Abstract
Finding novel targets in non-small cell lung cancer (NSCLC) is highly needed and identification of synthetic lethality between two genes is a new approach to target NSCLC. We previously found that TMPRSS4 promotes NSCLC growth and constitutes a prognostic biomarker. Here, through large-scale analyses across 5 public databases we identified consistent co-expression between TMPRSS4 and DDR1. Similar to TMPRSS4, DDR1 promoter was hypomethylated in NSCLC in 3 independent cohorts and hypomethylation was an independent prognostic factor of disease-free survival. Treatment with 5-azacitidine increased DDR1 levels in cell lines, suggesting an epigenetic regulation. Cells lacking TMPRSS4 were highly sensitive to the cytotoxic effect of the DDR1 inhibitor dasatinib. TMPRSS4/DDR1 double knock-down (KD) cells, but not single KD cells suffered a G0/G1 cell cycle arrest with loss of E2F1 and cyclins A and B, increased p21 levels and a larger number of cells in apoptosis. Moreover, double KD cells were highly sensitized to cisplatin, which caused massive apoptosis (~40%). In vivo studies demonstrated tumor regression in double KD-injected mice. In conclusion, we have identified a novel vulnerability in NSCLC resulting from a synthetic lethal interaction between DDR1 and TMPRSS4.
Subject terms: Non-small-cell lung cancer, Prognostic markers
Introduction
Progression of non-small cell lung cancer (NSCLC) is a consequence of both genetic and epigenetic changes that alter intracellular pathways leading to proliferation and invasion1,2. DNA promoter hypomethylation can cause expression of oncogenes, whereas hypermethylation has been associated with silencing of tumor suppressor genes. These epigenetic changes have been used as biomarkers for diagnostic or prognostic purposes3.
Changes in cell-cell and cell-extracellular matrix (ECM) interactions are crucial for metastasis development4, where proteases play a key role in the modification of tumor cells and ECM properties, a reason whereby dysregulation of protease activity is considered as a hallmark of cancer5. TMPRSS4 is a membrane-bound serine protease whose overexpression causes cell growth and metastasis in several cancer types6,7. We have previously shown that high levels of TMPRSS4 are significantly associated with worse prognosis in patients with squamous NSCLC and that increased expression of this protein is induced by hypomethylation of the TMPRSS4 DNA promoter8. Moreover, hypomethylation serves as a prognostic biomarker, as it is significantly associated with reduced disease-free survival (DFS)8. Previous experiments in vitro and in vivo led us to demonstrate that TMPRSS4 enhances tumor growth and metastasis, and confers both epithelial to mesenchymal transition (EMT) and cancer stem cell (CSC) features in lung cancer cells9.
In order to get more insights about TMPRSS4-associated pathways in NSCLC patients we sought in this study to identify genes co-expressed with TMPRSS4 that may be functionally related and cooperate to establish a malignant phenotype. An increasing number of studies are undertaking genome-wide co-expression approaches using microarray data to identify interconnected regulatory pathways and functional relationships between genes10,11. Using this strategy in NSCLC in the present study, we have found that TMPRSS4 is co-overexpressed with Discoidin Domain Receptor tyrosine kinase 1 (DDR1), a membrane protein that promotes cancer cell growth and dissemination12. We have also found that both TMPRSS4 and DDR1 are co-regulated by promoter hypomethylation, which is associated with poor prognosis. Moreover, we show here that both genes are functionally related to maintain cell proliferation and survival.
Results
Expression of TMPRSS4 correlates with expression of genes involved in tumor cell-ECM interactions in NSCLC
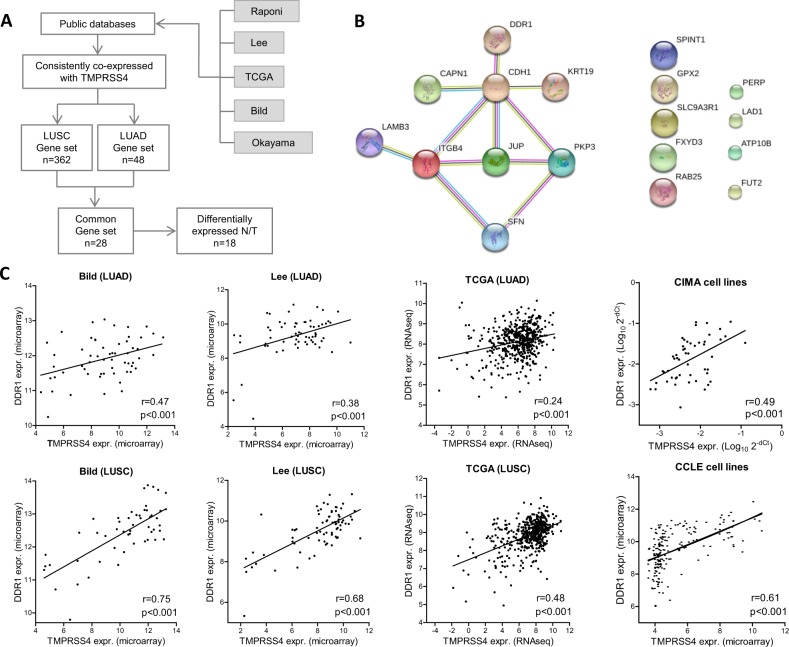
Our first goal was to identify genes that were consistently correlated with TMPRSS4 expression and differentially expressed in lung cancer patients. The strategy for the identification of the TMPRSS4-associated gene signature is shown in Fig. 1A. To this end, we carried out large-scale correlation analyses across 5 public databases and found that 362 genes were significantly coexpressed with TMPRSS4 in lung squamous carcinoma (LUSC) and 48 in the case of lung adenocarcinoma (LUAD), in all databases. Comparison of both gene sets identified a common 28-gene signature. Next, this signature was filtered out by considering just those genes that showed significantly different expression between normal and tumor lung samples in TCGA, which narrowed down the list to 18 (Supplementary Table 1). Heat map analysis of the 18-gene signature showed that most of these genes were up-regulated in tumors (Supplementary Fig. 1A). GO and IPA bioinformatic analyses revealed that most of these genes were related to cell adhesion and interaction with the ECM (Supplementary Table 1). Protein-protein network interactions using STRING13 showed that nine of the genes were significantly interconnected (FDR < 0.05; enrichment value p < 0.001) (Fig. 1B). This suggests that tumors with high TMPRSS4 expression may be associated with pathways involving cancer cell-ECM crosstalk in NSCLC, in agreement with the prometastatic role of TMPRSS4.
Figure 1.
(A) Schematic representation of the strategy used to identify genes coexpressed with TMPRSS4 in public databases. (B) Protein-protein network interactions analysis using STRING. Nine of the genes were significantly interconnected (FDR < 0.05). (C) Significant positive correlation between TMPRSS4 and DDR1 expression in LUAD and LUSC patient samples from Bild, Lee and TCGA databases, and in Cancer Cell Line Encyclopedia (CCLE) and CIMA cell lines.
One of the genes, Discoidin Domain Receptor tyrosine kinase 1 (DDR1), is a tyrosine kinase membrane-bound receptor with a role in invasion, remodeling of the ECM and metastasis. High DDR1 expression has been associated with poor prognosis in NSCLC14. A significant positive correlation between TMPRSS4 and DDR1 expression was found in all databases analyzed. Figure 1C shows results of 3 representative datasets in LUAD and LUSC: Bild, Lee and TCGA. Correlation was also found in cancer lines from CCLE and by qPCR in the CIMA lung cancer cell lines (Fig. 1C). We addressed whether the expression of TMPRSS4 and DDR1 would be mutually regulated in lung cancer cells. To this end we used H358 cells (with high expression of both genes) where we knocked-down either TMPRSS4 or DDR1. DDR1 levels were not reduced in clones where TMPRSS4 expression was depleted. Similarly, knock-down of DDR1 did not change levels of TMPRSS4 expression (Supplementary Fig. 1B,C).
DDR1 is overexpressed and epigenetically regulated in NSCLC
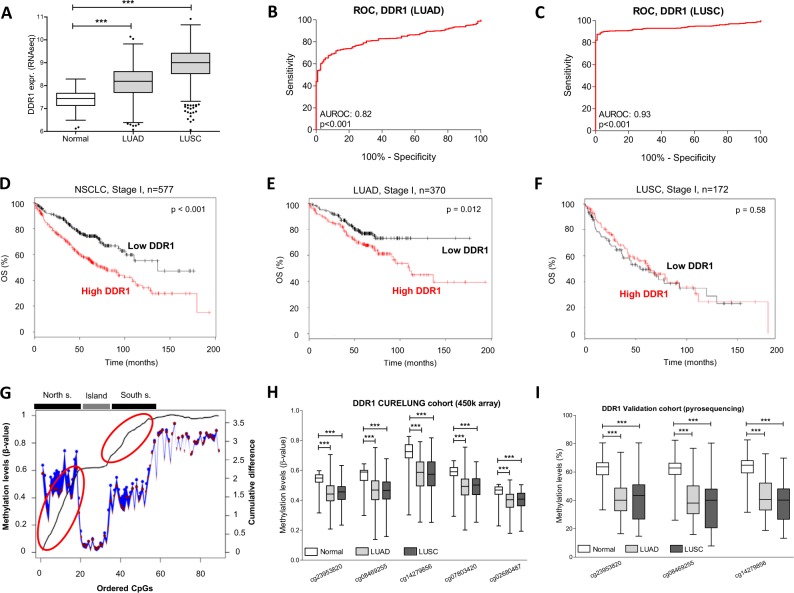
To study DDR1 expression, RNAseq values for DDR1 were evaluated in both LUAD and LUSC from TCGA (Fig. 2A). A significant increase (p < 0.001) in DDR1 levels was observed for both histological NSCLC types when compared to non-malignant lung, with AUROC values of 0.82, p < 0.001 (LUAD, Fig. 2B) and 0.93, p < 0.001 (LUSC, Fig. 2C). To study the prognostic value of DDR1 in early stage NSCLC we used Gyorffy’s database15, which includes data from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov), Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and Cancer Biomedical Informatics Grid (caBIG, http://cabig.cancer.gov/). Expression was categorized in high or low according to the mean value of the data in our study population. High levels of DDR1 were associated with reduced OS in stage I NSCLC (Fig. 2D). When separated by histology, patients with LUAD and high levels of DDR1 showed reduced OS (Fig. 2E), whereas those with LUSC did not show such association (Fig. 2F).
Figure 2.
(A) Expression of DDR1 was higher in NSCLC than in normal lung (TCGA). (B) Area under the ROC (AUROC) for LUAD (TCGA). (C) AUROC for LUSC (TCGA). (D) Kaplan Meier curves showing that high DDR1 levels were associated with reduced overall survival (OS) in stage I NSCLC. Considering LUAD samples only (E), the same result was obtained; on the contrary, DDR1 levels had no prognostic value in LUSC (F). (G) Representation of the methylation status of DDR1 promoter (CURELUNG data). The blue line and dots represent the percentage of methylation in normal lung, whereas the red dots correspond to tumors. The black line represents the cumulative difference in the percentage of methylation between consecutive CpGs comparing normal and malignant tissues. A steep slope was found in the north and south shores (red circles), thus showing strong DDR1 hypomethylation in tumors in this area. (H) Comparison of methylation status of CpGs from cg23953820 to cg02680487 (north shore) between normal versus tumor samples (CURELUNG data). (I) Validation of DDR1 promoter hypomethylation by pyrosequencing of cg23953820, cg08469255 and cg14279856 in the CUN-HGUV series of patients.
We also evaluated whether a combined index that included expression (median values) of both DDR1 and TMPRSS4 in the Gyorffy’s database or the CUN cohort (that included 50 NSCLC patients where expression values for the two genes were available) was superior than the prognostic value of each gene alone. However, the combined index was not better in predicting survival than each of the individual genes in any of the analyses (results not shown).
DNA promoter hypomethylation is responsible for overexpression of TMPRSS4 in NSCLC8. Because of the high correlation that we found between TMPRSS4 and DDR1 genes, we wondered whether DDR1 expression could also be epigenetically regulated. The DDR1 promoter contains a CpG island (from cg23001000 to cg26858073), flanked by a 5′ upstream shore (north) and a 3′ downstream shore (south). In the CURELUNG cohort, representation of cumulative difference in the percentage of methylation between 89 consecutive CpGs in the aforementioned promoter regions comparing normal and malignant tissues showed a steep slope in the north shore and, in a lesser extent, in the south shore (Fig. 2G). This reflects the presence of consistent hypomethylation in NSCLC. On the contrary, the island was characterized by a flat segment of the graphic without differences between malignant and healthy tissues (Fig. 2G). When separating the data by histological type, the same pattern was observed for both LUAD and LUSC (Supplementary Fig. 2A). Analysis performed in TCGA gave similar results (Supplementary Fig. 2B).
A specific region comprising 5 consecutive CpGs (from cg23953820 to cg02680487) with consistent hypomethylation in cancer specimens but not in non-malignant lung was identified in the north shore. Statistical comparisons confirmed that all these CpGs were significantly hypomethylated in LUAD and LUSC with respect to normal samples, for both CURELUNG (Fig. 2H) and TCGA (Supplementary Fig. 3A) cohorts. We then validated the hypomethylation of 3 CpGs by pyrosequencing in the CUN-HGUV cohort (Fig. 2I).
Given that both DDR1 and TMPRSS4 were co-expressed in tumors and their promoters hypomethylated, we assessed whether there would be a correlation between their methylation status. Indeed, Supplementary Fig. 3B shows that there was a very strong positive correlation in the promoter methylation status between TMPRSS4 and DDR1 in patients (CURELUNG cohort, r = 0.82, p < 0.001). As shown in Supplementary Fig. 3C, a similar correlation was observed for the CURELUNG cell lines (r = 0.65, p < 0.001). These results suggest that DDR1 and TMPRSS4 are co-regulated by hypomethylation.
Inverse correlation between DDR1 expression and promoter methylation status. Prognostic value of DDR1 in NSCLC
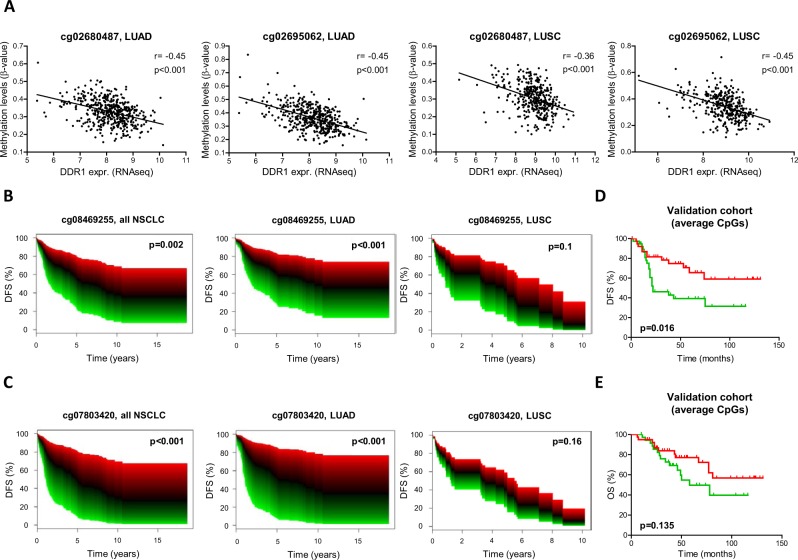
An inverse correlation between DDR1 promoter methylation and DDR1 expression was found for the 5 CpGs previously studied for both LUAD and LUSC (TCGA cohort). Examples of cg02680487 and cg02695062 are shown in Fig. 3A.
Figure 3.
(A) DDR1 expression showed significant inverse correlation with methylation status in patients. Data from TCGA for two representative CpGs in the DDR1 promoter (cg02680487 and cg02695062) in LUAD and LUSC. (B,C) Kaplan Meier curves in patients from the CURELUNG cohort showing continuous survival analysis. Significantly lower DFS in patients with low DDR1 methylation levels (green color), in comparison with patients with high methylation levels (red) was found. (D,E) Survival analyses in the CUN-HGUV cohort analyzed by pyrosequencing. Kaplan Meier curves showed that, levels below the median significantly predicted reduced DFS (D). In the case of OS (E), the trend was the same but results were not statistically significant.
The prognostic value of DDR1 methylation status was assessed in the CURELUNG cohort by multivariant Cox regression models considering the 5 CpGs mentioned above. Smoking, age and sex were considered as the confounding variables for the analysis. When considering all NSCLC patients, hypomethylation was associated with worse DFS for all the CpGs. Examples for cg08469255 and cg07803420 are shown in Fig. 3B,C, depicted as continuous survival curves. When analyzed by histology, the prognostic value was maintained in the case of LUAD but not for LUSC (Fig. 3B,C). Prognosis was also evaluated by pyrosequencing of cg23953820, cg08469255 and cg14279856 in the CUN-HGUV cohort. The 3 CpGs rendered similar results and data corresponding to the average value of these CpGs are represented in Fig. 3D, which shows that DFS was significantly lower in patients with DDR1 hypomethylation (p = 0.016). In the case of OS, the trend was the same but no statistical significance was found (p = 0.135) (Fig. 3E). We also evaluated the prognostic value of the combination between DDR1 and TMPRSS4 methylation status in the CURELUNG cohort of patients. Results of the likelihood ratio test between the Cox regression model including both genes and the model including only one of them was non-significant (p = 0.72). This was interpreted as non-evidence for a synergistic effect in survival prediction.
Demethylation of the DDR1 promoter reactivates DDR1 mRNA and protein expression
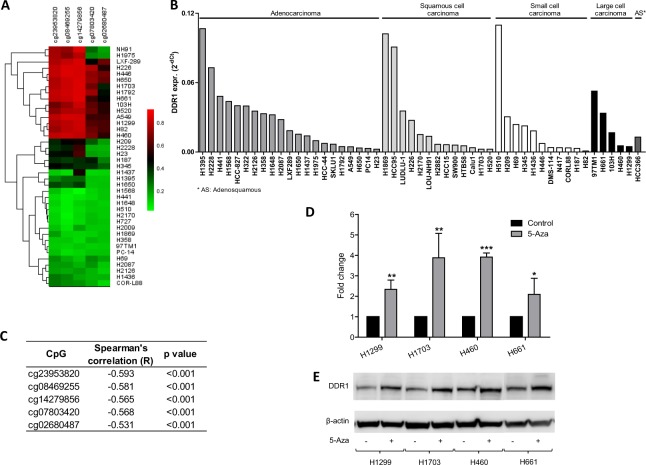
Methylation status of the 5 selected DDR1 CpGs for CURELUNG cell lines is shown in Fig. 4A, and DDR1 expression levels in the CIMA cell lines in Fig. 4B. DDR1 expression was inversely correlated with DDR1 promoter methylation (Fig. 4C). Treatment with the demethylating agent 5′-azacitidine in some cells with DDR1 methylated promoter increased DDR1 levels (Fig. 4D,E), indicating that expression is controlled by DNA methylation.
Figure 4.
(A) Cluster analysis of lung cancer cells (CURELUNG) based on DDR1 promoter methylation patterns: red, methylated; green, non-methylated. (B) Expression of DDR1 in the CIMA lung cancer cell lines analyzed by qPCR. (C) Inverse correlation between DDR1 expression and methylation in the cell lines. (D,E) Increased expression of DDR1 in cell lines upon treatment with the demethylating agent 5-azacitidine (5-aza) analyzed by qPCR (D) and Western blotting (E).
Co-inhibition of DDR1 and TMPRSS4 causes synergistic reduction of cell proliferation and tumor growth
Having demonstrated that both DDR1 and TMPRSS4 are co-expressed and epigenetically co-regulated in NSCLC, we wondered whether these genes could cooperate functionally to maintain a malignant cell behavior.
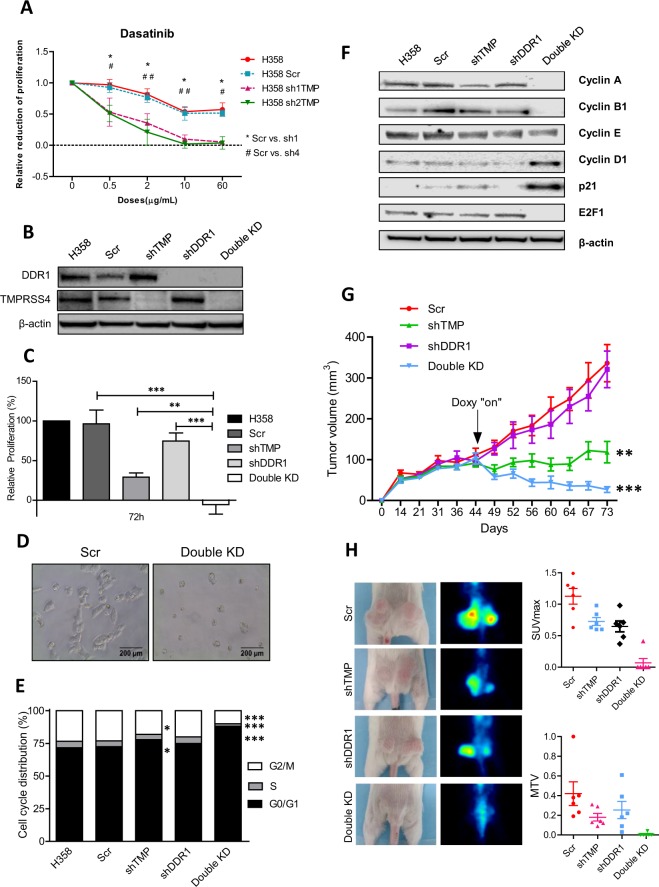
H358 cells, which express high levels of both DDR1 and TMPRSS4, were used for functional experiments. We developed clones with reduced levels of TMPRSS4 (conditional repression activated by doxycycline). Cell growth was first quantified upon treatment with the DDR1 inhibitor dasatinib at different doses in control cells and shTMPRSS4 clones (clones #1 and #2). Figure 5A shows that the cytotoxic effect was significantly higher in cells lacking TMPRSS4. We then developed clones with constitutive repression of DDR1 and clones lacking expression of both DDR1 and TMPRSS4 (Fig. 5B). Proliferation was reduced by ~25% in DDR1 KD cells and by (~70%) in TMPRSS4 KD cells (Fig. 5C). In the double KD clone we found not only a complete blockade of proliferation, but also cell death, as the total number of cells alive after 72 h in culture was lower than the number of cells seeded (negative values of cell proliferation) (Fig. 5C). The morphology of the double KD cells also suggested cell death (Fig. 5D). To confirm the antiproliferative effect of the double inhibition, we established H2170 single or double KD cells. MTT assays also showed a synergistic growth inhibitory effect in these cells when both DDR1 and TMPRSS4 were depleted (Supplementary Fig. 3D).
Figure 5.
(A) Dasatinib treatment in H358 with or without TMPRSS4. Cells lacking TMPRSS4 are significantly more sensitive to dasatinib than controls. (B) Western blot analysis of DDR1 and TMPRSS4 in H358 cell clones. (C) Proliferation analysis by MTT in single and double KD in comparison with controls. (D) Morphological observation of double KD cells compared to controls. (E) Cell cycle analysis. The double KD clone showed reduced proportion of cells in the G2/M and S phases and arrest in the G0/G1 phase. (F) Western blot analysis showing cyclins and cell cycle-related proteins. Double KD cells lacked cyclin A, cyclin B1 and E2F1 protein expression. A remarkable increase in p21 was observed in these cells. (G) Tumor volume in mice injected with shDDR1 cells did not change with respect to controls. Upon administration of doxycycline, tumors lacking TMPRSS4 were significantly smaller than controls. Tumors from double KD cells underwent tumor regression. (H) Representative images of macroscopic and microPET images from the different groups (left panel). Quantification by microPET of maximum standardized uptake value (SUVmax) and metabolic tumor volume (MTV). In vitro experiments were repeated 3 times. Statistical analysis for SUVmax and MTV was not performed because most values in the double KD group were “0”. TMP: TMPRSS4.
Based on these results, we evaluated the cell cycle. In TMPRSS4 KD cells, a significant increase (p < 0.05) in the proportion of cells in the G0/G1 phase and a reduction in the G2/M phase was found. No significant changes in the cell cycle were observed for DDR1 KD cells. On the contrary, double KD cells displayed very significant alterations in the G0/G1, S and G2/M phases (Fig. 5E). Regarding cell cycle related proteins, whereas there were little changes in shDDR1 and shTMPRSS4 clones, a total disappearance of cyclins A and B1 (both involved in S and G2/M transition) was found in double KD cells (Fig. 5F). In contrast, cyclins E and D1 levels were not reduced. Moreover, cyclin D1 levels were higher in the double KD clone, which could be related with a compensatory mechanism resulting from impaired cell proliferation. These changes were accompanied by an increase in p21 (inhibitor of cyclins A and E) and a total loss of E2F1 in double KD cells.
To evaluate whether these effects were translated into alteration in tumor growth, H358 parental cells and shRNA clones were injected into Rag2 mice. As shown in Fig. 5G, tumors resulting from shDDR1 cell injection were similar in size than controls. Tumors from the shTMPRSS4 group started to decrease after the administration of doxycycline and were significantly smaller at the end of the experiment (60% reduction, p < 0.01). Strikingly, tumors of the double KD group regressed rapidly after administration of doxycycline and were 93% smaller (p < 0.001) than controls at the end of the experiment. In fact, tumors were hardly measurable at this time. Moreover, by microPET analysis (Fig. 5H), a dramatic drop in 18-FDG uptake was found in animals injected with double KD cells, as evaluated by maximum standardized uptake value (SUVmax) and metabolic tumor volume (MTV).
Cisplatin enhances dramatically apoptosis in double KD cells
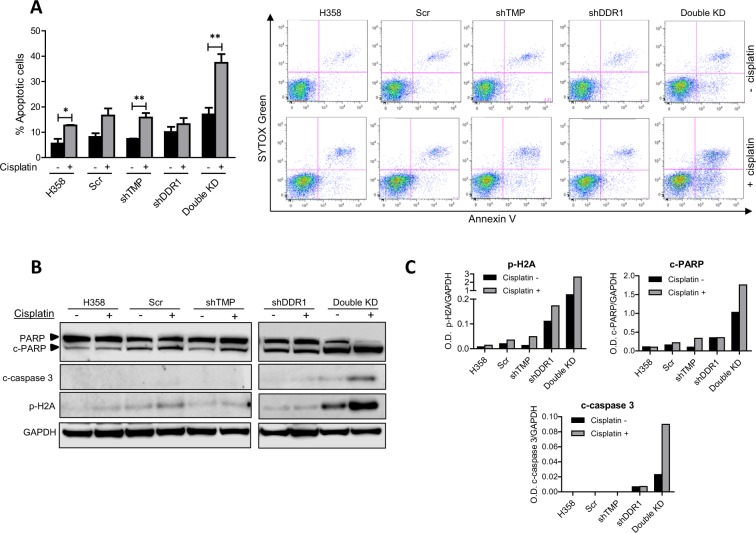
Because double KD cells were less proliferative and had alterations in the cell cycle, we hypothesized that they would be more sensitive to chemotherapy-induced cell death. In the double KD, basal apoptosis (without cisplatin) was higher than that found for the single KD and control cells (black bars in Fig. 6A). After cisplatin treatment, apoptosis in the double KD reached almost 40% of the cell population and was significantly higher than levels achieved for the controls or single KD, which was below 20% in all cases (Fig. 6A).
Figure 6.
(A) Apoptosis analysis of H358 cells treated or untreated with cisplatin. While untreated single KD cells did not show relevant changes in apoptosis in comparison with controls, double KD cells had more apoptotic cells than the rest of the groups. Upon treatment with cisplatin, levels of apoptotic cells were very significantly increased in double KD cells, showing chemosensitization. (B) Western blot analysis reveals that levels of c-PARP and c-caspase 3 were strongly induced in double KD cells treated with cisplatin in comparison with the other groups. A dramatic increase in p-H2A levels was also found in these cells. (C) Quantification of bands corresponding to the Western blots shown in Figure B. Experiments were repeated 3 times. TMP: TMPRSS4.
The double KD clone showed higher levels of cleaved (c-)-PARP, c-caspase 3 and phospho (p)-H2A than the rest of the cells in the absence of treatment (Fig. 6B) as shown in the densitometric quantification of the bands (Fig. 6C). Administration of cisplatin increased further levels of these apoptotic/DNA damage markers. Original full-size images for all the blots are shown in Supplementary Fig. 4.
This result demonstrates that simultaneous targeting of TMPRSS4 and DDR1 sensitizes H358 cells to cisplatin-mediated apoptotic cell death.
Discussion
Malignancy of cancer cells is usually maintained by multiple genetic and epigenetic alterations, some of which may cooperate in order to sustain tumor growth. Based on the hypothesis that these cooperative genes could be consistently co-expressed in tumors, we searched for TMPRSS4-correlated genes in publicly available genomic data and identified DDR1 among other genes related to cancer cell-ECM interaction. We have previously shown that levels of TMPRSS4 in NSCLC are much higher than those found in normal lung and that overexpression associates with worse prognosis8. Moreover, increased TMPRSS4 levels are due to DNA hypomethylation, which is also associated with reduced DFS8. We have found in the present study that, similar to TMPRSS4, hypomethylation of DDR1 causes DDR1 overexpression and that both expression and methylation status can be used as prognostic indicators in NSCLC. It is worth noticing that the degree of promoter methylation found for DDR1 is highly correlated with that of TMPRSS4 in NSCLC patients and cancer cell lines. Importantly, treatment of cells with 5-azacitidine increased levels of DDR1, a result that was also reported for TMPRSS48. All these data suggest that these genes are co-regulated by epigenetic mechanisms. Our results also show that expression levels of both genes are not mutually regulated. Therefore, it is possible that they share transcription factors and epigenetic regulators that control expression, although this hypothesis will have to be addressed in future studies.
Both TMPRSS4 and DDR1 have been shown to promote proliferation, migration, invasion and metastasis in NSCLC cells and in other cell types, depending on the cell context6,16,17. TMPRSS4 has been reported to stimulate proliferation in prostate and thyroid cancer cells18,19. In vivo studies have also shown that TMPRSS4 increases subcutaneous tumor growth and metastasis7,20,21. DDR1 may promote cell division depending on the cell type: in H460 NSCLC cells, abrogation of DDR1 levels did not modify proliferation in vitro, but affected invasion and reduced bone metastasis in vivo14. In pancreatic cells, shRNAs targeting DDR1 decreased clonogenicity and migration22. Genetic targeting of DDR1 inhibited cell proliferation and subcutaneous tumor growth in glioma23. Moreover, in human colon carcinoma cells DDR1 depletion caused cell death in response to induced DNA damage24. In our study, genetic targeting showed ~70% and ~25% reduction in proliferation after depletion of TMPRSS4 or DDR1 levels, respectively. Interestingly, lack of TMPRSS4 in these cells caused cisplatin-mediated apoptosis, which was not the case for DDR1.
Considering the hypothesis of mutual functional cooperation based on co-expression, we treated shTMPRSS4 clones with the DDR1 inhibitor dasatinib and knocked-down both TMPRSS4 and DDR1 in H358 and H2170 cells. First, dasatinib was significantly more effective at inhibiting proliferation in cells with no TMPRSS4 than in controls. In addition, co-silencing completely blocked proliferation and triggered apoptosis. We have demonstrated that double KD cells suffered G0/G1 cell cycle arrest accompanied by a decrease in the percentage of the cell population in the S and G2/M phases, with loss of cyclins A and B1, but not in cyclins D1 and E. In addition, double KD cells were characterized by loss of E2F1 and increase in p21. In in vivo experiments, simultaneous abrogation of both genes caused tumor regression and lack of 18-FDG uptake. Moreover, when double KD cells were challenged with cisplatin, a dramatic increase in apoptosis and DNA damage was found.
Our results show that co-targeting TMPRSS4 and DDR1 produces a synergistic antitumor effect, which fits in the concept of synthetic lethal interaction. The synthetic lethality has been defined as the inability of cells to proliferate when co-targeting two genes, with a synergistically superior inhibition than that found for each individual gene25. Consistent co-expression of two genes involved in a similar function is a predictor of synthetic lethality25, a strategy that is being applied to find out novel cancer vulnerabilities. The typical example of synthetic lethality is applied to mutations affecting a similar pathway, such as BRCA/PARP, where co-targeting leaves cells unprotected from DNA damage and repair. The synthetic interaction concept (appearance of a new phenotype) has now been expanded and large scale siRNA strategies are being currently used to identify combinations of loss of function leading to more-than-additive cell death/sickness25.
The possible mechanism of this synthetic lethal interaction is at this point unknown. As a hypothesis we can speculate that alteration in MAPK and PI3K/AKT signaling pathways might be responsible for this effect. Indeed, signaling triggered by both TMPRSS4 and DDR1 in cancer cells activate common pathways, including phosphorylation in ERK1/2 and AKT depending on the cellular context6,26–28. In addition, both proteins play a role in resistance to chemotherapy28,29. As synthetic lethality frequently occurs when targeting genes from parallel pathways involved in similar functions25, it is possible that blockade of both TMPRSS4 and DDR1 makes cells succumb to insufficient proliferation/survival signals. In fact, it has been demonstrated in cancer cells that co-targeting of the Ras/ERK1/2 and PI3K/AKT pathways with kinase inhibitors or genetic knockdown approaches targeting members of these pathways results in synthetic lethality30.
Co-inhibition of TMPRSS4 and DDR1 could then constitute a novel therapeutic strategy for NSCLC. Several DDR1 inhibitors have been developed including small molecules (dasatinib, imatinib) and monoclonal antibodies31. Dasatinib is currently being tested in clinical trials for the treatment of cancer patients (NCT02389309, NCT03216070). Combined targeting of DDR1 and Notch has also been shown as a new effective strategy to treat KRAS driven lung adenocarcinomas32. Regarding TMPRSS4, no drugs are currently available against this protein. A family of 2-hydroxydiarylamide derivatives that inhibit the catalytic activity of TMPRSS4 has been described33. In this study, leader compounds inhibited migration of colon cancer cells, but whether they exert in vivo antitumor effects remains to be elucidated.
In summary, through high-throughput correlation analysis using public databases we have discovered that TMPRSS4 and DDR1 are co-expressed, co-regulated by DNA methylation, and serve as prognostic indicators in NSCLC. In functional in vitro and in vivo experiments we have shown a novel cancer vulnerability based on a synthetic lethal interaction when both genes are absent. This suggests a novel therapeutic strategy for TMPRSS4/DDR1-positive lung tumors.
Materials and Methods
Patients
The CURELUNG cohort, which includes lung cancer patients (n = 444) and non-tumor lung tissue samples (n = 25) was used to assess promoter methylation with the Infinium 450 k array. Data on DFS is available in a subset of this cohort that includes 198 surgically resected NSCLC samples, which were used to study the prognostic value of DDR1 promoter methylation. The clinical characteristics of these NSCLC patients have been previously published34.
For validation of methylation studies we used 136 tumor samples from patients (59 of which included non-malignant adjacent tissue) of the Clinica Universidad de Navarra (CUN, Pamplona, Spain) and Hospital General Universitario de Valencia (HGUV, Valencia, Spain) (CUN-HGUV cohort). All patients were diagnosed with NSCLC and did not receive neoadjuvant therapy. Detailed clinical characteristics are shown in Supplementary Table 2. REMARK criteria for tumor biomarkers35 were followed in our study.
DNA isolation and methylation analysis by bisulfite pyrosequencing
DNA was extracted with the NucleoSpin Tissue® kit (Macherey-Nagel) following manufacturer’s instructions. Quantitative DNA methylation analysis of the DDR1 promoter was performed by bisulfite pyrosequencing of the following CpG’s: cg23953820, cg08469255 and cg14279856. Five hundred ng of each DNA sample was treated with sodium bisulfite using the EZ-96 DNA Methylation-Lightning Kit (Zymo Research). Primers for PCR amplification and pyrosequencing are shown in Supplementary Table 3. Pyrosequencing reactions and methylation quantification were carried out in a PyroMark Q24 System version 2.0.6 (Qiagen).
Cell culture and establishment of shRNA clones
Lung cancer cell lines from the CURELUNG consortium and the CIMA collection were used to evaluate DDR1 promoter methylation status using the Infinium 450 k array36 and DDR1 expression, respectively. Originally, all cells were obtained from ATCC and were cultured in standard culture conditions. To reduce gene expression we used lentiviral shRNAs: pLKO-puro targeting DDR1 and Tet-pLKO-puro targeting TMPRSS4. Controls consisted of shScramble-pLKO-puro (Scr) and shGFP-Tet-pLKO-puro. Conditional knock-down of TMPRSS4 was obtained by treatment with 1 μg/mL doxycycline. Upon cell infection of H358 and H2170 cells with lentiviral particles, clones were selected with 5 μg/mL puromycin. Several shRNA clones were generated for each gene. shTMPRSS4(1) and shDDR1(1) clones were used to establish the double knock-down (KD) clone.
Treatment with a demethylating agent
For demethylation experiments, cells were treated with 5-azacitidine (Sigma) at 5 μM for 72 h in quadruplicates. After this period of time, both total RNA and proteins were extracted to perform qPCRs and Western blotting.
qPCR and western blotting
qPCR and Western blots were performed as previously described9. Sequences of the primers are shown in Supplementary Table 3. An Applied Biosystems 7500 Real-time PCR equipment was used for the qPCRs. Antibodies and dilutions used for Western blotting are shown in Supplementary Table 4. Blot signals were captured with an Odyssey-Fc imaging system (Li-Cor Bioscience) and quantified by calculating the optical density (O.D.) after normalization with GAPDH O.D. using the Image Studio Lite software v5.2.
MTTs, cell cycle and apoptosis assays
For MTTs, experiments were conducted as previously described9. Cell cycle and apoptosis analysis were evaluated with a FACSCanto II cytometer (BD Bioscience) and the FlowJo® software v9.3. Experimental details are shown in Supplementary Materials and Methods. Dasatinib for the MTTs experiments was a kind donation from Bristol Myers Squibb.
In vivo tumor growth and microPET
All animal procedures were approved by the ethics committee/institutional review of the University of Navarra (Pamplona, Spain), under the protocol number 044/16.
Cells (10 × 106 per cell type) were subcutaneously injected into Rag2 mice (n = 8 per group; Harlan) and animals were administered with doxycycline in the drinking water (1 μg/mL) when tumors had reached 100 mm3. Tumor volume was measured with an electronic caliper using the following formula: V = (L × W2)/2, where L corresponds to tumor length and W to width. MicroPET was carried out by the Micro-PET Core Facility (CUN) as previously described37.
Genome-wide correlation methods, bioinformatics and statistical analysis
The Cancer Genome Atlas (TCGA) data were downloaded from Genomics Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov) and analyzed as previously described8. CEL files from Gene Expression Omnibus (GEO) were downloaded from the following datasets in the case of lung squamous carcinoma (LUSC): GSE4573, GSE3141, and GSE8894. For lung adenocarcinoma (LUAD) we used GSE3141, GSE8894 and GSE31210. Pearson analysis was performed to identify correlations between TMPRSS4 expression and expression of genes in the datasets. Methylation patterns were evaluated with the Infinium 450k array from TCGA and CURELUNG datasets using LIMMA. To classify patients according to their methylation levels, a β-value threshold of 0.5 was considered. Comparisons of methylation status were evaluated with the Mann-Whitney U test and by calculation of the cumulative differences in the percentage of methylation for each consecutive CpG. Association between methylation and expression levels was tested with Pearson’s or Spearman’s correlation. Prognostic value of methylation status was assessed by Elastic Net penalized Cox regression models, which are able to adjust regression models and perform variable selection at the same time, thus providing a more reliable method of CpGs selection for development of the prediction model. Additionally, Cox regression models were fitted to estimate the effects of the different selected CpGs on survival times. Internal validation of the models was assessed using bootstrap with 1000 replicates. The prognostic value of the combination of both genes (DDR1 and TMPRSS4) was assessed by performing a likelihood ratio test between a Cox regression model including both genes and a Cox regression model including only one of them. Prognostic studies on DDR1 expression were evaluated using public data from Gyorffy et al.15. Disease-free survival (DFS) was estimated as the time from surgery to recurrence and overall survival (OS) as the time from diagnosis to the date of death or the patient’s last follow-up.
For in vitro and in vivo studies, Student’s t test (for two groups) or ANOVA (for several groups) were used for comparisons. Cells were clustered based on their methylation status by hierarchical clustering analysis using Genesis (http://genome.tugraz.at).
Statistical analyses were performed with R (version 3.4.2), R packages glmnet (version 2.0–13) and Survival (version 2.41–3), or with Prism 5 software (GraphPad). Statistical significance was defined as (*)p < 0.05, (**)p < 0.01 and (***)p < 0.001.
Statement on use of human samples and experiments in animals
In the studies carried out with human samples, written informed consent was obtained from each patient. The study protocol was approved by the ethical committee of the Clinica Universidad de Navarra (CUN, Pamplona, Spain) and Hospital General Universitario de Valencia (HGUV, Valencia, Spain) (CUN-HGUV). The study was conducted according to the Declaration of Helsinki.
All animal procedures were approved by the ethics committee/institutional review of the University of Navarra (Pamplona, Spain), under the protocol number 044/16.
Supplementary information
Acknowledgements
We thank Diana Garcia (Health Research Unit, La Fe, Valencia) for technical support. This work has been funded by: ISC-III (PI16/01352, to A.C.; PI17/0041 to R.P.; PI16/0295 to J.S.; PI15/00753; PI16/01821 to L.M. and CB16/12/00350 to E.J.L.; “Juan Rodés” JR17/00016 contract to A.D.L.; “Miguel Servet” contract CP13/00055 to J.S.; CIBERONC CB16/12/00443 to L.M.M.), Juan Serrano, AECC and Ramón Areces Foundations (to LMM). E.J.L. was also funded by Fundación Arnal Planelles. Fellowships support: F.E., “Amigos de la Universidad de Navarra”; M.V. and E.R., “FPU, Spanish Ministry of Education”.
Author contributions
M.V., E.R., J.S., L.M.M., R.P., C.A. and A.C. conceived and designed the study. E.G. and D.H. performed the bioinformatics analyses. M.V., E.R., F.E., A.D.L., C.r.C., M.R., K.V., E.J.L., C.S., M.J.P., J.S. and A.C. performed experiments or analyzed data. All authors wrote or revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Juan Sandoval and Alfonso Calvo.
Contributor Information
Juan Sandoval, Email: epigenomica@iislafe.es.
Alfonso Calvo, Email: acalvo@unav.es.
Supplementary information
is available for this paper at 10.1038/s41598-019-51066-3.
References
- 1.Reck, M. & Rabe, K. F. Precision Diagnosis and Treatment for Advanced Non–Small-Cell Lung Cancer. Longo, D. L., editor. New England Journal of Medicine. 377:849–61 (2017). [DOI] [PubMed]
- 2.Langevin SM, Kratzke RA, Kelsey KT. Epigenetics of lung cancer. Translational Research. 2015;165:74–90. doi: 10.1016/j.trsl.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duruisseaux M, Esteller M. Lung cancer epigenetics: From knowledge to applications. Seminars in Cancer Biology. 2018;51:116–28.. doi: 10.1016/j.semcancer.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell. 2017;168:670–91.. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–74.. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.de Aberasturi AL, Calvo A. TMPRSS4: an emerging potential therapeutic target in cancer. British Journal of Cancer. 2015;112:4–8. doi: 10.1038/bjc.2014.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C-H, et al. TMPRSS4 facilitates epithelial-mesenchymal transition of hepatocellular carcinoma and is a predictive marker for poor prognosis of patients after curative resection. Scientific Reports. 2015;5:12366. doi: 10.1038/srep12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villalba M, et al. Epigenetic alterations leading to TMPRSS4 promoter hypomethylation and protein overexpression predict poor prognosis in squamous lung cancer patients. Oncotarget. 2014;7:22752–69. doi: 10.18632/oncotarget.8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Aberasturi AL, et al. TMPRSS4 induces cancer stem cell-like properties in lung cancer cells and correlates with ALDH expression in NSCLC patients. Cancer letters. 2016;370:165–76. doi: 10.1016/j.canlet.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Liu R, Cheng Y, Yu J, Lv Q-L, Zhou H-H. Identification and validation of gene module associated with lung cancer through coexpression network analysis. Gene. 2015;563:56–62. doi: 10.1016/j.gene.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Tian F, Zhao J, Fan X, Kang Z. Weighted gene co-expression network analysis in identification of metastasis-related genes of lung squamous cell carcinoma based on the Cancer Genome Atlas database. Journal of thoracic disease. 2017;9:42–53. doi: 10.21037/jtd.2017.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rammal H, et al. Discoidin Domain Receptors: Potential Actors and Targets in Cancer. Frontiers in Pharmacology. 2016;7:55. doi: 10.3389/fphar.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids research. 2019;47:D607–13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valencia K, et al. Inhibition of collagen receptor discoidin domain receptor-1 (DDR1) reduces cell survival, homing, and colonization in lung cancer bone metastasis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:969–80. doi: 10.1158/1078-0432.CCR-11-1686. [DOI] [PubMed] [Google Scholar]
- 15.Győrffy, B., Surowiak, P., Budczies J. & Lánczky, A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. Chellappan, S. P. editor. PloS one. 8, e82241 (2013). [DOI] [PMC free article] [PubMed]
- 16.Rammal H, et al. Discoidin Domain Receptors: Potential Actors and Targets in Cancer. Frontiers in pharmacology. 2016;7:55. doi: 10.3389/fphar.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanabe LM, List K. The role of type II transmembrane serine protease-mediated signaling in cancer. The FEBS Journal. 2017;284:1421–36.. doi: 10.1111/febs.13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y, et al. TMPRSS4 induces invasion and proliferation of prostate cancer cells through induction of Slug and cyclin D1. Oncotarget. 2016;7:50315–32.. doi: 10.18632/oncotarget.10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan H, et al. Transmembrane Protease Serine 4 Promotes Thyroid Cancer Proliferation via CREB Phosphorylation. Thyroid. 2015;25:85–94. doi: 10.1089/thy.2014.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larzabal L, et al. Overexpression of TMPRSS4 in non-small cell lung cancer is associated with poor prognosis in patients with squamous histology. British Journal of Cancer. 2011;105:1608–14. doi: 10.1038/bjc.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung H, et al. TMPRSS4 promotes invasion, migration and metastasis of human tumor cells by facilitating an epithelial–mesenchymal transition. Oncogene. 2008;27:2635–47. doi: 10.1038/sj.onc.1210914. [DOI] [PubMed] [Google Scholar]
- 22.Aguilera KY, et al. Inhibition of Discoidin Domain Receptor 1 Reduces Collagen-mediated Tumorigenicity in Pancreatic Ductal Adenocarcinoma. Molecular cancer therapeutics. 2017;16:2473–85.. doi: 10.1158/1535-7163.MCT-16-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamanaka R, et al. Identification of expressed genes characterizing long-term survival in malignant glioma patients. Oncogene. 2006;25:5994–6002. doi: 10.1038/sj.onc.1209585. [DOI] [PubMed] [Google Scholar]
- 24.Ongusaha PP, et al. p53 induction and activation of DDR1 kinase counteract p53-mediated apoptosis and influence p53 regulation through a positive feedback loop. The EMBO Journal. 2003;22:1289–301. doi: 10.1093/emboj/cdg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nijman SMB. Synthetic lethality: general principles, utility and detection using genetic screens in human cells. FEBS letters. 2011;585:1–6. doi: 10.1016/j.febslet.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, et al. TMPRSS4 induces invasion and epithelial-mesenchymal transition through upregulation of integrin alpha5 and its signaling pathways. Carcinogenesis. 2010;31:597–606. doi: 10.1093/carcin/bgq024. [DOI] [PubMed] [Google Scholar]
- 27.El Azreq M-A, et al. Discoidin domain receptor 1 promotes Th17 cell migration by activating the RhoA/ROCK/MAPK/ERK signaling pathway. Oncotarget. 2016;7:44975–90.. doi: 10.18632/oncotarget.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vehlow A, et al. Interaction of Discoidin Domain Receptor 1 with a 14-3-3-Beclin-1-Akt1 Complex Modulates Glioblastoma Therapy Sensitivity. Cell reports. 2019;26:3672–3683.e7. doi: 10.1016/j.celrep.2019.02.096. [DOI] [PubMed] [Google Scholar]
- 29.Exposito F, et al. Targeting of TMPRSS4 sensitizes lung cancer cells to chemotherapy by impairing the proliferation machinery. Cancer Letters. 2019;453:21–33. doi: 10.1016/j.canlet.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Guenther MK, Graab U, Fulda S. Synthetic lethal interaction between PI3K/Akt/mTOR and Ras/MEK/ERK pathway inhibition in rhabdomyosarcoma. Cancer Letters. 2013;337:200–9. doi: 10.1016/j.canlet.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Lu X, Ren X, Ding K. Small Molecule Discoidin Domain Receptor Kinase Inhibitors and Potential Medical Applications. Journal of Medicinal Chemistry. 2015;58:3287–301. doi: 10.1021/jm5012319. [DOI] [PubMed] [Google Scholar]
- 32.Ambrogio C, et al. Combined inhibition of DDR1 and Notch signaling is a therapeutic strategy for KRAS-driven lung adenocarcinoma. Nature Medicine. 2016;22:270–7. doi: 10.1038/nm.4041. [DOI] [PubMed] [Google Scholar]
- 33.Kang S, Min H-J, Kang M-S, Jung M-G, Kim S. Discovery of novel 2-hydroxydiarylamide derivatives as TMPRSS4 inhibitors. Bioorganic & Medicinal Chemistry Letters. 2013;23:1748–51. doi: 10.1016/j.bmcl.2013.01.055. [DOI] [PubMed] [Google Scholar]
- 34.Sandoval J, et al. A Prognostic DNA Methylation Signature for Stage I Non–Small-Cell Lung Cancer. Journal of Clinical Oncology. 2013;31:4140–7. doi: 10.1200/JCO.2012.48.5516. [DOI] [PubMed] [Google Scholar]
- 35.Sauerbrei W, Taube SE, McShane LM, Cavenagh MM, Altman DG. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): An Abridged Explanation and Elaboration. JNCI: Journal of the National Cancer Institute. 2018;110:803–11. doi: 10.1093/jnci/djy088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iorio F, et al. A Landscape of Pharmacogenomic Interactions in. Cancer. Cell. 2016;166:740–54. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castañón E, et al. The inhibitor of differentiation-1 (Id1) enables lung cancer liver colonization through activation of an EMT program in tumor cells and establishment of the pre-metastatic niche. Cancer Letters. 2017;402:43–51. doi: 10.1016/j.canlet.2017.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.