs.jproteome.5b00543. [DOI] [PubMed] [Google Scholar]
- 23.Gillet, L. C. et al. Targeted Data Extraction of the MS/MS Spectra Generated by Data-independent Acquisition: A New Concept for Consistent and Accurate Proteome Analysis. Molecular & Cellular Proteomics11 (2012). [DOI] [PMC free article] [PubMed]
- 24.Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, A Human Retinal Pigment Epithelial Cell Line with Differentiated Properties. Experimental Eye Research. 1996;62:155–170. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- 25.Meyer JS, et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16698–16703. doi: 10.1073/pnas.0905245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamba DA, Karl MO, Ware CB, Reh TA. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proceedings of the National Academy of Sciences. 2006;103:12769–12774. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehat MS, et al. Transplantation of Human Embryonic Stem Cell-Derived Retinal Pigment Epithelial Cells in Macular Degeneration. Ophthalmology. 2018;125:1765–1775. doi: 10.1016/j.ophtha.2018.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz SD, et al. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: follow-up of two open-label phase 1/2 studies. The Lancet. 2015;385:509–516. doi: 10.1016/S0140-6736(14)61376-3. [DOI] [PubMed] [Google Scholar]
- 29.Bruderer R, et al. Extending the Limits of Quantitative Proteome Profiling with Data-Independent Acquisition and Application to Acetaminophen-Treated Three-Dimensional Liver Microtissues. Molecular & Cellular Proteomics: MCP. 2015;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cochemé HM, Murphy MP. Complex I Is the Major Site of Mitochondrial Superoxide Production by Paraquat. Journal of Biological Chemistry. 2008;283:1786–1798. doi: 10.1074/jbc.M708597200. [DOI] [PubMed] [Google Scholar]
- 31.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullins RF, Olvera MA, Clark AF, Stone EM. Fibulin-5 distribution in human eyes: Relevance to age-related macular degeneration. Experimental Eye Research. 2007;84:378–380. doi: 10.1016/j.exer.2006.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stone EM, et al. Missense Variations in the Fibulin 5 Gene and Age-Related Macular Degeneration. New England Journal of Medicine. 2004;351:346–353. doi: 10.1056/NEJMoa040833. [DOI] [PubMed] [Google Scholar]
- 34.Auer-Grumbach M, et al. Fibulin-5 mutations link inherited neuropathies, age-related macular degeneration and hyperelastic skin. Brain. 2011;134:1839–1852. doi: 10.1093/brain/awr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang B, et al. Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex. Nature Genetics. 2003;34:220–225. doi: 10.1038/ng1153. [DOI] [PubMed] [Google Scholar]
- 36.Nogami S, et al. Taxilin; a novel syntaxin-binding protein that is involved in Ca2+-dependent exocytosis in neuroendocrine cells. Genes to Cells. 2003;8:17–28. doi: 10.1046/j.1365-2443.2003.00612.x. [DOI] [PubMed] [Google Scholar]
- 37.Schreiber R, et al. Expression and Function of Epithelial Anoctamins. Journal of Biological Chemistry. 2010;285:7838–7845. doi: 10.1074/jbc.M109.065367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levy O, et al. APOE Isoforms Control Pathogenic Subretinal Inflammation in Age-Related Macular Degeneration. The Journal of Neuroscience. 2015;35:13568–13576. doi: 10.1523/JNEUROSCI.2468-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKay GJ, et al. Evidence of association of APOE with age-related macular degeneration - a pooled analysis of 15 studies. Human Mutation. 2011;32:1407–1416. doi: 10.1002/humu.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards D, Handsley M, Pennington C. The ADAM metalloproteinases. Molecular Aspects of Medicine. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guaiquil V, et al. ADAM9 Is Involved in Pathological Retinal Neovascularization. Molecular and Cellular Biology. 2009;29:2694–2703. doi: 10.1128/MCB.01460-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, et al. Tetranectin as a Potential Biomarker for Stable Coronary Artery Disease. Scientific Reports. 2015;5:17632. doi: 10.1038/srep17632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nielsen H, Clemmensen L, Kharazmi A. Tetranectin: a Novel Secretory Protein from Human Monocytes. Scandinavian Journal of Immunology. 1993;37:39–42. doi: 10.1111/j.1365-3083.1993.tb01662.x. [DOI] [PubMed] [Google Scholar]
- 44.Borregaard N, Christensen L, Bejerrum OW, Birgens HS, Clemmensen I. Identification of a highly mobilizable subset of human neutrophil intracellular vesicles that contains tetranectin and latent alkaline phosphatase. J. Clin. Invest. 1990;85:408–416. doi: 10.1172/JCI114453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berry DC, O’Byrne SM, Vreeland AC, Blaner WS, Noy N. Cross Talk between Signaling and Vitamin A Transport by the Retinol-Binding Protein Receptor STRA6. Molecular and Cellular Biology. 2012;32:3164–3175. doi: 10.1128/MCB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levin E, Lopez-Martinez G, Fane B, Davidowitz G. Hawkmoths use nectar sugar to reduce oxidative damage from flight. Science. 2017;355:733. doi: 10.1126/science.aah4634. [DOI] [PubMed] [Google Scholar]
- 47.Gil-Yarom N, et al. CD74 is a novel transcription regulator. Proceedings of the National Academy of Sciences. 2017;114:562–567. doi: 10.1073/pnas.1612195114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bootcov MR, et al. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF- superfamily. Proceedings of the National Academy of Sciences. 1997;94:11514–11519. doi: 10.1073/pnas.94.21.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gemenetzi M, Lotery AJ. Complement pathway biomarkers and age-related macular degeneration. Eye. 2016;30:1–14. doi: 10.1038/eye.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zipfel, P. F., Lauer, N. & Skerka, C. The Role of Complement in AMD. in Inflammation and Retinal Disease: Complement Biology and Pathology (eds Lambris, J. D. & Adamis, A. P.) 703, 9–24 (Springer New York, 2010). [DOI] [PubMed]
- 51.Amin R, Puklin JE, Frank RN. Growth factor localization in choroidal neovascular membranes of age-related macular degeneration. Investigative ophthalmology & visual science. 1994;35:3178–3188. [PubMed] [Google Scholar]
- 52.Malek G, et al. Apolipoprotein E allele-dependent pathogenesis: A model for age-related retinal degeneration. Proceedings of the National Academy of Sciences. 2005;102:11900–11905. doi: 10.1073/pnas.0503015102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toops, K. A., Tan, L. X. & Lakkaraju, A. Apolipoprotein E Isoforms and AMD. In Retinal Degenerative Diseases (eds Bowes Rickman, C. et al.) 854, 3–9 (Springer International Publishing, 2016). [DOI] [PubMed]
- 54.Yang Z, et al. A Variant of the HTRA1 Gene Increases Susceptibility to Age-Related Macular Degeneration. Science. 2006;314:992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 55.Kanda A, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proceedings of the National Academy of Sciences. 2007;104:16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iejima D, et al. HTRA1 (High Temperature Requirement A Serine Peptidase 1) Gene Is Transcriptionally Regulated by Insertion/Deletion Nucleotides Located at the 3′ End of the ARMS2 (Age-related Maculopathy Susceptibility 2) Gene in Patients with Age-related Macular Degeneration. Journal of Biological Chemistry. 2015;290:2784–2797. doi: 10.1074/jbc.M114.593384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kita T, et al. Role of TGF- in proliferative vitreoretinal diseases and ROCK as a therapeutic target. Proceedings of the National Academy of Sciences. 2008;105:17504–17509. doi: 10.1073/pnas.0804054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X, et al. LRG1 promotes angiogenesis by modulating endothelial TGF-β signalling. Nature. 2013;499:306–311. doi: 10.1038/nature12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ueki Y, Reh TA. Activation of BMP-Smad1/5/8 Signaling Promotes Survival of Retinal Ganglion Cells after Damage In Vivo. PLoS ONE. 2012;7:e38690. doi: 10.1371/journal.pone.0038690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gong D, et al. TGFβ signaling plays a critical role in promoting alternative macrophage activation. BMC Immunology. 2012;13:31. doi: 10.1186/1471-2172-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Werner F, et al. Transforming Growth Factor-β1 Inhibition of Macrophage Activation Is Mediated via Smad3. Journal of Biological Chemistry. 2000;275:36653–36658. doi: 10.1074/jbc.M004536200. [DOI] [PubMed] [Google Scholar]
- 62.Talati M, et al. BMP Pathway Regulation of and by Macrophages. PLoS ONE. 2014;9:e94119. doi: 10.1371/journal.pone.0094119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singla DK, Singla R, Wang J. BMP-7 Treatment Increases M2 Macrophage Differentiation and Reduces Inflammation and Plaque Formation in Apo E−/− Mice. PLOS ONE. 2016;11:e0147897. doi: 10.1371/journal.pone.0147897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Natoli R, et al. Retinal Macrophages Synthesize C3 and Activate Complement in AMD and in Models of Focal Retinal Degeneration. Investigative Opthalmology & Visual Science. 2017;58:2977. doi: 10.1167/iovs.17-21672. [DOI] [PubMed] [Google Scholar]
- 65.Grassmann F, Fritsche LG, Keilhauer CN, Heid IM, Weber BHF. Modelling the Genetic Risk in Age-Related Macular Degeneration. PLoS ONE. 2012;7:e37979. doi: 10.1371/journal.pone.0037979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mello, E. et al. Rapid detection of CFH (p.Y402H) and ARMS2 (p.A69S) polymorphisms in age-related macular degeneration using high-resolution melting analysis. Clinical Chemistry and Laboratory Medicine50 (2012). [DOI] [PubMed]
- 67.Lamba DA, Gust J, Reh TA. Transplantation of Human Embryonic Stem Cell-Derived Photoreceptors Restores Some Visual Function in Crx-Deficient Mice. Cell Stem Cell. 2009;4:73–79. doi: 10.1016/j.stem.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lamba DA, Reh TA. Microarray Characterization of Human Embryonic Stem Cell–Derived Retinal Cultures. Investigative Opthalmology & Visual Science. 2011;52:4897. doi: 10.1167/iovs.10-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lamba DA, et al. Generation, Purification and Transplantation of Photoreceptors Derived from Human Induced Pluripotent Stem Cells. PLoS ONE. 2010;5:e8763. doi: 10.1371/journal.pone.0008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maminishkis, A. & Miller, S. S. Experimental Models for Study of Retinal Pigment Epithelial Physiology and Pathophysiology. Journal of Visualized Experiments, 10.3791/2032 (2010). [DOI] [PMC free article] [PubMed]
- 71.Croze RH, et al. ROCK Inhibition Extends Passage of Pluripotent Stem Cell-Derived Retinal Pigmented Epithelium: ROCK Inhibition Extends Passage of Stem Cell-Derived RPE. STEM CELLS Translational Medicine. 2014;3:1066–1078. doi: 10.5966/sctm.2014-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakayasu, E. S. et al. MPLEx: a Robust and Universal Protocol for Single-Sample Integrative Proteomic, Metabolomic, and Lipidomic Analyses. mSystems1 (2016). [DOI] [PMC free article] [PubMed]
- 73.Matsumoto E, et al. Fabricating retinal pigment epithelial cell sheets derived from human induced pluripotent stem cells in an automated closed culture system for regenerative medicine. PLOS ONE. 2019;14:e0212369. doi: 10.1371/journal.pone.0212369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tezel TH, Del Priore LV. Serum-free Media for Culturing and Serial-Passaging of Adult Human Retinal Pigment Epithelium. Experimental Eye Research. 1998;66:807–815. doi: 10.1006/exer.1998.0492. [DOI] [PubMed] [Google Scholar]
- 75.Collins BC, et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nature Communications. 2017;8:291. doi: 10.1038/s41467-017-00249-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.R Development Core Team. R: A language and environment for statistical computing. (R Foundation for Statistical Computing, 2008).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw mass spectrometry data files are available from massive.ucsd.edu under dataset identifier MSV000083398 (password: intrpesec).