Abstract
Transposable elements (TEs) are genomic sequences that can move, multiply, and often form sizable fractions of vertebrate genomes. Fish belong to a unique group of vertebrates, since their karyotypes and genome sizes are more diverse and complex, with probably higher diversity and evolution specificity of TE. To investigate the characteristics of fish TEs, we compared the mobilomes of 39 species, and observed significant variation of TE content in fish (from 5% in pufferfish to 56% in zebrafish), along with a positive correlation between fish genome size and TE content. In different classification hierarchies, retrotransposons (class), long terminal repeat (order), as well as Helitron, Maverick, Kolobok, CMC, DIRS, P, I, L1, L2, and 5S (superfamily) were all positively correlated with fish genome size. Consistent with previous studies, our data suggested fish genomes to not always be dominated by DNA transposons; long interspersed nuclear elements are also prominent in many species. This study suggests CR1 distribution in fish genomes to be obviously regular, and provides new clues concerning important events in vertebrate evolution. Altogether, our results highlight the importance of TEs in the structure and evolution of fish genomes and suggest fish species diversity to parallel transposon content diversification.
Subject terms: Evolutionary genetics, Mobile elements
Introduction
In addition to functional genes, the genome contains numerous scattered repeats known as transposable elements (TEs). Published first in the 1950s by Barbara McClintock1, TEs can ‘jump’ from one chromosomal site to another. Transposable elements are either retrotransposons or DNA transposons based on whether they use RNA or DNA for mobilization, respectively. Below this broader categorization, TEs are grouped into five classes according to enzymology, structure, and sequences: long terminal repeat (LTR) retrotransposons, non-LTR retrotransposons (e.g., long and short interspersed nuclear elements, or LINEs and SINEs), cut-and-paste DNA transposons, rolling-circle DNA transposons (Helitrons), and self-synthesizing DNA transposons (Polintons)2.
Initially, TEs did not attract much scientific attention because they were erroneously treated as “junk DNA.” However, they are now recognized as critical functional and evolutionary components of the genome3,4, involved in processes such as speciation5, sex determination6–9, new gene creation10,11, and chromosome rearrangement12. Overall, TEs appear to drive genetic diversification and provide genetic material during genome evolution13,14. Thus, evaluating TEs is essential to the investigation of genome evolution dynamics.
Fishes are the oldest and largest group of vertebrates. Their long evolutionary history includes multiple rounds of whole genome duplication and re-diploidization events that increased their genetic complexity, so, fishes have more complex karyotypes and more diverse genome sizes than any other vertebrate taxon15. Notably, this taxonomic diversity is paralleled by extensive variation in genetic and phenotypic characteristics, as well as by the presence of TEs. It would be important to stress that cytological haploid genome size of fish ranges very widely (0.35–133 Gb)16, C value paradox arises from widely different C-values like these. Genome size has been implicated in several phenotypic traits, including cell size17,18 and metabolic rate19,20. Thus, disentangling the forces and mechanisms that regulate genome size is critical for a better understanding of piscine molecular evolution. Unfortunately, data concerning fish TEs are limited, and a detailed systematic comparative study is yet to be attempted. Rapid advancements in sequencing technology, however, has resulted in the publication of several fish genomes, providing a means to comprehensively study fish TEs. In a previous study, we created a fish-specific TE database (FishTEDB)21 to facilitate research on TE function and evolution in fish genomes, but we have not applied the database for systematic evaluation of TE diversity.
Therefore, this study expanded on the original FishTEDB through the addition of TE data from nine fish species. The updated database contains 39 species genomes, including 35 from Actinopterygii (14 orders), 1 from Chondrichthyes, 1 from Sarcopterygii, 1 from Agnatha, and 1 from Chordata. We used these data to evaluate correlations between TE content and genome size across different classification hierarchies, with the aim of exploring how different TE categories contribute to genome-size evolution. Furthermore, based on TE diversity, we attempted to clarify TE effects on fish evolution and explain TE specificity in fishes that occupy key positions in the evolutionary tree.
Results
TE content diversity and its contribution to fish genome size
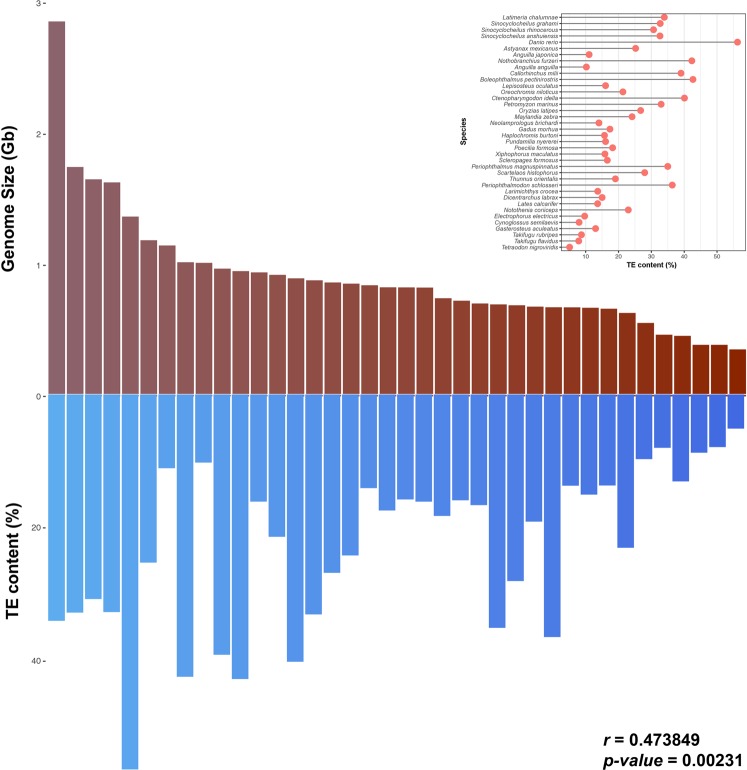
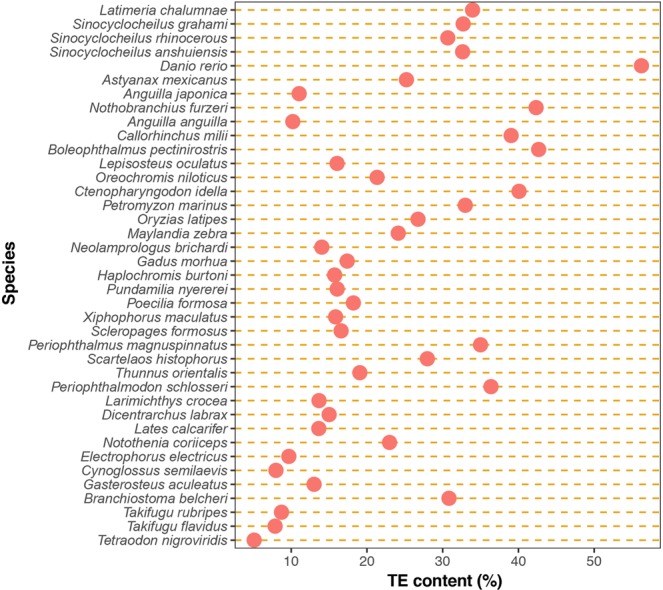
In global level (Fig. 1, Supplementary Table S2), we found TE content to be variable, ranging from 5% in pufferfish to 56% in zebrafish, and was positively correlated (Pearson correlation r = 0.47, p-value = 0.002) with fish genome size (Fig. 2, Supplementary Table S6).
Figure 1.
Total TE content of all species analysed in this study, sorted by genome size.
Figure 2.
Correlation between genome size and TE content of fish. The histogram above the graph (in red) shows the distribution of genome size (unit, 1000 megabases) while that below the graph (in blue) shows TE distribution. Correlation analysis was performed by the Pearson method using the R program.
Similar to the results of previous studies22–24, our data showed (Fig. 3, Supplementary Table S4) fish genomes to not always be dominated by DNA transposons, but also by LINEs in many species, as in the elephant shark (Callorhinchus milii), which had very few DNA transposons. In addition, most of the fish genomes studied appeared to be particularly poor in SINEs24. We then tested the relationship across genome size, DNA transposon, and retrotransposon (including LTR, LINE, and SINE) content; results of the analysis showed a positive correlation between retrotransposon content and genome size. This finding was statistically supported by our correlation analysis (Pearson correlation r = 0.39, p-value = 0.013), and LTR content was positively correlated (Pearson correlation r = 0.43, p-value = 0.006) with fish genome size (Supplementary Table S6).
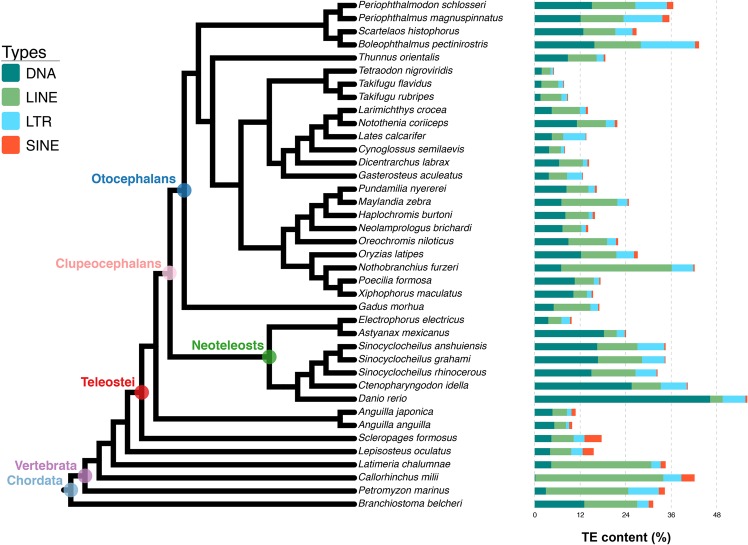
Figure 3.
DNA transposon and retrotransposon levels in various fish genomes (Lancelet added). The percentages of DNA transposons, LTR, LINE, and SINE retrotransposons are presented.
Further, to analyse TE content and distribution in fish, we calculated the levels of each TE superfamily in each species (Fig. 4, Supplementary Table S5). Our results showed Tc/mariner, hAT, L1, L2, and Gypsy to be widespread and the most predominant TE superfamilies in the fish genomes included in this study; distribution of other superfamilies was more erratic and species-dependent. Notably, the Cyprinidae lineage fish species (Sinocyclocheilus anshuiensis, S. graham, S. rhinocerous, Ctenopharyngodon idella, and Danio rerio) had the highest level of TE diversity among the species studied. Among the early diverging fishes (C. milii, Latimeria chalumnae, Lepisosteus oculatus, and Petromyzon marinus) and Branchiostoma belcheri, without teleost-specific whole genome duplication event, TEs of the CR1 superfamily were predominant, although the abundance of CR1 was very low in the fishes that diverged more recently. The levels of each TE superfamily appeared to be highly specific and species-dependent. This was particularly true for Gypsy in Boleophthalmus pectinirostris, L2 and RTE in Nothobranchius furzeri, Tc/mariner in Astyanax mexicanus, hAT in D. rerio, CR1, L1, and L2 in L. chalumnae, and CR1 and L2 in C. milii. We also evaluated the relationship between genome size and superfamily content. Our results showed that the higher levels of Helitron, Maverick, Kolobok, CMC, P, DIRS, I, L1, L2, and 5S superfamilies positively correlated with genome size (Supplementary Table S6).
Figure 4.
Diversity and abundance of TE superfamily in all fish genomes investigated in this study (Lancelet added). Results are presented using a heat map; content of superfamilies is shown by squares with colour gradient.
TE transposition history and activity during fish evolution
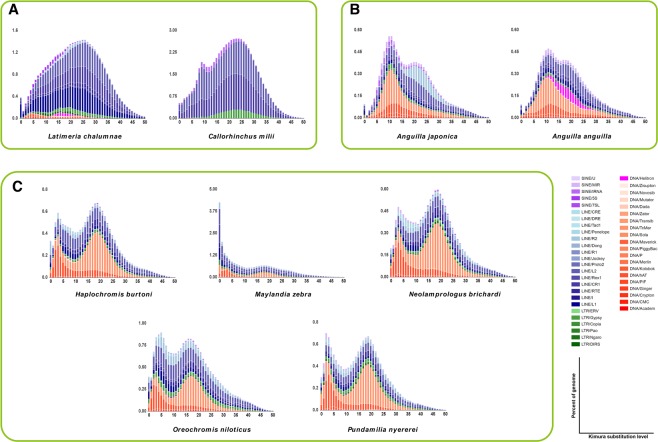
The percentages of TE in the genome of each species were clustered based on their K-values (Fig. 5, Supplementary Fig. S1). Notably, copy divergence appeared to be correlated with activity age, with very similar copies (low K-values) being indicative of somewhat recent activity (shown on the left side of the graph) while divergent copies (high K-values) were likely generated by older transposition events (shown on the right side of the graph)22.
Figure 5.
Kimura distance-based copy divergence analyses of transposable elements in coelacanth and elephant shark (A), eels (B), and Africa cichlids (C). The graphs represent genome coverage for each TE superfamily in the different genomes analysed. Clustering was performed according to their Kimura distances (K-value from 0 to 50). Copies clustering on the left side of the graph did not greatly diverge from the consensus sequence and potentially corresponded to recent events while sequences on the right side likely corresponded to older divergence.
Indeed, each peak in the graph indicated a transposition/TE burst. Transposition bursts are common in fish, and they generally have at least one or two of them. In this process, there is usually a continuous increase in the number of active transposons before transposon “explosion”, and a continuous decline in the number of active transposons after transposon explosion. In most fish genomes, the rate at which the number of active transposons increases is smaller than the rate at which the number of active transposons declines; therefore, most of the fish genomes contain fewer ancient copies (K-values > 25) than recent copies (K-values < 25). However, we observed an opposite trend in C. milii and L. chalumnae (Fig. 5A). Interestingly, there were also some notable superfamily-dependent differences, occurring even between closely related species with similar TE landscapes. For example, in Japanese and European eels there were obvious differences in R2 and Helitron transposon bursts, respectively (Fig. 5B). In African cichlids, which generally have two transposon bursts across all TE superfamilies, we observed a recent burst in the Maylandia zebra (Fig. 5C).
Discussion
Transposable elements are important evolutionary components of the genome. Although various studies have been published addressing TE function and diversity24,25, the distribution and role of TEs in some species, especially fish, remain largely unknown owing to their complicated genomes24. In this study, we used species-specific TE libraries in FishTEDB, along with that of additional nine species, to analyse different classifications of TEs. Abundance, diversity, activity, and evolution of TE were explored and related to genome size and evolutionary history of fish.
With over 33,900 known species (FishBase, http://www.fishbase.org/, version 02/2018), fish comprise the majority of vertebrates. It is, therefore, not surprising that remarkable differences in morphology, population structure, and genome size have been observed across fish species. Differences in genome size, in particular, can be up to 379.5 times (0.35–133 Gb)16. In this study, we observed a considerable variation in TE content (5–56%) across the fish species analysed. Such variation was not only restricted to the overall levels; according to our statistical analysis of the different classifications, diversity may also be more variable and complex. Fish genomes predominantly contained DNA transposons and LINEs, whereas SINEs were the least abundant. Focusing on TE superfamilies, Tc/mariner, hAT, L1, L2, and Gypsy were found to be the most widespread among the fish genomes analysed. Most TEs show patchy distribution, indicating multiple events of loss and gain. However, there were some exceptions to this trend in TE diversity. In the elephant shark (Chondrichthyes), for example, the most prevalent TEs were the LINE superfamilies L2 and CR1, rather than Tc/mariner and hAT. SINEs were also well represented, whereas only a few DNA transposons were detected. Therefore, our results also hinted that the TE landscapes in cartilaginous fish might be more similar to that of jawless fish rather than of bony fish26. In the African coelacanth (Sarcopterygii) genome, CR1, L1, and L2 (LINEs) were predominant. However, in this species, the DNA transposons appeared to have recently undergone transposition, and SINEs were not well represented. These features also existed in some tetrapods, for example, Squamata, Testudines, Crocodilia, and Aves24. The data, therefore, indicate that TE landscape in African coelacanth might be similar to that of tetrapods. This is consistent with previous studies on the phylogenetic relationships of these species27. In conclusion, fish TEs were regularly distributed, and the relationship across species with similar distribution regularity was consistent with the phylogenetic relationship. This indicated that TEs play a vital role in fish evolution.
Our analysis of TE superfamilies suggested a critical role of the CR1 superfamily in vertebrate evolution. In fact, among the earliest-diverging fishes (C. milii, L. chalumnae, L. oculatus, and P. marinus), B. belcheri, and tetrapods (terrestrial animals), CR1 elements appeared to have a strong genomic contribution and were often widely distributed22,24. However, teleost contains fewer of these elements, hence suggesting that the CR1 superfamily existed in ancestral vertebrates, and a significant loss occurred during the evolution of fish. Nevertheless, these elements were preserved, and proliferated from aquatic to terrestrial transition in tetrapods. Although previous studies indicated that TEs are important to genome evolution and could influence piscine adaptation to various habitats28, additional studies would be necessary to uncover the full function and evolutionary role of CR1 superfamily in fish and other species.
With the exception of elephant sharks and African coelacanths, the presence and levels of some superfamilies appeared to be highly species-specific. For example, we observed Gypsy in B. pectinirostris, L2 and RTE in N. furzeri, Tc/mariner in A. mexicanus, and hAT in D. rerio. In fact, the losses and gains of specific TEs during evolution appeared to primarily determine the content and distribution of different superfamilies in each species. Because genome defense machinery (e.g., DNA methylation, Piwi-interacting small RNAs) regulates TEs, the loss-and-gain process must be associated with host genomes29. Previous studies had indicated that TEs could have biological significance owing to their interaction with the host genome, akin to how species interact with ecosystems. The similarity between genomes and ecosystems was first drawn in 1989 by Holmquist30, who suggested that genome components may be different niches. These niches include the darkly stained, heterochromatic bands of chromosomes and TEs. Like organisms in their habitats, TEs proliferate and use resources inside the genome environment while interacting with each other31. Similarly, the Red Queen paradigm applies to interactions between host genomes and TEs, describing an antagonistic relationship that is continually evolving. Researchers have proposed these analogies to be useful in understanding TE abundance and diversity32. Combined with the theory of natural selection, TEs that compete with the host genome (harmful TEs) are more likely to be eliminated, whereas TEs beneficial to the host genome are more likely to be conserved. Therefore, superfamilies that are highly specific in some fish species should be considered important players in genome evolution and may be related to the biological characteristics of the species itself.
Like gene number and intron number, TE content is also a crucial genomic parameter. Many studies have described a positive relationship between TE levels and genome size23,33–35, and TEs have been universally recognised as a driver of genome size. Our analysis supports this conclusion in fish. Through further correlation analysis, we also confirmed that the effect of retrotransposons (Class I) on genome size was higher than that of DNA transposons (Class II). Of the various types of retrotransposons, LTRs appeared to be significantly correlated with genome size. However, despite the various DNA transposons (Helitron, Maverick, Kolobok, CMC, and P), LTRs (DIRS), LINEs (L1, L2, and I), and SINEs (5S) being positively correlated with genome size, whether these TEs drive genome size remains unclear, since most of them were present only at low levels in the fish genomes. Thus, while understanding the full function of TEs would require further study, there was indeed a general trend in fish, whereby the TE content increased with increase of genome size.
In addition to analysing the relationship between TE content and genome size, we also evaluated TE evolution and activity concerning transposition bursts. Transposition bursts occur at least once or twice, if not more, over the evolutionary history of a fish. In fact, there are active and inactive periods of TE throughout the TE ‘lifecycle’, which begins with the invasion of TE into a new genome (via a horizontal transfer event) or the evolution of a new, distinct TE lineage from a previously existing one (via a genetic mutation). Although the new element can establish itself into the genome, the host can also mount a defence against this change and proliferation can be curtailed. However, if the insertion is in some way beneficial to the host, then the TE will be conserved, and co-evolution of the element and the host will occur36–39. Thus, transposition bursts are likely to be associated with significant evolutionary events, as is supported by previous studies that linked speciation with a high TE activity5,40,41. In the present study, the content of M. zebra transposition burst was the highest across all the species studied. We also observed an unusually high proportion of recent bursts in the M. zebra of African cichlids. African cichlids are famous for their large, diverse, and replicated adaptive radiations in the Great Lakes of East Africa42. The activity of TE is closely related to species formation and adaptive radiation41,43,44. Therefore, based on our data, we believe that TEs may have the potential for continued differentiation. However, the phenomenon of TE burst is not unique; Lates calcarifer appears to have undergone a similar process. However, we could not speculate whether this phenomenon is related to adaptive radiation, since the existence of adaptive radiation in the evolutionary process of L. calcarifer has not yet been reported. Additionally, we could not rule out other factors such as environmental adaptation45–47. In Japanese and European eels, there are obvious differences in the R2 and Helitron transposon bursts, respectively, despite their similar TE landscapes. This may have occurred during or after differentiation of their common ancestors, and preserved henceforth.
Conclusions
In this report, we present an overview of TE abundance, diversity, activity, and evolution in fish with varying genome sizes and positions in the fish tree of life. High levels of diversity and patchy distribution were the main characteristics of TEs in the fish genomes analysed. In combination with ‘genomic ecology’ and TE ‘lifecycle’ theory, our data suggested that differential TE bursts may have actively contributed to essential evolutionary events. The CR1 TE superfamily also appeared to play an important role in the differentiation of aquatic and terrestrial animals. Although further studies would be required to explore the relationship between TE burst/activity and vertebrate evolution, this study provides significant insight into the role of TE activity, specificity, and diversity in fish evolution and genome size, and highlights the application of FishTEDB.
Methods
Data collection and species-specific TE library construction
All genomes used in this study were downloaded from public databases (Supplementary Table S1). We directly used the zebrafish TE library in Repbase (http://www.girinst.org/repbase/). TE libraries of 30 species were downloaded from FishTEDB (http://www.fishtedb.org/) (Supplementary Table S1). Other TE libraries were generated using de-novo, homology-based, and structure-based methods. De-novo identification of TEs was performed using RepeatModeler (http://www.repeatmasker.org/RepeatModeler/, version 1.0.7). For the structure-based method, we used MGEScan-non-LTR48, LTR_STRUC49, MGEScan-LTR (http://darwin.informatics.indiana.edu/cgi-bin/evolution/daphnia_ltr.pl), and TESeeker50. REPCLASS (https://github.com/feschottelab/REPCLASS, version 1.0) and TEclass51 were used to classify TEs. A more detailed pipeline had been described in our previous study21.
TE annotation and statistical analysis
RepeatMasker version 4.0.5 (http://www.repeatmasker.org/RMDownload.html) was used to mask the genomes. Notably, the “-a” and “-lib” default parameters were applied. Pearson correlation analysis via “cor.test ()” function in R language was applied to analyse the correlation between genome size and TE content.
Phylogenetic tree construction
Since genome analysis of the species used in this study had already been conducted, most of their phylogenetic relationships are clear. Therefore, the phylogenetic tree was constructed by combining NCBI Taxonomy (https://www.ncbi.nlm.nih.gov/taxonomy/?term=) with existing literature52–58.
TE divergence distribution
To estimate TE “age” and transposition history in fish, we performed a copy-divergence analysis of the TE superfamilies, based on their Kimura 2-parameter distances (K-values)59. Kimura distances between genome copies and TE consensus from the library were determined using buildSummary.pl, calcDivergenceFromAlign.pl, and createRepeatLandscape.pl (in RepeatMasker util directory) on alignment files (.align files) after genome masking. Transition and transversion rates were calculated for these alignments, and then transformed to Kimura distances59 with the following equation: K = −1/2 ln(1 − 2p − q) − 1/4 ln(1 − 2q), where q is the proportion of sites with transversions, and p is the proportion of sites with transitions.
Supplementary information
Supplementary information: Evolution and diversity of transposable elements in fish genomes
Supplementary Table S1, Table S2, Table S3, Table S4, Table S5, Table S6
Acknowledgements
We thank Vinay Mittal, Ryan Kennedy, and Xiangfeng Li in the use of software. We also thank Huahao Zhang and Hongen Xu for their help in the identification of TEs. This work was supported by the grants from the National Natural Science Foundation of China (31572254 & 31872204).
Author contributions
F.S. performed the major part of data analysis and drafted the manuscript. Z.P. and M.H. contributed to research design and final edits to the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-51888-1.
References
- 1.Pontarotti, P. Evolutionary biology: self/nonself evolution, species and complex traits evolution, methods and concepts. (Springer International Publishing, 2017).
- 2.Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet. 2008;9:411–412. doi: 10.1038/nrg2165-c1. [DOI] [PubMed] [Google Scholar]
- 3.Biemont C. A brief history of the status of transposable elements: from junk DNA to major players in evolution. Genetics. 2010;186:1085–1093. doi: 10.1534/genetics.110.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 2017;18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auvinet J, et al. Mobilization of retrotransposons as a cause of chromosomal diversification and rapid speciation: the case for the Antarctic teleost genus Trematomus. BMC Genomics. 2018;19:339. doi: 10.1186/s12864-018-4714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalopin D, Volff JN, Galiana D, Anderson JL, Schartl M. Transposable elements and early evolution of sex chromosomes in fish. Chromosome Res. 2015;23:545–560. doi: 10.1007/s10577-015-9490-8. [DOI] [PubMed] [Google Scholar]
- 7.Kvikstad EM, Makova KD. The (r)evolution of SINE versus LINE distributions in primate genomes: sex chromosomes are important. Genome Res. 2010;20:600–613. doi: 10.1101/gr.099044.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schartl M, et al. Sox5 is involved in germ-cell regulation and sex determination in medaka following co-option of nested transposable elements. BMC Biol. 2018;16:16. doi: 10.1186/s12915-018-0485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sliwinska EB, Martyka R, Tryjanowski P. Evolutionary interaction between W/Y chromosome and transposable elements. Genetica. 2016;144:267–278. doi: 10.1007/s10709-016-9895-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennetzen JL. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 2005;15:621–627. doi: 10.1016/j.gde.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Long M, Betrán E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 12.Lonnig WE, Saedler H. Chromosome rearrangements and transposable elements. Annu. Rev. Genet. 2002;36:389–410. doi: 10.1146/annurev.genet.36.040202.092802. [DOI] [PubMed] [Google Scholar]
- 13.Bennetzen JL. Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 2000;42:251–269. doi: 10.1023/A:1006344508454. [DOI] [PubMed] [Google Scholar]
- 14.Platt RN, Vandewege MW, Ray DA. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 2018;26:1–19. doi: 10.1007/s10577-017-9570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volff JN. Genome evolution and biodiversity in teleost fish. Heredity. 2005;94:280–294. doi: 10.1038/sj.hdy.6800635. [DOI] [PubMed] [Google Scholar]
- 16.Kapusta A, Suh A, Feschotte C. Dynamics of genome size evolution in birds and mammals. Proc. Natl. Acad. Sci. USA. 2017;114:E1460–E1469. doi: 10.1073/pnas.1616702114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavalier-Smith T. Skeletal DNA and the evolution of genome size. Annu. Rev. Biophys. Bioeng. 1982;11:273–302. doi: 10.1146/annurev.bb.11.060182.001421. [DOI] [PubMed] [Google Scholar]
- 18.Gregory TR, Hebert PD. The modulation of DNA content: proximate causes and ultimate consequences. Genome Res. 1999;9:317–324. [PubMed] [Google Scholar]
- 19.Andrews CB, Mackenzie SA, Gregory TR. Genome size and wing parameters in passerine birds. Proc. Biol. Sci. 2009;276:55–61. doi: 10.1098/rspb.2008.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright NA, Gregory TR, Witt CC. Metabolic ‘engines’ of flight drive genome size reduction in birds. Proc. Biol. Sci. 2014;281:20132780. doi: 10.1098/rspb.2013.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao F, Wang J, Xu H, Peng Z. FishTEDB: a collective database of transposable elements identified in the complete genomes of fish. Database. 2018;2018:bax106. doi: 10.1093/database/bax106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chalopin D, Naville M, Plard F, Galiana D, Volff JN. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol. Evol. 2015;7:567–580. doi: 10.1093/gbe/evv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao B, et al. The contribution of transposable elements to size variations between four teleost genomes. Mob. DNA. 2016;7:4. doi: 10.1186/s13100-016-0059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sotero-Caio CG, Platt RN, Suh A, Ray DA. Evolution and diversity of transposable elements in vertebrate genomes. Genome Biol. Evol. 2017;9:161–177. doi: 10.1093/gbe/evw264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishihara H, et al. Coordinately co-opted multiple transposable elements constitute an enhancer for wnt5a expression in the mammalian secondary palate. PLoS Genet. 2016;12:e1006380. doi: 10.1371/journal.pgen.1006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogiwara I, Miya M, Ohshima K, Okada N. Retropositional parasitism of SINEs on LINEs: identification of SINEs and LINEs in elasmobranchs. Mol. Biol. Evol. 1999;16:1238–1250. doi: 10.1093/oxfordjournals.molbev.a026214. [DOI] [PubMed] [Google Scholar]
- 27.Amemiya CT, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan Z, et al. Comparative genome analysis of 52 fish species suggests differential associations of repetitive elements with their living aquatic environments. BMC Genomics. 2018;19:141. doi: 10.1186/s12864-018-4516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmquist GP. Evolution of chromosome bands: molecular ecology of noncoding DNA. J. Mol. Evol. 1989;28:469–486. doi: 10.1007/BF02602928. [DOI] [PubMed] [Google Scholar]
- 31.Leonardo TE, Nuzhdin SV. Intracellular battlegrounds: conflict and cooperation between transposable elements. Genet. Res. 2002;80:155–161. doi: 10.1017/S0016672302009710. [DOI] [PubMed] [Google Scholar]
- 32.Venner S, Feschotte C, Biemont C. Dynamics of transposable elements: towards a community ecology of the genome. Trends Genet. 2009;25:317–323. doi: 10.1016/j.tig.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kidwell MG. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 2002;115:49–63. doi: 10.1023/A:1016072014259. [DOI] [PubMed] [Google Scholar]
- 34.Pellicer J, Hidalgo O, Dodsworth S, Leitch IJ. Genome size diversity and its impact on the evolution of land plants. Genes. 2018;9:88. doi: 10.3390/genes9020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talla V, et al. Rapid increase in genome size as a consequence of transposable element hyperactivity in wood-white (Leptidea) butterflies. Genome Biol. Evol. 2017;9:2491–2505. doi: 10.1093/gbe/evx163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elliott, T. A. Conceptual and empirical investigations of eukaryotic transposable element evolution, http://hdl.handle.net/10214/10154 (2016).
- 37.Hua-Van A, Le Rouzic A, Maisonhaute C, Capy P. Abundance, distribution and dynamics of retrotransposable elements and transposons: similarities and differences. Cytogenet. Genome. Res. 2005;110:426–440. doi: 10.1159/000084975. [DOI] [PubMed] [Google Scholar]
- 38.Kidwell MG, Lisch DR. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 39.Schaack S, Choi E, Lynch M, Pritham EJ. DNA transposons and the role of recombination in mutation accumulation in Daphnia pulex. Genome Biol. 2010;11:R46. doi: 10.1186/gb-2010-11-4-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dion-Cote AM, Renaut S, Normandeau E, Bernatchez L. RNA-seq reveals transcriptomic shock involving transposable elements reactivation in hybrids of young lake whitefish species. Mol. Biol. Evol. 2014;31:1188–1199. doi: 10.1093/molbev/msu069. [DOI] [PubMed] [Google Scholar]
- 41.Oliver KR, Greene WK. Mobile DNA and the TE-Thrust hypothesis: supporting evidence from the primates. Mob. DNA. 2011;2:8. doi: 10.1186/1759-8753-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brawand D, et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513:375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grahn RA, Rinehart TA, Cantrell MA, Wichman HA. Extinction of LINE-1 activity coincident with a major mammalian radiation in rodents. Cytogenet. Genome Res. 2005;110:407–415. doi: 10.1159/000084973. [DOI] [PubMed] [Google Scholar]
- 44.Platt RN, et al. Large numbers of novel miRNAs originate from DNA transposons and are coincident with a large species radiation in bats. Mol. Biol. Evol. 2014;31:1536–1545. doi: 10.1093/molbev/msu112. [DOI] [PubMed] [Google Scholar]
- 45.Casacuberta E, Gonzalez J. The impact of transposable elements in environmental adaptation. Mol. Ecol. 2013;22:1503–1517. doi: 10.1111/mec.12170. [DOI] [PubMed] [Google Scholar]
- 46.Lanciano S, Mirouze M. Transposable elements: all mobile, all different, some stress responsive, some adaptive? Curr. Opin. Genet. Dev. 2018;49:106–114. doi: 10.1016/j.gde.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 47.McGraw JE, Brookfield JF. The interaction between mobile DNAs and their hosts in a fluctuating environment. J. Theor. Biol. 2006;243:13–23. doi: 10.1016/j.jtbi.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 48.Rho M, Tang H. MGEScan-non-LTR: computational identification and classification of autonomous non-LTR retrotransposons in eukaryotic genomes. Nucleic Acids Res. 2009;37:e143. doi: 10.1093/nar/gkp752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCarthy EM, McDonald JF. LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics. 2003;19:362–367. doi: 10.1093/bioinformatics/btf878. [DOI] [PubMed] [Google Scholar]
- 50.Kennedy RC, Unger MF, Christley S, Collins FH, Madey GR. An automated homology-based approach for identifying transposable elements. BMC Bioinformatics. 2011;12:130. doi: 10.1186/1471-2105-12-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abrusan G, Grundmann N, DeMester L, Makalowski W. TEclass–a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics. 2009;25:1329–1330. doi: 10.1093/bioinformatics/btp084. [DOI] [PubMed] [Google Scholar]
- 52.Betancur RR, et al. Phylogenetic classification of bony fishes. BMC Evol. Biol. 2017;17:162. doi: 10.1186/s12862-017-0958-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hughes LC, et al. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc. Natl. Acad. Sci. USA. 2018;115:6249–6254. doi: 10.1073/pnas.1719358115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawahara R, et al. Interrelationships of the 11 gasterosteiform families (sticklebacks, pipefishes, and their relatives): a new perspective based on whole mitogenome sequences from 75 higher teleosts. Mol. Phylogenet. Evol. 2008;46:224–236. doi: 10.1016/j.ympev.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 55.Tine M, et al. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nature Commun. 2014;5:5770. doi: 10.1038/ncomms6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vij S, et al. Chromosomal-level assembly of the Asian seabass genome using long sequence reads and multi-layered scaffolding. PLoS Genet. 2016;12:e1005954. doi: 10.1371/journal.pgen.1005954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang J, et al. The Sinocyclocheilus cavefish genome provides insights into cave adaptation. BMC Biol. 2016;14:1. doi: 10.1186/s12915-015-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.You X, et al. Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nature Commun. 2014;5:5594. doi: 10.1038/ncomms6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information: Evolution and diversity of transposable elements in fish genomes
Supplementary Table S1, Table S2, Table S3, Table S4, Table S5, Table S6