Abstract
The search for biomarkers associated with obesity-related diseases is ongoing, but it is not clear whether plasma and serum can be used interchangeably in this process. Here we used high-throughput screening to analyze 358 proteins and 76 lipids, selected because of their relevance to obesity-associated diseases, in plasma and serum from age- and sex-matched lean and obese humans. Most of the proteins/lipids had similar concentrations in plasma and serum, but a subset showed significant differences. Notably, a key marker of cardiovascular disease PAI-1 showed a difference in concentration between the obese and lean groups only in plasma. Furthermore, some biomarkers showed poor correlations between plasma and serum, including PCSK9, an important regulator of cholesterol homeostasis. Collectively, our results show that the choice of biofluid may impact study outcome when screening for obesity-related biomarkers and we identify several markers where this will be the case.
Subject terms: Translational immunology, Biomarkers, Metabolic disorders
Introduction
Obesity-related illness is an increasingly important global health issue that places a tremendous economic burden on society1. The negative health effects of prolonged obesity are partly fuelled by chronic low-grade inflammation, which contributes to cardiometabolic and kidney pathophysiology2–4. However, the exact mechanisms that link obesity with cardiometabolic and kidney diseases are unclear and remain a subject of intensive research. The search for biomarkers that assist in the identification of novel disease-related pathways is critical to develop new therapies that are tailored to subpopulations particularly prone to obesity-related pathophysiology.
Disease-related biomarkers are often identified and quantified in blood-derived plasma or serum5,6. Preparation of plasma and serum requires the removal of cellular components by centrifugation. Generation of plasma is preceded by the addition of an anti-coagulant (e.g. EDTA, heparin or citrate) to the whole blood. By contrast, the blood used for serum is allowed to clot before centrifugation, resulting in lower concentrations of clotting factors (such as fibrinogen and coagulation cascade proteins) in serum than in plasma. The World Health Organization generally recommends using plasma as this more accurately reflects the physiological and/or pathophysiological state of the patient7. However, biomarkers are often reported to have better detectability in serum8 despite the fact that serum has a slightly lower total protein concentration than plasma9. Indeed, some intracellularly stored proteins and lipids are only detectable upon coagulation-induced release from leukocytes and platelets, and serum is preferred in assays detecting, for example, cardiac troponins10–12. Importantly, the choice of biofluid is not merely a question of detectability, but it may also affect the conclusions drawn from a study. For example, Alsaif et al. showed that of 16 proteins (identified in either plasma or serum) that were differentially expressed between healthy controls and subjects with bipolar disorder, only two showed differential expression in both serum and plasma13.
The aim of our study was to determine whether the use of plasma or serum would yield different results when screening for obesity-related biomarkers. We analyzed proteins and lipids that have previously been suggested to play a role in obesity-related cardiometabolic diseases in plasma and serum from age- and sex-matched groups of lean and obese humans. Our results show that the use of plasma or serum may have an effect on study outcome when screening for obesity-related biomarkers and we identify key markers that highlight this issue.
Results and Discussion
Detectability of proteins in plasma versus serum
We used four Olink multiplex protein panels (inflammation, cardiometabolic, cardiovascular II, cardiovascular III) selected on the basis of their relevance to obesity-related diseases to measure protein concentrations in plasma and serum from 11 obese subjects and 11 age- and sex-matched lean controls. The characteristics of the human cohort are presented in Table 1. Of the 368 proteins analyzed (10 of which were measured in duplicate panels, see Supplementary Table S1 for the full list), one protein (BDNF) was excluded due to technical issues, nine proteins (IL-1 alpha, IL-2, TSLP, IL-22 RA1, IL-13, TNF, IL-20, IL-33, IFN-gamma) were excluded because they were undetectable in both plasma and serum, and 23 additional proteins were excluded because values were missing in >30% of the samples in all of the four groups (lean plasma, lean serum, obese plasma, obese serum; Supplementary Table S1). Detectability issues with one of the excluded proteins, NT-proBNP, have previously been reported14. In total, 335 proteins were included in the comparative analyses (Supplementary Fig. S1).
Table 1.
Summary of cohort demographics.
| Lean | Obese | |||||
|---|---|---|---|---|---|---|
| Men | Women | All | Men | Women | All | |
| Sex | 3 | 8 | 3♂/8♀ | 3 | 8 | 3♂/8♀ |
| BMI (kg/m2) | 23.3 ± 0.9 | 22.0 ± 1.0 | 22.4 ± 2.4 | 41.0 ± 2.1 | 44.4 ± 1.5 | 43.5 ± 4.1 |
| Age (years) | 42.3 ± 4.3 | 40.0 ± 5.3 | 40.6 ± 13.0 | 45.0 ± 3.1 | 40.4 ± 5.6 | 41.6 ± 13.6 |
| Hormone replacement therapy | — | 0/8 | — | — | 0/8 | — |
| Hormonal contraceptive pill | — | 2/8 | — | — | 0/8 | — |
| Intrauterine contraceptive device | — | 0/8 | — | — | 2/8 | — |
Data are shown as mean ± SEM.
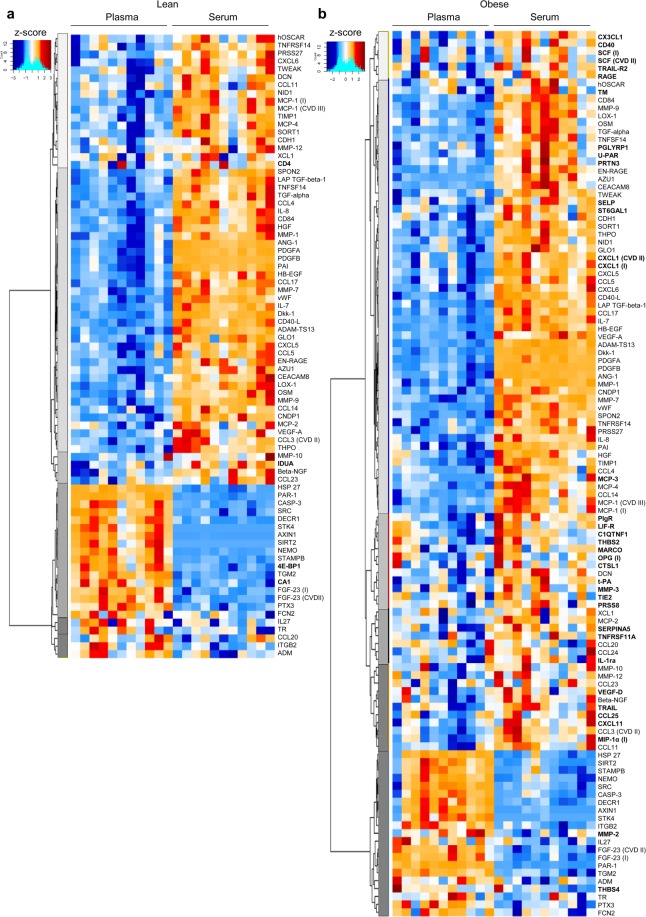
For the majority of proteins, their concentrations were similar between plasma and serum (Supplementary Fig. S2a,b). After adjusting for multiple comparisons using the stringent Holm-Bonferroni test, we found significantly different concentrations between plasma and serum for 23.5% and 33.4% of proteins in the lean and obese cohorts, respectively [adjusted (adj.) p < 0.05, Fig. 1]. Most of these proteins were present at higher concentrations in serum, which may partly be explained by the clotting-induced volume displacement effect15,16 and by the fact that coagulation elicits release of platelet granules and intracellularly stored cytokines17–19. The intracellularly stored protein MCP-1, for example, exhibited significantly higher concentrations in serum compared with plasma (in both the inflammation and the cardiovascular III panels) in the lean and obese groups. Of note, we did not record female menstruation cycle and/or menopausal state, which may affect platelet activation, although conflicting results have been shown20–25.
Figure 1.
Detectability of proteins in plasma versus serum. Heatmaps showing protein biomarkers that exhibited significantly different concentrations in plasma versus serum in (a) lean subjects (n = 11) and (b) obese subjects (n = 11) after adjustment for multiple comparisons using the method of Holm-Bonferroni at adj. p < 0.05. Proteins that are significantly different in only one of the groups (lean or obese) are marked in bold. For proteins that are present in duplicate protein panels, the panel is indicated in parentheses: I, inflammation; CVII, cardiovascular II; and CVIII, cardiovascular III. Relative protein concentrations are reported as z-scores.
A subset of proteins with significantly different concentrations in plasma and serum (including HSP-27, PAR-1, 4E-BP1 and SRC) exhibited lower concentrations in serum (Fig. 1). HSP-27 has been proposed as a biomarker for both cardiometabolic disease and cancer26, although controversial results have been reported27. Of note, a recent study showed that the concentration of HSP-27 increased by about three-fold with just one freeze-thaw cycle in plasma but was more stable in serum28. All of our samples underwent two freeze-thaw cycles, which could explain the higher detectability of HSP-27 in plasma. During coagulation, PAR-1 is cleaved29 and SRC30 and 4-EBP131,32 become prone to degradation through proteolytic pathways, which likely explains their lower concentrations in serum. Furthermore, two proteins, AXIN1 and STK4, passed our cut-off criteria for detection in plasma but not in serum (Supplementary Table S1). Enrichment analysis of all the proteins that were significantly altered between plasma and serum confirmed the enrichment of pathways involved in neutrophil chemotaxis and platelet activation (Supplementary Fig. S2c, Supplementary Table S3).
Sensitivity of plasma versus serum when screening for obesity-related protein biomarkers
Most of the biomarkers that showed significantly different concentrations between the obese and lean groups were present at higher levels in the obese group; however, a small number (including IGFBP-1 and GH) showed lower concentrations in the obese group, with significant differences observed in both plasma and serum (Table 2). The number of proteins with significantly different concentrations between the lean and obese groups was greater in serum (Table 2), in agreement with (1) an earlier study that reported higher sensitivity of serum to detect diabetes-associated differences in metabolite concentration33 and (2) the fact that obesity is associated with higher leukocyte and platelet counts and increased platelet activation34,35. MCP-3 was present at higher concentrations in the obese versus lean group in serum but not plasma (Table 2), and showed low detectability in plasma in both groups (Supplementary Table S1). However, concentrations of PAI-1 were only significantly higher in the obese versus lean group in plasma despite showing higher detectability in serum (Table 2). This difference in sensitivity versus detectability for PAI-1 was confirmed by ELISA (Supplementary Fig. S3). PAI-1 inhibits fibrinolysis and has been proposed to be an important biomarker in cardiometabolic and diabetes research, although, as recently reviewed, conflicting results have been reported36. A possible explanation for this discrepancy, at least in part, may be due to the interchangeable use of plasma versus serum; indeed, studies comparing lean versus obese and/or diabetic groups have reported differences in PAI-1 levels when using plasma37 but not serum38. Coagulation-induced secretion of intracellular PAI-1 is likely responsible for the high serum levels of PAI-1, which may mask the differences between the lean and obese groups.
Table 2.
Proteins that exhibited significant differences in concentrations between obese and lean groups in plasma and/or serum.
| Protein | Plasma | Serum | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lean | Obese | Obese vs lean log2 ratio | H-B Adj. p value | FDR Adj. p value | Lean | Obese | Obese vs lean log2 ratio | H-B Adj. p value | FDR Adj. p value | |
| NPX (mean ± SEM) | NPX (mean ± SEM) | |||||||||
| 4E-BP1 | 6.57 ± 0.21 | 7.54 ± 0.10 | 0.97 | ns | 0.0106 | 4.57 ± 0.10 | 6.18 ± 0.33 | 1.62 | ns | 0.0081 |
| ADAM-TS13 | 5.17 ± 0.06 | 4.94 ± 0.04 | −0.22 | ns | ns | 6.38 ± 0.05 | 6.19 ± 0.03 | −0.18 | ns | 0.0336 |
| ADM | 6.33 ± 0.06 | 7.11 ± 0.09 | 0.78 | 0.0002 | 0.0001 | 6.03 ± 0.08 | 6.82 ± 0.11 | 0.79 | 0.0032 | 0.0004 |
| AGRP | 3.64 ± 0.11 | 3.12 ± 0.07 | −0.52 | ns | 0.0136 | 3.74 ± 0.13 | 3.10 ± 0.08 | −0.64 | ns | 0.0081 |
| AMBP | 5.52 ± 0.02 | 5.72 ± 0.04 | 0.20 | ns | 0.0037 | 5.53 ± 0.05 | 5.78 ± 0.03 | 0.26 | ns | 0.0058 |
| CCL3 (CVD II) | 3.18 ± 0.04 | 3.67 ± 0.05 | 0.49 | 0.0002 | 0.0001 | 3.65 ± 0.07 | 4.13 ± 0.07 | 0.48 | 0.0241 | 0.0018 |
| CCL4 | 6.04 ± 0.07 | 6.67 ± 0.13 | 0.63 | ns | 0.0095 | 7.33 ± 0.12 | 7.76 ± 0.19 | 0.43 | ns | ns |
| CCL18 | 5.28 ± 0.22 | 6.09 ± 0.19 | 0.81 | ns | ns | 5.36 ± 0.21 | 6.25 ± 0.18 | 0.89 | ns | 0.0259 |
| CCL19 | 8.98 ± 0.15 | 9.66 ± 0.13 | 0.68 | ns | 0.0246 | 9.16 ± 0.15 | 9.82 ± 0.12 | 0.66 | ns | 0.0222 |
| CDCP1 | 1.56 ± 0.14 | 2.16 ± 0.17 | 0.60 | ns | ns | 1.64 ± 0.14 | 2.37 ± 0.18 | 0.73 | ns | 0.0274 |
| CES1 | 1.41 ± 0.05 | 2.04 ± 0.20 | 0.63 | ns | ns | 1.31 ± 0.04 | 2.02 ± 0.16 | 0.71 | ns | 0.0136 |
| CHI3L1 | 5.27 ± 0.21 | 6.21 ± 0.27 | 0.94 | ns | ns | 5.60 ± 0.13 | 6.61 ± 0.27 | 1.01 | ns | 0.0266 |
| CHL1 | 2.47 ± 0.09 | 2.14 ± 0.04 | −0.33 | ns | 0.0406 | 2.64 ± 0.12 | 2.20 ± 0.05 | −0.43 | ns | 0.0366 |
| CSF-1 | 7.03 ± 0.06 | 7.30 ± 0.07 | 0.27 | ns | ns | 7.13 ± 0.05 | 7.43 ± 0.08 | 0.30 | ns | 0.0349 |
| CSTB | 3.65 ± 0.14 | 4.27 ± 0.12 | 0.62 | ns | 0.0321 | 3.65 ± 0.13 | 4.54 ± 0.21 | 0.89 | ns | 0.0155 |
| CTSD | 3.76 ± 0.09 | 4.61 ± 0.11 | 0.85 | 0.0025 | 0.0003 | 4.14 ± 0.05 | 4.94 ± 0.14 | 0.79 | 0.0400 | 0.0026 |
| CTSZ | 3.81 ± 0.13 | 4.33 ± 0.12 | 0.52 | ns | ns | 3.98 ± 0.06 | 4.46 ± 0.14 | 0.47 | ns | 0.0373 |
| CXCL10 | 8.82 ± 0.17 | 9.51 ± 0.15 | 0.69 | ns | ns | 8.70 ± 0.18 | 9.59 ± 0.15 | 0.89 | ns | 0.0127 |
| CXCL11 | 6.36 ± 0.16 | 6.96 ± 0.18 | 0.60 | ns | ns | 6.87 ± 0.16 | 7.94 ± 0.26 | 1.07 | ns | 0.0213 |
| ENG | 1.47 ± 0.07 | 1.40 ± 0.06 | −0.07 | ns | ns | 1.54 ± 0.05 | 1.31 ± 0.03 | −0.23 | ns | 0.0183 |
| FABP4 | 3.65 ± 0.30 | 5.57 ± 0.15 | 1.93 | 0.0115 | 0.0010 | 3.78 ± 0.26 | 5.72 ± 0.16 | 1.93 | 0.0024 | 0.0004 |
| FCN2 | 4.48 ± 0.16 | 5.22 ± 0.10 | 0.74 | ns | 0.0106 | 4.04 ± 0.14 | 4.87 ± 0.10 | 0.83 | 0.0422 | 0.0026 |
| FGF-21 (CVD II) | 4.56 ± 0.43 | 7.12 ± 0.48 | 2.56 | ns | 0.0105 | 4.54 ± 0.43 | 7.03 ± 0.48 | 2.49 | ns | 0.0105 |
| FGF-21 (I) | 3.50 ± 0.41 | 5.98 ± 0.44 | 2.48 | ns | 0.0086 | 3.59 ± 0.40 | 6.03 ± 0.44 | 2.44 | ns | 0.0081 |
| Gal-9 | 6.96 ± 0.04 | 7.51 ± 0.07 | 0.55 | 0.0019 | 0.0003 | 7.07 ± 0.06 | 7.64 ± 0.08 | 0.57 | 0.0045 | 0.0005 |
| GH | 9.51 ± 0.69 | 6.38 ± 0.66 | −3.13 | ns | 0.0318 | 9.63 ± 0.69 | 6.46 ± 0.64 | −3.17 | ns | 0.0222 |
| GLO1 | 3.29 ± 0.10 | 3.78 ± 0.18 | 0.49 | ns | ns | 4.66 ± 0.17 | 5.56 ± 0.25 | 0.90 | ns | 0.0415 |
| HAOX1 | 2.90 ± 0.30 | 4.71 ± 0.41 | 1.80 | ns | 0.0220 | 2.97 ± 0.31 | 4.81 ± 0.42 | 1.84 | ns | 0.0183 |
| HB-EGF | 3.84 ± 0.09 | 3.94 ± 0.08 | 0.10 | ns | ns | 5.32 ± 0.13 | 6.52 ± 0.16 | 1.20 | 0.0030 | 0.0004 |
| HGF | 6.76 ± 0.07 | 7.62 ± 0.14 | 0.86 | 0.0249 | 0.0019 | 7.58 ± 0.09 | 8.60 ± 0.14 | 1.02 | 0.0026 | 0.0004 |
| IGFBP-1 | 3.85 ± 0.20 | 1.47 ± 0.26 | −2.38 | 0.0002 | 0.0001 | 3.98 ± 0.18 | 1.55 ± 0.27 | −2.44 | 0.0002 | 0.0002 |
| IGFBP-2 | 6.65 ± 0.25 | 5.81 ± 0.12 | −0.85 | ns | ns | 6.84 ± 0.21 | 5.95 ± 0.11 | −0.89 | ns | 0.0146 |
| IL-1ra | 5.33 ± 0.09 | 7.05 ± 0.21 | 1.72 | 0.0013 | 0.0003 | 5.80 ± 0.10 | 7.47 ± 0.20 | 1.67 | 0.0009 | 0.0004 |
| IL-6 | 2.36 ± 0.17 | 4.17 ± 0.34 | 1.81 | ns | 0.0044 | 2.46 ± 0.16 | 4.22 ± 0.33 | 1.75 | ns | 0.0043 |
| IL-10RB | 6.34 ± 0.09 | 6.70 ± 0.08 | 0.36 | ns | 0.0473 | 6.53 ± 0.08 | 6.94 ± 0.08 | 0.41 | ns | 0.0146 |
| IL-18 | 7.75 ± 0.14 | 8.47 ± 0.20 | 0.72 | ns | ns | 7.88 ± 0.15 | 8.67 ± 0.22 | 0.79 | ns | 0.0396 |
| IL-18R1 | 6.61 ± 0.11 | 7.16 ± 0.13 | 0.55 | ns | 0.0360 | 6.80 ± 0.09 | 7.37 ± 0.13 | 0.57 | ns | 0.0188 |
| KIT | 3.31 ± 0.08 | 2.84 ± 0.09 | −0.47 | ns | 0.0095 | 3.29 ± 0.08 | 2.99 ± 0.10 | −0.30 | ns | ns |
| LAP TGF-β−1 | 5.64 ± 0.11 | 6.01 ± 0.08 | 0.38 | ns | ns | 6.94 ± 0.09 | 7.28 ± 0.08 | 0.34 | ns | 0.0417 |
| LEP | 4.06 ± 0.32 | 6.66 ± 0.11 | 2.60 | 0.0019 | 0.0003 | 4.09 ± 0.34 | 6.81 ± 0.11 | 2.72 | 0.0018 | 0.0004 |
| LILRB2 | 2.18 ± 0.09 | 2.62 ± 0.08 | 0.44 | ns | 0.0225 | 2.11 ± 0.11 | 2.68 ± 0.08 | 0.58 | ns | 0.0083 |
| LTBR | 1.75 ± 0.12 | 2.02 ± 0.08 | 0.27 | ns | ns | 1.84 ± 0.03 | 2.10 ± 0.05 | 0.26 | ns | 0.0043 |
| MCP-1 | 9.35 ± 0.07 | 9.87 ± 0.05 | 0.52 | 0.0058 | 0.0005 | 10.52 ± 0.13 | 11.02 ± 0.13 | 0.51 | ns | ns |
| MCP-3 | 1.39 ± 0.00 | 1.48 ± 0.04 | 0.09 | ns | ns | 1.42 ± 0.02 | 2.07 ± 0.10 | 0.65 | 0.0254 | 0.0018 |
| MCP-4 | 2.19 ± 0.16 | 2.72 ± 0.12 | 0.53 | ns | ns | 3.56 ± 0.17 | 4.36 ± 0.22 | 0.80 | ns | 0.0450 |
| MIP-1 alpha (I) | 3.35 ± 0.03 | 3.83 ± 0.06 | 0.48 | 0.0027 | 0.0003 | 3.72 ± 0.07 | 4.28 ± 0.08 | 0.56 | 0.0107 | 0.0009 |
| MPO | 2.30 ± 0.23 | 2.91 ± 0.08 | 0.61 | ns | ns | 2.92 ± 0.13 | 3.52 ± 0.12 | 0.59 | ns | 0.0222 |
| NCAM1 | 2.27 ± 0.09 | 1.89 ± 0.06 | −0.38 | ns | 0.0191 | 2.26 ± 0.10 | 1.90 ± 0.08 | −0.36 | ns | ns |
| NEMO | 3.40 ± 0.17 | 3.76 ± 0.22 | 0.36 | ns | ns | 1.58 ± 0.04 | 2.27 ± 0.18 | 0.69 | ns | 0.0223 |
| OSM | 2.40 ± 0.12 | 3.39 ± 0.21 | 1.00 | ns | 0.0106 | 3.95 ± 0.16 | 5.14 ± 0.29 | 1.19 | ns | 0.0208 |
| PAI-1 | 3.49 ± 0.31 | 5.80 ± 0.23 | 2.31 | 0.0030 | 0.0003 | 7.21 ± 0.09 | 7.60 ± 0.07 | 0.39 | ns | 0.0274 |
| PLC | 5.00 ± 0.14 | 5.45 ± 0.07 | 0.45 | ns | ns | 5.23 ± 0.06 | 5.61 ± 0.03 | 0.38 | 0.0064 | 0.0006 |
| PON3 | 5.45 ± 0.26 | 4.29 ± 0.27 | −1.17 | ns | 0.0406 | 5.43 ± 0.21 | 4.22 ± 0.26 | −1.22 | ns | 0.0146 |
| PRCP | 0.78 ± 0.07 | 1.11 ± 0.06 | 0.33 | ns | 0.0231 | 0.68 ± 0.05 | 1.11 ± 0.06 | 0.43 | 0.0067 | 0.0006 |
| PRSS8 | 8.75 ± 0.08 | 9.20 ± 0.08 | 0.45 | ns | 0.0106 | 8.93 ± 0.09 | 9.43 ± 0.08 | 0.50 | ns | 0.0073 |
| RARRES2 | 9.60 ± 0.13 | 10.22 ± 0.07 | 0.62 | ns | 0.0106 | 9.97 ± 0.09 | 10.44 ± 0.05 | 0.48 | ns | 0.0036 |
| SCGB3A2 | 2.12 ± 0.26 | 0.96 ± 0.12 | −1.15 | ns | 0.0130 | 2.23 ± 0.26 | 0.98 ± 0.13 | −1.25 | ns | 0.0081 |
| SELE | 2.10 ± 0.13 | 2.81 ± 0.12 | 0.70 | ns | 0.0095 | 2.26 ± 0.12 | 2.93 ± 0.14 | 0.67 | ns | 0.0146 |
| SPON2 | 9.78 ± 0.04 | 10.01 ± 0.04 | 0.24 | ns | 0.0036 | 10.31 ± 0.05 | 10.54 ± 0.03 | 0.23 | ns | 0.0105 |
| STAMPB | 3.16 ± 0.12 | 3.56 ± 0.15 | 0.41 | ns | ns | 1.84 ± 0.04 | 2.38 ± 0.16 | 0.54 | ns | 0.0450 |
| t-PA | 4.06 ± 0.20 | 5.23 ± 0.09 | 1.17 | 0.0276 | 0.0019 | 4.28 ± 0.24 | 5.89 ± 0.09 | 1.61 | 0.0082 | 0.0007 |
| TGM2 | 6.03 ± 0.13 | 6.35 ± 0.08 | 0.32 | ns | ns | 3.85 ± 0.08 | 4.66 ± 0.17 | 0.81 | ns | 0.0082 |
| TNF-R1 | 4.69 ± 0.14 | 5.27 ± 0.06 | 0.58 | ns | 0.0220 | 5.01 ± 0.06 | 5.53 ± 0.06 | 0.52 | 0.0019 | 0.0004 |
| TNF-R2 | 3.24 ± 0.13 | 3.64 ± 0.04 | 0.41 | ns | ns | 3.42 ± 0.07 | 3.77 ± 0.07 | 0.35 | ns | 0.0146 |
| TNFRSF10A | 2.06 ± 0.05 | 2.27 ± 0.07 | 0.22 | ns | ns | 2.13 ± 0.07 | 2.42 ± 0.07 | 0.29 | ns | 0.0462 |
| TNFRSF11A | 4.04 ± 0.09 | 4.60 ± 0.08 | 0.56 | ns | 0.0036 | 4.40 ± 0.12 | 4.94 ± 0.08 | 0.54 | ns | 0.0146 |
| TNFSF14 | 2.96 ± 0.08 | 3.73 ± 0.08 | 0.77 | 0.0004 | 0.0001 | 4.23 ± 0.12 | 4.97 ± 0.19 | 0.74 | ns | 0.0274 |
| TR-AP | 3.40 ± 0.14 | 3.89 ± 0.08 | 0.49 | ns | ns | 3.54 ± 0.14 | 4.12 ± 0.10 | 0.58 | ns | 0.0266 |
| TRAIL-R2 | 4.40 ± 0.09 | 4.66 ± 0.05 | 0.26 | ns | ns | 4.59 ± 0.09 | 4.89 ± 0.05 | 0.30 | ns | 0.0481 |
| TRAIL | 7.25 ± 0.09 | 7.61 ± 0.08 | 0.36 | ns | ns | 7.49 ± 0.09 | 7.92 ± 0.09 | 0.43 | ns | 0.0213 |
| U-PAR | 3.28 ± 0.15 | 3.64 ± 0.06 | 0.36 | ns | ns | 3.78 ± 0.07 | 4.26 ± 0.10 | 0.48 | ns | 0.0104 |
| VEGF-A | 9.19 ± 0.06 | 9.58 ± 0.05 | 0.40 | 0.0196 | 0.0016 | 9.85 ± 0.13 | 10.40 ± 0.09 | 0.55 | ns | 0.0213 |
| vWF | 3.18 ± 0.18 | 3.55 ± 0.11 | 0.37 | ns | ns | 6.21 ± 0.15 | 6.95 ± 0.18 | 0.73 | ns | 0.0349 |
Differences in mean normalized protein expression (NPX) values between obese and lean groups are reported as a log2 ratio. p values were adjusted for multiple comparisons using either Holm-Bonferroni (H-B) or false discovery rate (FDR); ns, not significant (adj. p > 0.05). For proteins that are present in duplicate protein panels, the panel is indicated in parentheses: I, inflammation; CVII, cardiovascular II; and CVIII, cardiovascular III.
Protein correlations in plasma versus serum
For the correlation analysis, 316 proteins survived the cut-off criteria (Supplementary Fig. S1). We observed significant correlations between plasma and serum samples for most (68.8%) of the proteins analyzed in the lean and obese groups combined (Table 3), although fewer significant correlations were seen when dividing the cohort into obese and lean (Supplementary Table S4). Of the 10 proteins that were measured in duplicate panels, eight displayed similar correlations between plasma and serum. However, MCP-1 and uPA only showed a significant correlation between plasma and serum in one of the duplicate panels.
Table 3.
Correlations of protein concentrations in plasma versus serum in all subjects.
| Inflammation | Cardiometabolic | Cardiovascular II | Cardiovascular III | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein | r | H-B Adj. p value | Protein | r | H-B Adj. p value | Protein | r | H-B Adj. p value | Protein | r | H-B Adj. p value |
| FGF-21 | 1.00 | 1.57E-20 | MBL2 | 0.99 | 2.79E-15 | GH | 1.00 | 1.47E-25 | IGFBP-1 | 0.98 | 4.93E-13 |
| FGF-19 | 0.99 | 1.11E-15 | FCGR2A | 0.98 | 6.92E-14 | FGF-21 | 1.00 | 1.82E-22 | Ep-CAM | 0.97 | 1.47E-11 |
| CCL20 | 0.99 | 7.03E-15 | LILRB5 | 0.97 | 5.69E-12 | LEP | 1.00 | 2.85E-21 | FABP4 | 0.96 | 1.32E-10 |
| IL-18 | 0.98 | 3.69E-14 | FCN2 | 0.97 | 9.41E-11 | HAOX1 | 1.00 | 8.14E-20 | CHIT1 | 0.96 | 1.85E-10 |
| MMP-10 | 0.98 | 1.01E-13 | LYVE1 | 0.96 | 6.02E-10 | SERPINA12 | 0.99 | 1.00E-17 | TFF3 | 0.96 | 2.79E-10 |
| CXCL9 | 0.98 | 2.95E-13 | CCL18 | 0.95 | 1.43E-09 | IL-6 | 0.99 | 1.90E-16 | SCGB3A2 | 0.95 | 3.62E-09 |
| CDCP1 | 0.97 | 2.68E-11 | COMP | 0.95 | 6.47E-09 | FABP2 | 0.99 | 8.91E-16 | IGFBP-2 | 0.94 | 2.47E-08 |
| TRANCE | 0.97 | 6.59E-11 | TIMD4 | 0.95 | 9.36E-09 | KIM-1 | 0.99 | 2.38E-14 | CCL24 | 0.94 | 2.85E-08 |
| MCP-2 | 0.97 | 1.05E-10 | THBS4 | 0.94 | 1.10E-08 | IL-18 | 0.98 | 3.44E-13 | PON3 | 0.94 | 3.53E-08 |
| CCL19 | 0.96 | 4.09E-10 | IGLC2 | 0.94 | 2.73E-08 | GIF | 0.98 | 1.35E-12 | TR | 0.93 | 9.55E-08 |
| IL-12B | 0.96 | 1.30E-09 | REG1A | 0.94 | 3.16E-08 | CTRC | 0.98 | 2.26E-12 | CCL22 | 0.92 | 1.88E-07 |
| OPG | 0.95 | 4.06E-09 | CR2 | 0.94 | 3.88E-08 | MMP-12 | 0.98 | 4.02E-12 | t-PA | 0.92 | 2.80E-07 |
| PD-L1 | 0.95 | 4.80E-09 | FCGR3B | 0.93 | 5.93E-08 | REN | 0.97 | 1.18E-11 | CPA1 | 0.92 | 5.11E-07 |
| CXCL10 | 0.95 | 9.69E-09 | PRSS2 | 0.93 | 7.84E-08 | IL-1ra | 0.97 | 1.28E-11 | DLK-1 | 0.91 | 8.47E-07 |
| IL-18R1 | 0.94 | 1.68E-08 | ANGPTL3 | 0.93 | 1.56E-07 | SCF | 0.97 | 4.73E-11 | CPB1 | 0.91 | 9.00E-07 |
| Flt3L | 0.93 | 7.43E-08 | SAA4 | 0.93 | 1.61E-07 | ADM | 0.96 | 1.70E-10 | TNFRSF10C | 0.90 | 1.99E-06 |
| uPA | 0.93 | 8.58E-08 | TNC | 0.92 | 2.47E-07 | ACE2 | 0.96 | 1.05E-09 | CHI3L1 | 0.89 | 4.47E-06 |
| CD6 | 0.93 | 1.38E-07 | NRP1 | 0.92 | 2.75E-07 | LPL | 0.95 | 2.47E-09 | CCL15 | 0.89 | 6.88E-06 |
| SCF | 0.93 | 1.45E-07 | DPP4 | 0.92 | 5.55E-07 | MMP-7 | 0.95 | 2.92E-09 | MMP-3 | 0.89 | 7.27E-06 |
| CCL23 | 0.92 | 2.06E-07 | CRTAC1 | 0.90 | 2.52E-06 | PRSS8 | 0.95 | 3.92E-09 | TIMP4 | 0.88 | 1.43E-05 |
| TNFB | 0.92 | 2.87E-07 | APOM | 0.89 | 5.45E-06 | XCL1 | 0.95 | 7.68E-09 | LDL receptor | 0.87 | 2.59E-05 |
| TRAIL | 0.91 | 6.72E-07 | GP1BA | 0.89 | 6.21E-06 | VEGF-D | 0.95 | 8.75E-09 | SELE | 0.87 | 3.29E-05 |
| CD244 | 0.91 | 6.72E-07 | LILRB2 | 0.89 | 7.09E-06 | TNFRSF13B | 0.94 | 1.13E-08 | ST2 | 0.84 | 0.0002 |
| CCL25 | 0.90 | 2.94E-06 | FETUB | 0.89 | 8.64E-06 | HO-1 | 0.94 | 1.23E-08 | IL-6RA | 0.84 | 0.0002 |
| OSM | 0.87 | 2.65E-05 | CDH1 | 0.88 | 1.36E-05 | BMP-6 | 0.93 | 6.58E-08 | CTSZ | 0.83 | 0.0003 |
| CCL11 | 0.87 | 3.92E-05 | TIE1 | 0.88 | 1.95E-05 | IgG Fc R II-b | 0.93 | 7.84E-08 | SHPS-1 | 0.81 | 0.0008 |
| CST5 | 0.86 | 5.29E-05 | NCAM1 | 0.87 | 2.06E-05 | IL16 | 0.93 | 8.80E-08 | CTSD | 0.81 | 0.0009 |
| CCL28 | 0.85 | 0.0001 | TCN2 | 0.87 | 2.34E-05 | RAGE | 0.93 | 1.07E-07 | CCL16 | 0.80 | 0.001 |
| IL-10RB | 0.85 | 0.0001 | AOC3 | 0.87 | 3.12E-05 | TIE2 | 0.93 | 1.26E-07 | GDF-15 | 0.80 | 0.001 |
| HGF | 0.85 | 0.0001 | VCAM1 | 0.85 | 0.0001 | MERTK | 0.92 | 3.06E-07 | Gal-4 | 0.79 | 0.002 |
| CCL4 | 0.84 | 0.0001 | TGFBI | 0.84 | 0.0002 | TF | 0.92 | 4.20E-07 | CD93 | 0.79 | 0.002 |
| TNFRSF9 | 0.84 | 0.0002 | F7 | 0.84 | 0.0002 | TRAIL-R2 | 0.92 | 4.51E-07 | CD163 | 0.78 | 0.003 |
| CSF-1 | 0.83 | 0.0003 | C2 | 0.84 | 0.0002 | IL27 | 0.92 | 4.63E-07 | RARRES2 | 0.77 | 0.004 |
| IL-8 | 0.83 | 0.0004 | ANG | 0.84 | 0.0002 | Gal-9 | 0.91 | 6.07E-07 | IL2-RA | 0.75 | 0.007 |
| MIP-1α | 0.82 | 0.0005 | SERPINA7 | 0.83 | 0.0004 | IL1RL2 | 0.91 | 9.77E-07 | RETN | 0.75 | 0.008 |
| CXCL11 | 0.79 | 0.002 | OSMR | 0.83 | 0.0004 | AGRP | 0.91 | 1.06E-06 | BLM hydrol. | 0.75 | 0.008 |
| ADA | 0.78 | 0.003 | IGFBP6 | 0.82 | 0.0005 | CTSL1 | 0.91 | 1.53E-06 | MPO | 0.74 | 0.009 |
| TWEAK | 0.78 | 0.003 | ICAM3 | 0.81 | 0.0008 | TNFRSF11A | 0.90 | 2.86E-06 | IL-17RA | 0.73 | 0.01 |
| MCP-4 | 0.78 | 0.003 | PROC | 0.81 | 0.0008 | CD4 | 0.89 | 7.67E-06 | TNF-R1 | 0.73 | 0.01 |
| CD5 | 0.77 | 0.004 | ICAM1 | 0.80 | 0.001 | TM | 0.88 | 1.09E-05 | ICAM-2 | 0.73 | 0.01 |
| CD40 | 0.77 | 0.004 | QPCT | 0.79 | 0.002 | MARCO | 0.88 | 1.12E-05 | TLT-2 | 0.73 | 0.02 |
| 4E-BP1 | 0.76 | 0.006 | PRCP | 0.79 | 0.002 | FS | 0.88 | 1.15E-05 | IL-18BP | 0.72 | 0.02 |
| LIF-R | 0.76 | 0.007 | IL7R | 0.79 | 0.002 | DCN | 0.87 | 3.66E-05 | IL-1RT2 | 0.72 | 0.02 |
| DNER | 0.75 | 0.007 | C1QTNF1 | 0.78 | 0.003 | SOD2 | 0.86 | 5.53E-05 | PI3 | 0.71 | 0.02 |
| IL-10 | 0.75 | 0.008 | CHL1 | 0.78 | 0.003 | CCL3 | 0.85 | 9.10E-05 | PAI-1 | 0.71 | 0.02 |
| MMP-1 | 0.73 | 0.01 | SERPINA5 | 0.77 | 0.004 | hOSCAR | 0.84 | 0.0002 | COL1A1 | 0.71 | 0.02 |
| MCP-1 | 0.70 | 0.03 | SPARCL1 | 0.77 | 0.004 | PD-L2 | 0.83 | 0.0004 | MEPE | 0.71 | 0.02 |
| TNFSF14 | 0.70 | 0.03 | IGFBP3 | 0.76 | 0.005 | THBS2 | 0.82 | 0.0005 | TFPI | 0.71 | 0.02 |
| EN-RAGE | 0.69 | 0.04 | NID1 | 0.76 | 0.006 | PlGF | 0.82 | 0.0005 | OPG | 0.70 | 0.03 |
| β-NGF | 0.69 | 0.04 | SELL | 0.76 | 0.007 | Protein BOC | 0.82 | 0.0005 | MB | 0.69 | 0.04 |
| VEGF-A | 0.68 | 0.048 | PCOLCE | 0.75 | 0.008 | PAR-1 | 0.81 | 0.0009 | TR-AP | 0.69 | 0.04 |
| SLAMF1 | 0.65 | ns | CST3 | 0.75 | 0.008 | PRELP | 0.77 | 0.004 | PLC | 0.64 | ns |
| CXCL6 | 0.62 | ns | CD59 | 0.74 | 0.01 | AMBP | 0.76 | 0.005 | vWF | 0.64 | ns |
| LAP TGF-β-1 | 0.59 | ns | GAS6 | 0.74 | 0.01 | SORT1 | 0.76 | 0.006 | MMP-9 | 0.63 | ns |
| CXCL1 | 0.56 | ns | CFHR5 | 0.73 | 0.01 | VSIG2 | 0.76 | 0.006 | CDH5 | 0.63 | ns |
| CXCL5 | 0.56 | ns | ST6GAL1 | 0.72 | 0.02 | SPON2 | 0.73 | 0.01 | PSP-D | 0.63 | ns |
| CX3CL1 | 0.55 | ns | LILRB1 | 0.71 | 0.03 | CCL17 | 0.73 | 0.01 | uPA | 0.62 | ns |
| STAMPB | 0.47 | ns | F11 | 0.64 | ns | CD84 | 0.70 | 0.03 | GRN | 0.62 | ns |
| FGF-23 | 0.43 | ns | CA4 | 0.62 | ns | THPO | 0.64 | ns | ITGB2 | 0.62 | ns |
| FGF-5 | 0.42 | ns | TIMP1 | 0.62 | ns | IDUA | 0.64 | ns | Gal-3 | 0.61 | ns |
| CASP-8 | 0.25 | ns | LCN2 | 0.62 | ns | FGF-23 | 0.63 | ns | AXL | 0.60 | ns |
| ST1A1 | 0.23 | ns | PAM | 0.56 | ns | GLO1 | 0.63 | ns | PGLYRP1 | 0.60 | ns |
| IL-7 | 0.18 | ns | VASN | 0.54 | ns | PSGL-1 | 0.61 | ns | CSTB | 0.59 | ns |
| TGF-α | 0.18 | ns | KIT | 0.52 | ns | CXCL1 | 0.60 | ns | PECAM-1 | 0.59 | ns |
| ARTN | # | # | CNDP1 | 0.51 | ns | PRSS27 | 0.53 | ns | TNF-R2 | 0.58 | ns |
| AXIN1 | # | # | TNXB | 0.51 | ns | ANG-1 | 0.52 | ns | U-PAR | 0.58 | ns |
| GDNF | # | # | ENG | 0.50 | ns | LOX-1 | 0.52 | ns | TNFRSF14 | 0.58 | ns |
| IL-10RA | # | # | MET | 0.49 | ns | ADAM-TS13 | 0.48 | ns | CNTN1 | 0.56 | ns |
| IL-15RA | # | # | GNLY | 0.48 | ns | TGM2 | 0.48 | ns | Notch 3 | 0.53 | ns |
| IL-17A | # | # | TGFBR3 | 0.48 | ns | PTX3 | 0.48 | ns | FAS | 0.53 | ns |
| IL-17C | # | # | CD46 | 0.47 | ns | CEACAM8 | 0.44 | ns | IGFBP-7 | 0.52 | ns |
| IL-20RA | # | # | CA1 | 0.45 | ns | GDF-2 | 0.42 | ns | TNFSF13B | 0.51 | ns |
| IL-22RA1 | # | # | PLXNB2 | 0.44 | ns | CD40-L | 0.39 | ns | IL-1RT1 | 0.51 | ns |
| IL-24 | # | # | EFEMP1 | 0.43 | ns | Dkk-1 | 0.36 | ns | AP-N | 0.51 | ns |
| IL-2RB | # | # | CCL14 | 0.39 | ns | HB-EGF | 0.31 | ns | SELP | 0.50 | ns |
| IL-4 | # | # | CA3 | 0.38 | ns | PDGF-B | 0.31 | ns | CXCL16 | 0.49 | ns |
| IL-5 | # | # | COL18A1 | 0.36 | ns | SRC | 0.26 | ns | MMP-2 | 0.49 | ns |
| IL-6 | # | # | NOTCH1 | 0.35 | ns | PIgR | 0.24 | ns | OPN | 0.46 | ns |
| LIF | # | # | PTPRS | 0.32 | ns | IL-17D | 0.24 | ns | PRTN3 | 0.44 | ns |
| MCP-3 | # | # | CCL5 | 0.22 | ns | HSP 27 | 0.20 | ns | LTBR | 0.38 | ns |
| NRTN | # | # | MFAP5 | 0.21 | ns | BNP | # | # | SPON1 | 0.36 | ns |
| NT3 | # | # | MEGF9 | 0.11 | ns | CA5A | # | # | MCP-1 | 0.34 | ns |
| SIRT2 | # | # | CES1 | # | # | DECR1 | # | # | ALCAM | 0.28 | ns |
| IFN-γ | nd | nd | DEFA1 | # | # | GT | # | # | PCSK9 | 0.27 | ns |
| IL-1α | nd | nd | FAP | # | # | IL-4RA | # | # | JAM-A | 0.24 | ns |
| IL-13 | nd | nd | ITGAM | # | # | ITGB1BP2 | # | # | CASP-3 | 0.20 | ns |
| IL-2 | nd | nd | LTBP2 | # | # | NEMO | # | # | PDGF-A | 0.16 | ns |
| IL-20 | nd | nd | PLA2G7 | # | # | PAPPA | # | # | KLK6 | 0.15 | ns |
| IL-33 | nd | nd | PLTP | # | # | PARP-1 | # | # | AZU1 | 0.10 | ns |
| TNF | nd | nd | REG3A | # | # | SLAMF7 | # | # | EGFR | −0.02 | ns |
| TSLP | nd | nd | SOD1 | # | # | STK4 | # | # | EPHB4 | # | # |
| BDNF | Ϯ | Ϯ | UMOD | # | # | TNFRSF10A | # | # | NT-Pro-BNP | # | # |
Pearson correlations (r) between NPX values in plasma and serum samples from the total cohort (n = 22) are shown. p values were adjusted by the Holm-Bonferroni (H-B) multiple comparison test; ns, not significant (adj. p > 0.05). nd, not detected. #Excluded due to too many missing values. Ϯ, removed due to technical issue.
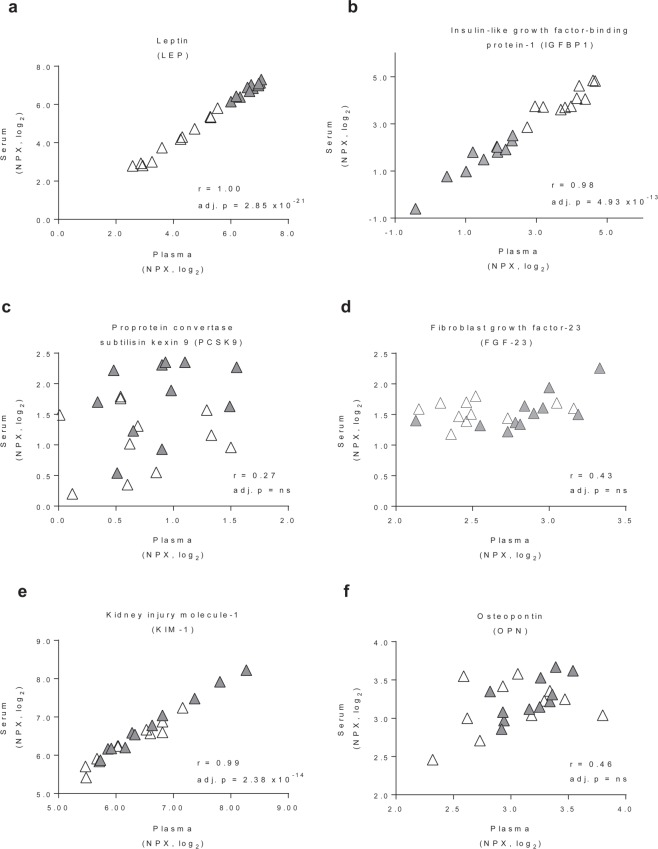
We observed good correlations between plasma and serum samples for leptin (r = 1.00, adj. p < 0.001) and IGFBP-1 (r = 0.98, adj. p < 0.001), which are proteins that exhibited obesity-associated differences in concentration (Fig. 2a,b). Some proteins showed poor correlations, such as PCSK9 (r = 0.27, ns) and FGF-23 (r = 0.43 and 0.64 in the inflammation and cardiovascular II panels, respectively, both ns) (Fig. 2c,d). PCSK9 binds to the receptor for low-density lipoprotein and PCSK9 inhibitors are therefore of intense interest to pharmaceutical companies39,40. Studies interchangeably measure PCSK9 in plasma41,42 and serum43,44, but our result indicates that the choice of biofluid could potentially have a significant impact on the conclusions drawn. Our panels also included the FDA-approved biomarkers KIM-1 and osteopontin, which are used to monitor kidney disease45,46. KIM-1 was well correlated between plasma and serum (r = 0.99, adj. p < 0.001) but osteopontin displayed a poor correlation (r = 0.46, ns) (Fig. 2e,f).
Figure 2.
Protein correlations in plasma versus serum. Pearson correlations (r) between normalized protein expression (NPX) values for proteins in plasma and serum samples. Each data point is from one individual (open triangles: obese; closed triangles: lean). p values were adjusted by the Holm-Bonferroni multiple comparison test.
Lipids in plasma versus serum, and in lean versus obese groups
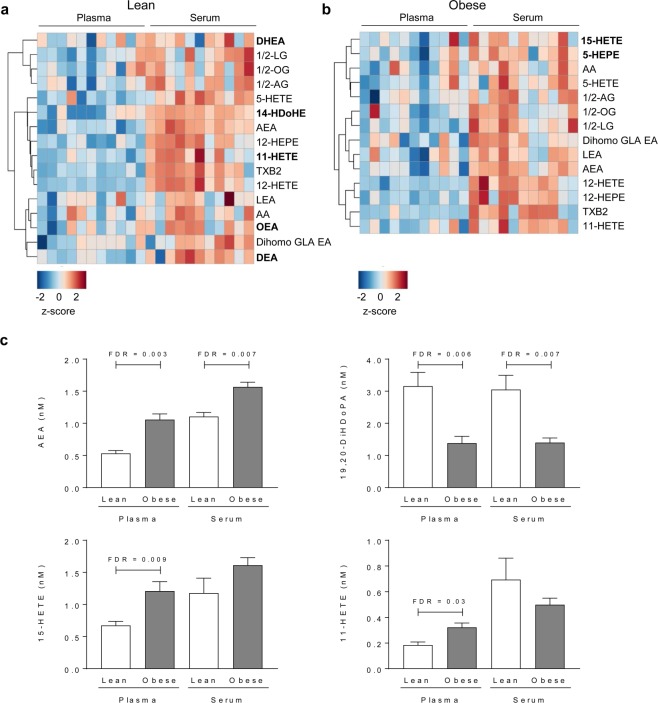
We also performed targeted lipidomics of inflammation-related lipids in plasma and serum from the lean and obese groups. Of the 76 lipids analyzed (see Supplementary Table S2), two were excluded as they did not survive the cut-off criteria for the comparative analysis (Supplementary Fig. S4). For most of the lipids, there were no major differences in concentration between plasma and serum (Supplementary Fig. S5). We observed that concentrations of 21.6% of the lipids in the lean cohort and 18.9% of the lipids in the obese cohort were significantly higher in serum than in plasma (after FDR adjustment, adj. p < 0.05); none of the lipids showed lower concentrations in serum (Fig. 3a,b). In total, 73 lipids survived the cut-off for the correlation analyses; we observed significant correlations between plasma and serum for 64% of the analyzed lipids when analyzed in the lean and obese groups combined (Table 4), and fewer significant correlations were seen when dividing the cohort into obese and lean (Supplementary Table S5).
Figure 3.
Oxylipins in plasma versus serum, and in lean versus obese groups. Heatmaps showing lipids that exhibited significantly different concentrations in plasma versus serum in (a) lean subjects (n = 11) and (b) obese subjects (n = 11) after adjustment for multiple comparisons using the false discovery rate (FDR) test at adj. p < 0.05. Lipids that are significantly different in only one of the groups (lean or obese) are marked in bold. Relative lipid concentrations are reported as z-scores. (c) Lipids that showed significantly different concentrations between the obese and lean groups in plasma and/or serum after FDR adjustment.
Table 4.
Correlations of lipid concentrations in plasma versus serum in all subjects.
| Lipid | r | H-B Adj. p value |
|---|---|---|
| 9,10-DiHOME | 0.99 | 1.78E-18 |
| 9,10-DiHODE | 0.99 | 9.18E-18 |
| 13-HODE | 0.99 | 2.59E-15 |
| 12,13-DiHOME | 0.99 | 3.83E-15 |
| 15,16-DiHODE | 0.98 | 4.77E-13 |
| 19,20-DiHDoPA | 0.97 | 1.26E-12 |
| 9-HOTE | 0.97 | 3.93E-12 |
| 9-HODE | 0.97 | 1.18E-11 |
| 13-HOTE | 0.96 | 1.48E-10 |
| 12(13)-EpOME | 0.96 | 2.04E-10 |
| 15(16)-EpODE | 0.94 | 7.71E-09 |
| DHA | 0.93 | 1.60E-08 |
| C16:1n7 | 0.93 | 3.84E-08 |
| EPA | 0.90 | 5.81E-07 |
| C18:2n6 | 0.90 | 6.28E-07 |
| C14:0 | 0.89 | 1.40E-06 |
| AA | 0.89 | 1.83E-06 |
| C18:1n9 | 0.89 | 2.17E-06 |
| C12:0 | 0.88 | 2.80E-06 |
| DHEA | 0.88 | 2.94E-06 |
| C18:1n7 | 0.87 | 5.36E-06 |
| ALA | 0.87 | 7.22E-06 |
| C14 Ceramide | 0.87 | 8.78E-06 |
| C18:3n3 | 0.85 | 2.07E-05 |
| C20:5n3 | 0.84 | 4.24E-05 |
| PGF2a | 0.84 | 4.75E-05 |
| LEA | 0.84 | 5.45E-05 |
| NA-Gly | 0.83 | 9.19E-05 |
| aLEA | 0.83 | 9.19E-05 |
| LA | 0.82 | 0.0001 |
| C16 Ceramide | 0.82 | 0.0001 |
| C24 dihydroceramide | 0.81 | 0.0002 |
| AEA | 0.79 | 0.0005 |
| 1/2-LG | 0.78 | 0.0008 |
| 17,18-DiHETE | 0.75 | 0.002 |
| C18:1 Ceramide | 0.75 | 0.002 |
| C24 Ceramide | 0.75 | 0.002 |
| C15:0 | 0.75 | 0.002 |
| 9(10)-EpOME | 0.72 | 0.005 |
| C18 Ceramide | 0.71 | 0.007 |
| 1/2-AG | 0.70 | 0.01 |
| C17:0 | 0.68 | 0.01 |
| 11,12-DiHETrE | 0.67 | 0.02 |
| C16:0 | 0.67 | 0.02 |
| 4-HDoHE | 0.66 | 0.02 |
| C20 Ceramide | 0.64 | 0.04 |
| 1/2-OG | 0.64 | 0.04 |
| C16:1n7t | 0.59 | ns |
| 14,15-DiHETrE | 0.59 | ns |
| Dihomo GLA EA | 0.56 | ns |
| C20:1n9 | 0.55 | ns |
| C20:2n6 | 0.54 | ns |
| 5-HETE | 0.53 | ns |
| 12(13)-Ep-9-KODE | 0.51 | ns |
| 5-HEPE | 0.50 | ns |
| DEA | 0.49 | ns |
| TXB2 | 0.45 | ns |
| 9c | 0.44 | ns |
| 15-HETE | 0.39 | ns |
| OEA | 0.37 | ns |
| 18:1 Sphingosine | 0.32 | ns |
| 12-HEPE | 0.31 | ns |
| 5,6-DiHETrE | 0.27 | ns |
| C20:4n6 | 0.25 | ns |
| C20:3n6 | 0.19 | ns |
| 9,10-e-DiHO | 0.13 | ns |
| C18:0 | 0.12 | ns |
| 11-HETE | 0.10 | ns |
| 12-HETE | 0.08 | ns |
| NO-Gly | 0.04 | ns |
| C20:0 | 0.04 | ns |
| 9-KODE | 0.01 | ns |
| 9,10-EpO | −0.27 | ns |
| C22:4n6 | # | # |
| C22:5n3 | # | # |
| 14-HDoHE | # | # |
Pearson correlations (r) between lipid concentrations in plasma and serum samples from the total cohort (n = 22) are shown. p values were adjusted by the Holm-Bonferroni (H-B) multiple comparison test; ns, not significant (adj. p > 0.05). #Excluded due to too many missing values.
Four lipids showed significantly different concentrations between the obese and lean groups in plasma and/or serum (Fig. 3c). Concentrations of AEA and 19,20-DiHDoPA were significantly different (higher for AEA and lower for 19,20-DiHDoPA in the obese group) in both plasma and serum, but concentrations of 15-HETE and 11-HETE were significantly different (both higher in the obese group) only in plasma (Fig. 3c).
Concluding remarks
In this study, we investigated whether the use of plasma or serum would yield different results when screening for obesity-related biomarkers. For most of the proteins and lipids, their concentrations showed good correlations between plasma and serum. However, it is important to note that PCSK9 concentrations did not correlate between plasma and serum, indicating that caution must be taken when comparing studies that use different biofluids. Although most of the protein and lipids had similar concentrations in plasma and serum, those that did differ were generally present at higher concentrations in serum. Importantly, we observed significantly higher concentrations of the key disease-associated biomarker PAI-1 in the obese group only in plasma and not in serum, despite the protein showing higher detectability in serum. This result highlights that sensitivity does not necessarily parallel detectability. Furthermore, some obesity-induced changes, for example of MCP-3 concentrations, were only detected in serum. Collectively, these findings show that care should be taken when choosing biofluids for the study of biomarkers, particularly those for which we report differences in sensitivity/detectability between plasma and serum.
Methods
Study participants
We recruited obese subjects [body mass index (BMI) 35–55 kg/m2, aged 18–65 years] from a cohort scheduled to undergo gastric bypass surgery, as well as age- and sex-matched lean subjects (BMI 18.5–24.9 kg/m2). Subjects were excluded if they were taking anti-inflammatory and/or immunosuppressive drugs, currently smoked, or had been diagnosed with significant gastrointestinal disease or inflammatory bowel disease. Study participants were enrolled in accordance with the Helsinki Declaration and provided written informed consent. The study was approved by the Gothenburg Ethical Review Board #682-14 (ClinicalTrials.gov NCT02322073).
Blood collection
Venous blood samples obtained from study participants after an overnight fast were collected in eitherplasma tubes spray coated with K2EDTA (Greiner Bio One) or serum tubes containing inert separator gel and silica particles as clot activator (Greiner Bio One). Plasma samples were centrifuged immediately whereas serum samples were allowed to clot for 30 min at room temperature before centrifugation (10 min at room temperature, 3,000 rpm Hettich EBA200). Samples were snap-frozen in liquid nitrogen and stored at −80 °C until analysis.
Multiplex protein assay
Protein biomarkers were analyzed using the proximity extension assay, using four protein panels (inflammation, cardiometabolic, cardiovascular II and cardiovascular III) (Olink Proteomics, Uppsala, Sweden) at the Clinical Biomarkers Facility at Science for Life Laboratory (Uppsala University, Sweden) according to the manufacturer’s instructions. Briefly, 1 µl plasma or serum was incubated with a mixture of 92 proximity antibody pairs tagged with oligonucleotides in a 96-well plate. In this assay, once a pair of antibodies binds to their corresponding antigens in close proximity, linked oligonucleotides hybridize into double stranded DNA, which is further extended and amplified, and ultimately quantified by high-throughput real-time PCR (BioMark™ HD System, Fluidigm Corporation). To avoid intra-assay variability, plasma and serum samples were analyzed on the same plate.
ELISA
Plasma and serum PAI-1 levels were measured using a commercially available ELISA for Human Total Serpin E1/PAI-1 (#DY9387-05, R&D), according to the manufacturer’s instructions. To ensure that the protein was quantified within the linear range of the standard curve, plasma and serum were diluted 1:100 and 1:500, respectively.
Measurements of oxylipins, endocannabinoids and ceramides
Oxylipins, endocannabinoids, and ceramides in plasma and serum were isolated and quantified using modifications of published protocols47–49. Briefly, plasma or serum aliquots (40 µl) were spiked with deuterated oxylipin, endocannabinoid and ceramide surrogates, mixed with butylated hydroxyl toluene and ethylene diamine tetraacetic acid, and extracted with 200 µl isopropanol containing the internal standards 1-cyclohexyl ureido, 3-dodecanoic acid and 1-phenyl ureido 3-hexanoic acid in isopropanol. The homogenate was then centrifuged (10 min, 4 °C, 15,000 g) and the isopropanol supernatant was collected and stored at −20 °C until analysis.
Analytes were separated using a Waters Acquity ultra-performance liquid chromatography (UPLC; Waters, Milford, MA) on a 2.1 mm × 150 mm, 1.7 µm BEH C18 column (Waters) for analysis of oxylipins and endocannabinoids, and 2.1 mm × 150 mm, 1.7 µm BEH C8 column (Waters) for analysis of ceramides. Separated analytes were detected by tandem mass-spectrometry, using electrospray ionization with multi reaction monitoring on an API 6500 QTRAP (Sciex, Redwood City, CA) for oxylipins and endocannabinoids, and an API 4000 QTRAP (Sciex) for ceramides. Analytes were quantified using internal standard methods and 7–9 point calibration curves of authentic standards.
Measurement of non-esterified fatty acids
Non-esterified fatty acids in plasma and serum were isolated and converted to fatty acid methyl esters (FAMEs) as previously reported47. Briefly, plasma or serum aliquots (50 µl) were spiked with lipid class surrogates, mixed with 410 µl isopropanol, followed by 520 µl cyclohexane and 570 µl 0.1 M ammonium acetate. Samples were then centrifuged (5 min, 4 °C, 15,000 g), the upper organic phase was collected, and the remainder was re-extracted with a second 520 µl cyclohexane aliquot. The samples were then dried by vacuum centrifugation and reconstituted in 100 µl toluene and 180 µl methanol. To prepare FAMEs, 280 µl of toluene/methanol extracts were enriched with 20 µl methanol containing 60 µM C15:1n5 and incubated with 45 µl 2 M TMS-diazomethane in hexane (Sigma-Aldrich, St. Louis MO) for 30 min at room temperature. Samples were dried under vacuum and the residue was dissolved in 100 µl hexane containing 4 µM C23:0, which acted as an internal standard. Samples were then stored at −20 °C until analysis.
FAMEs were separated on a 30 m × 0.25 mm × 0.25 µm DB-225 ms column in a 6890 gas chromatogram interfaced with a 5973A mass selective detector (Agilent Technologies, Santa Clara, CA). All fatty acids were quantified against a 7-point calibration curves of authentic standards. Peak identifications were based on retention times and m/z ratios, with peak confirmation by inspection of simultaneously acquired full scan spectra collected from 50–400 m/z. Calibrants and internal standards were purchased from NuchekPrep (Elysian, MN), Sigma-Aldrich, or Avanti Polar Lipids. Data were quantified using Chemstation vE.02.14 (Agilent Technologies) against 6–8 point calibration curves.
Statistical analysis
Data are reported for proteins and lipids that had <30% missing values in: (1) at least one of the four groups (lean plasma, lean serum, obese plasma, obese serum) for the comparative analyses or (2) all of the four groups for the plasma-serum correlations.
Statistical analysis of the protein multiplex data was done in the R environment (version 3.5.1) using packages gplots (3.0.1) and gdata (2.18.0)50. For the proteins reported, missing values were replaced with limit of detection (LOD) values. Hierarchical clustering with Pearson correlation distance and complete linkage confirmed that the dataset did not include outliers. Concentrations of proteins are reported as normalized protein expression (NPX) values, an arbitrary unit on a log2 scale. Heatmaps were generated using hierarchical clustering based on correlation distance and Ward’s (ward.D2) clustering. Comparisons of protein levels using Student’s t-test were paired when comparing individual donor plasma versus serum values and unpaired when comparing the lean versus obese groups; p values were adjusted for multiple comparisons using either the stringent Holm-Bonferroni test or the commonly used false discovery rate (FDR) test as indicated (adjusted p values < 0.05 were considered significant). Pearson coefficient of correlation (r) values were calculated and p values were adjusted by the Holm-Bonferroni multiple comparison test.
The pathway enrichment analysis for proteins was done using Metascape51. Briefly, Gene IDs corresponding to significantly altered proteins were analysed, using the 325 unique proteins that survived the cut-off criteria as the background list. A Gene Ontology category was deemed significantly enriched if the p value was lower than 0.01 and displayed a minimum enrichment of 1.5.
Statistical analysis of the lipidomics data was done in MetaboAnalyst52. For the lipids reported, missing values were replaced with half of the lowest reported value. Fatty acid data normalization was optimized in Jmp Pro v 12.0 and confirmed using the Shapiro-Wilk normality test. For statistical analysis, data points underwent log transformation and pareto scaling. Heatmaps were generated using hierarchical clustering based on Euclidean’s method of distance calculation and Ward’s clustering. Unadjusted p values were adjusted using FDR (adjusted p values < 0.05 were considered significant).
Supplementary information
Supplementary figures and table descriptions
Acknowledgements
Scientific input and technical support is acknowledged from the SciLifeLab, Uppsala, Sweden. The Börgeson laboratory is supported by the Wallenberg Centre for Molecular & Translational Medicine at University of Gothenburg and Knut & Alice Wallenberg Foundation, the Swedish Research Council (#2016/82), the Swedish Society for Medical Research (#S150086), Åke Wiberg’s Foundation (#M15-0058), and an ERC Starting Grant (#804418). Metabolomic analyses were supported by a NIH West Coast Metabolomics Center Pilot and Feasibility study to S.L. and E.B. funded through National Institutes of Health Grant U24 DK097154, NIH U2C 030158 and NIH U24 DK097154 (O.F.). Additional support was provided by USDA Intramural project #2032-51530-022-00D (J.W.N.). The USDA is an equal opportunity provider, and employer. S.L. is supported by grants from the NIH/NHLBI and the Swedish Heart Lung Foundation (HL128457, HL107744; Project Number: 20180199).
Author contributions
E.B., V.W. and S.L. conceived and designed the project, in consultation with O.F. and J.W.N. regarding metabolomic considerations. M.R., M.S., C.B., V.W. and E.B. collected samples. M.R., M.S., F.B., T.S., K.B. performed experiments and statistical analysis of the data. R.P., M.R. and E.B. wrote the paper. M.R., M.S., F.B., T.S., K.B., N.J.A., C.B., T.L.L., S.R., M.S., P.L., O.F., J.W.N., R.P., V.W., S.L. and E.B. analyzed and interpreted the data and commented on the manuscript. E.B. supervised this work.
Data availability
Protocols used to generate the findings of this study are available from the corresponding author upon request. In accordance with Swedish ethical regulations and GDPR, primary data from human subjects cannot be made publically available.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Meenu Rajan and Matus Sotak.
Supplementary information
is available for this paper at 10.1038/s41598-019-51673-0.
References
- 1.NCD Risk Factor Collaboration Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: A pooled analysis of 2416 population-based measurement studies in 128.9 million children, adolescents, and adults. Lancet. 2017;390:2627–2642. doi: 10.1016/S0140-6736(17)32129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borgeson E, Sharma K. Obesity, immunomodulation and chronic kidney disease. Curr Opin Pharmacol. 2013;13:618–624. doi: 10.1016/j.coph.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saltiel AR, Olefsky JM. Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest. 2017;127:1–4. doi: 10.1172/JCI92035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spite M, Claria J, Serhan CN. Resolvins, specialized proresolving lipid mediators, and their potential roles in metabolic diseases. Cell Metab. 2014;19:21–36. doi: 10.1016/j.cmet.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cominetti O, et al. Obesity shows preserved plasma proteome in large independent clinical cohorts. Sci Rep. 2018;8:16981. doi: 10.1038/s41598-018-35321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kraus VB. Biomarkers as drug development tools: Discovery, validation, qualification and use. Nat Rev Rheumatol. 2018;14:354–362. doi: 10.1038/s41584-018-0005-9. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization. Use of anticoagulants in diagnostic laboratory investigations in Diagnostic Imaging and Laboratory Technology. https://apps.who.int/iris/handle/10665/65957 (2002).
- 8.Henno LT, et al. Effect of of the anticoagulant, storage time and temperature of blood samples on the concentrations of 27 multiplex assayed cytokines - Consequences for defining reference values in healthy humans. Cytokine. 2017;97:86–95. doi: 10.1016/j.cyto.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Lima-Oliveira Gabriel, Monneret Denis, Guerber Fabrice, Guidi Gian Cesare. Sample management for clinical biochemistry assays: Are serum and plasma interchangeable specimens? Critical Reviews in Clinical Laboratory Sciences. 2018;55(7):480–500. doi: 10.1080/10408363.2018.1499708. [DOI] [PubMed] [Google Scholar]
- 10.Bruserud O. Bidirectional crosstalk between platelets and monocytes initiated by Toll-like receptor: An important step in the early defense against fungal infections? Platelets. 2013;24:85–97. doi: 10.3109/09537104.2012.678426. [DOI] [PubMed] [Google Scholar]
- 11.Brogren H, et al. Platelets synthesize large amounts of active plasminogen activator inhibitor 1. Blood. 2004;104:3943–3948. doi: 10.1182/blood-2004-04-1439. [DOI] [PubMed] [Google Scholar]
- 12.Thomas MR, Storey RF. The role of platelets in inflammation. Thromb Haemostasis. 2015;114:449–458. doi: 10.1160/Th14-12-1067. [DOI] [PubMed] [Google Scholar]
- 13.Alsaif M, et al. Analysis of serum and plasma identifies differences in molecular coverage, measurement variability, and candidate biomarker selection. Proteom Clin Appl. 2012;6:297–303. doi: 10.1002/prca.201100061. [DOI] [PubMed] [Google Scholar]
- 14.Siegbahn, A., Eriksson, N., Lindbäck, J. & Wallentin, L. A comparison of the proximity extension assay with established immunoassays in Advancing precision medicine: Current and future proteogenomic strategies for biomarker discovery and development. (Science/AAAS, Washington, DC), 22–25 (2017).
- 15.Lundblad, R. L. Considerations for the use of blood plasma and serum for proteomic analysis. Internet J Genomics Proteomics1, (2004).
- 16.Tammen H. Specimens collection of and handling: Standardization of blood sample collection. Methods Mol Biol (Clifton, N.J.) 2008;428:35–42. doi: 10.1007/978-1-59745-117-8_2. [DOI] [PubMed] [Google Scholar]
- 17.Yun SH, Sim EH, Goh RY, Park JI, Han JY. Platelet activation: The mechanisms and potential biomarkers. Biomed Res Int. 2016;2016:9060143. doi: 10.1155/2016/9060143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naldini, A., Sower, L., Bocci, V., Meyers, B. & Carney, D. H. Thrombin receptor expression and responsiveness of human monocytic cells to thrombin is linked to interferon-induced cellular differentiation. J Cell Physiol177, 76–84, doi:10.1002/(SICI)1097-4652(199810)177:1<76::AID-JCP8>3.0.CO;2-B (1998). [DOI] [PubMed]
- 19.Naldini A, Carney DH, Pucci A, Pasquali A, Carraro F. Thrombin regulates the expression of proangiogenic cytokines via proteolytic activation of protease-activated receptor-1. Gen Pharmacol. 2000;35:255–259. doi: 10.1016/s0306-3623(01)00113-6. [DOI] [PubMed] [Google Scholar]
- 20.Berlin G, Hammar M, Tapper L, Tynngard N. Effects of age, gender and menstrual cycle on platelet function assessed by impedance aggregometry. Platelets. 2019;30:473–479. doi: 10.1080/09537104.2018.1466387. [DOI] [PubMed] [Google Scholar]
- 21.Aldrighi JM, et al. Platelet activation status decreases after menopause. Gynecol Endocrinol. 2005;20:249–257. doi: 10.1080/09513590500097549. [DOI] [PubMed] [Google Scholar]
- 22.Roshan TM, Normah J, Rehman A, Naing L. Effect of menopause on platelet activation markers determined by flow cytometry. Am J Hematol. 2005;80:257–261. doi: 10.1002/ajh.20472. [DOI] [PubMed] [Google Scholar]
- 23.Markham SM, Dubin NH, Rock JA. The effect of the menstrual cycle and of decompression stress on arachidonic acid-induced platelet aggregation and on intrinsic platelet thromboxane production in women compared with men. Am J Obstet Gynecol. 1991;165:1821–1829. doi: 10.1016/0002-9378(91)90039-t. [DOI] [PubMed] [Google Scholar]
- 24.Melamed N, et al. The effect of menstrual cycle on platelet aggregation in reproductive-age women. Platelets. 2010;21:343–347. doi: 10.3109/09537101003770595. [DOI] [PubMed] [Google Scholar]
- 25.Robb AO, et al. The influence of the menstrual cycle, normal pregnancy and pre-eclampsia on platelet activation. Thromb Haemost. 2010;103:372–378. doi: 10.1160/TH08-12-0780. [DOI] [PubMed] [Google Scholar]
- 26.Vidyasagar A, Wilson NA, Djamali A. Heat shock protein 27 (HSP27): Biomarker of disease and therapeutic target. Fibrogenesis Tissue Repair. 2012;5:7. doi: 10.1186/1755-1536-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge H, He X, Guo L, Yang X. Clinicopathological significance of HSP27 in gastric cancer: A meta-analysis. Onco Targets Ther. 2017;10:4543–4551. doi: 10.2147/OTT.S146590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmermann M, et al. In vitro stability of heat shock protein 27 in serum and plasma under different pre-analytical conditions: Implications for large-scale clinical studies. Ann Lab Med. 2016;36:353–357. doi: 10.3343/alm.2016.36.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davie EW, Kulman JD. An overview of the structure and function of thrombin. Semin Thromb Hemost. 2006;32(Suppl 1):3–15. doi: 10.1055/s-2006-939550. [DOI] [PubMed] [Google Scholar]
- 30.Yang L, Li Y, Bhattacharya A, Zhang YS. A plasma proteolysis pathway comprising blood coagulation proteases. Oncotarget. 2016;7:40919–40938. doi: 10.18632/oncotarget.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weyrich AS, et al. Signal-dependent translation of a regulatory protein, Bcl-3, in activated human platelets. P Natl Acad Sci USA. 1998;95:5556–5561. doi: 10.1073/pnas.95.10.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elia A, Constantinou C, Clemens MJ. Effects of protein phosphorylation on ubiquitination and stability of the translational inhibitor protein 4E-BP1. Oncogene. 2008;27:811–822. doi: 10.1038/sj.onc.1210678. [DOI] [PubMed] [Google Scholar]
- 33.Yu Z, et al. Differences between human plasma and serum metabolite profiles. PloS One. 2011;6:e21230. doi: 10.1371/journal.pone.0021230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furuncuoglu Y, et al. How obesity affects the neutrophil/lymphocyte and platelet/lymphocyte ratio, systemic immune-inflammatory index and platelet indices: A retrospective study. Eur Rev Med Pharmacol Sci. 2016;20:1300–1306. [PubMed] [Google Scholar]
- 35.Santilli F, Vazzana N, Liani R, Guagnano MT, Davi G. Platelet activation in obesity and metabolic syndrome. Obes Rev. 2012;13:27–42. doi: 10.1111/j.1467-789X.2011.00930.x. [DOI] [PubMed] [Google Scholar]
- 36.Yarmolinsky J, et al. Plasminogen activator inhibitor-1 and type 2 diabetes: A systematic review and meta-analysis of observational studies. Sci Rep. 2016;6:17714. doi: 10.1038/srep17714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somodi S, et al. Plasminogen activator inhibitor-1 level correlates with lipoprotein subfractions in obese nondiabetic subjects. Int J Endocrinol. 2018;2018:9596054. doi: 10.1155/2018/9596054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polat SB, et al. Evaluation of serum fibrinogen, plasminogen, alpha2-anti-plasmin, and plasminogen activator inhibitor levels (PAI) and their correlation with presence of retinopathy in patients with type 1 DM. J Diabetes Res. 2014;2014:317292. doi: 10.1155/2014/317292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mullard A. Nine paths to PCSK9 inhibition. Nat Rev Drug Discov. 2017;16:299–301. doi: 10.1038/nrd.2017.83. [DOI] [PubMed] [Google Scholar]
- 40.Harper AR, Nayee S, Topol EJ. Protective alleles and modifier variants in human health and disease. Nat Rev Genet. 2015;16:689–701. doi: 10.1038/nrg4017. [DOI] [PubMed] [Google Scholar]
- 41.Lakoski SG, Lagace TA, Cohen JC, Horton JD, Hobbs HH. Genetic and metabolic determinants of plasma PCSK9 levels. J Clin Endocrinol Metab. 2009;94:2537–2543. doi: 10.1210/jc.2009-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cariou B, et al. Plasma PCSK9 concentrations during an oral fat load and after short term high-fat, high-fat high-protein and high-fructose diets. Nutr Metab. 2013;10:4. doi: 10.1186/1743-7075-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor BA, et al. Serum PCSK9 levels distinguish individuals who do not respond to high-dose statin therapy with the expected reduction in LDL-C. J Lipids. 2014;2014:140723. doi: 10.1155/2014/140723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eisenga MF, et al. High serum PCSK9 is associated with increased risk of new-onset diabetes after transplantation in renal transplant recipients. Diabetes Care. 2017;40:894–901. doi: 10.2337/dc16-2258. [DOI] [PubMed] [Google Scholar]
- 45.Beker BM, Corleto MG, Fieiras C, Musso CG. Novel acute kidney injury biomarkers: Their characteristics, utility and concerns. Int Urol Nephrol. 2018;50:705–713. doi: 10.1007/s11255-017-1781-x. [DOI] [PubMed] [Google Scholar]
- 46.Taub PR, Borden KC, Fard A, Maisel A. Role of biomarkers in the diagnosis and prognosis of acute kidney injury in patients with cardiorenal syndrome. Expert Rev Cardiovasc Ther. 2012;10:657–667. doi: 10.1586/erc.12.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedersen TL, Newman JW. Establishing and performing targeted multi-residue analysis for lipid mediators and fatty acids in small clinical plasma samples. Methods Mol Biol (Clifton, N. J.) 2018;1730:175–212. doi: 10.1007/978-1-4939-7592-1_13. [DOI] [PubMed] [Google Scholar]
- 48.Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39:82–91. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 49.Midtbo LK, et al. Intake of farmed Atlantic salmon fed soybean oil increases hepatic levels of arachidonic acid-derived oxylipins and ceramides in mice. J Nutr Biochem. 2015;26:585–595. doi: 10.1016/j.jnutbio.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 50.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, http://www.R-project.org/ (2013).
- 51.Tripathi S, et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chong J, Xia J. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics. 2018;34:4313–4314. doi: 10.1093/bioinformatics/bty528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and table descriptions
Data Availability Statement
Protocols used to generate the findings of this study are available from the corresponding author upon request. In accordance with Swedish ethical regulations and GDPR, primary data from human subjects cannot be made publically available.