Abstract
The recent report of RBS-Seq to map simultaneously the epitranscriptomic modifications N1-methyladenosine, 5-methylcytosine, and pseudouridine (Ψ) via bisulfite treatment of RNA provides a key advance to locate these important modifications. The locations of Ψ were found by a deletion signature generated during cDNA synthesis after bisulfite treatment for which the chemical details of the reaction are poorly understood. In the present work, the bisulfite reaction with Ψ was explored to identify six isomers of bisulfite adducted to Ψ. We found four of these adducts involved the heterocyclic ring, similar to the reaction with other pyrimidines. The remaining two adducts were bonded to the 1‵ carbon, which resulted in opening of the ribose ring. The utilization of complementary 1D- and 2D-NMR, Raman and electronic circular dichroism spectroscopies led to the assignment of the two ribose adducts being the constitutional isomers of an S- and an O-adduct of bisulfite to the ribose, and these are the final products after heating. A mechanistic proposal is provided to rationalize chemically the formation and stereochemistries of all six isomeric bisulfite adducts to Ψ; conversion of intermediate adducts to the two final products is proposed to involve E2, SN2’ and [2,3]-sigmatropic shift reactions. Lastly, a synthetic RNA template with Ψ at a known location was treated with bisulfite leading to a deletion signature after reverse transcription, supporting the RBS-Seq report. This classical bisulfite reaction used for epigenomic and epitranscriptomic sequencing diverges from the C nucleoside Ψ to form stable bisulfite end products that yield signatures for next-generation sequencing.
TOC Graphic

Introduction
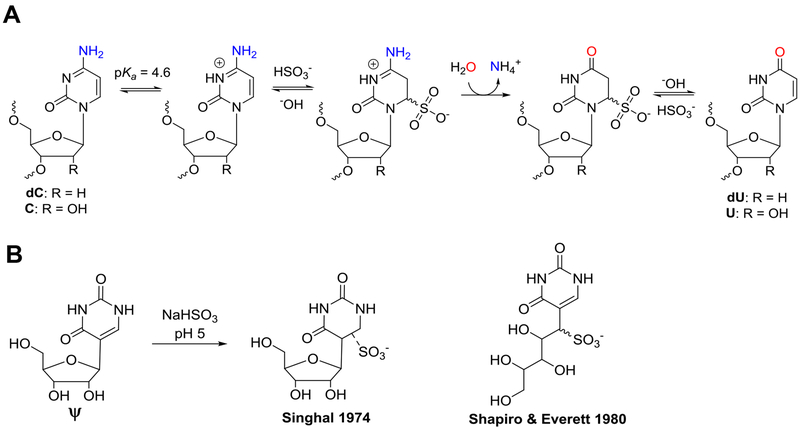
Bisulfite addition to carbonyl-containing compounds is a classical reaction in organic chemistry that has revolutionized our understanding of biology. Historically, reversible bisulfite addition to aldehydes or ketones was a convenient method for purification of organic compounds by recrystallization of the bisulfite adduct.1 While modern chromatographic techniques have displaced this reaction from organic chemistry textbooks, bisulfite chemistry finds application today in its conjugate addition reaction to carbon-6 of cytidine. In 1970, Shapiro and Hayatsu independently observed that under acidic conditions bisulfite adds to the heterocyclic ring in 2‵-deoxycytidine (dC) resulting in selective deamination of the exocyclic amine to furnish 2‵-deoxyuridine (dU) upon desulfonation (Scheme 1A).2,3 In 1980, Ehrlich demonstrated that 5-methyl-2‵-deoxycytidine (5mC) reacts ~50-fold slower with bisulfite than dC.4 Finally, in 1992 Frommer, et al. applied the bisulfite reaction to DNA to map the locations of the epigenetic mark 5mC.5 Bisulfite treatment of DNA enabled sequencing for 5mC because the dC bases would preferentially react and deaminate to be sequenced as thymidine (dU equivalent) in the PCR amplicons, while the 5mCs continued to code like dC. The relative ease in conducting the bisulfite reaction has resulted in this method being applied to high-throughput sequencing.6,7 Hybrid approaches using the bisulfite reaction were developed to sequence the TET-mediated oxidation products of 5mC that include 5-hydroxymethyl-2`-deoxycytidine,8,9 5-formyl-2`-deoxycytidine,10,11 and 5-carboxyl-2`-deoxycytidine.12 Many additional chemoselective tags have been developed for sequencing the 5mC oxidation products in DNA.13,14 Thus, bisulfite chemistry opened the door for many discoveries to be made regarding how epigenetic modification of dC in particular regions of the genome impacts cellular phenotype.15
Scheme 1.
(A) The bisulfite reaction with dC/C yields dU/U that codes like T. (B) The bisulfite adduct to Ψ as reported by Singhal16 or Shapiro and Everett.17
Decoration of RNA with site-specific modifications is well established.18–20 More recent findings have identified that RNA modifications, particularly in mRNA, can alter the coding potential, cellular half-life, alternative splicing, and compartmentalization of these coding strands, opening the field of epitranscriptomics.21–23 Chemical modifications to RNA alter its physical properties and impact secondary structure or change the coding potential of the bases.24–26 Prominent regulatory modifications in mRNA include N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-methylcytidine (m5C), inosine (I), and pseudouridine (Ψ); all of these appear to function together at various stages in regulating RNA biogenesis.18,21–23,27 Enabling chemical tools for sequencing RNA modifications include glyoxal and RNase T1 to generate site-specific strand cleavage at I that is identified during sequencing;28 metabolic incorporation of photoreactive 5-thiouridine to trap m6A-specific antibodies for RNA enrichment and sequencing;29 bisulfite chemistry to sequence m5C in RNA or other chemoselective tags;14,30 and N-cyclohexyl-N‵-(2-morpholinoethyl)-carbodiimide (CMC) or its derivatives react specifically with Ψ to generate a stop in cDNA synthesis observed during sequencing.31–35 Recently, we assisted the Cairns laboratory to discover RBS-Seq, a bisulfite-based reaction on RNA that permits sequencing of three RNA modifications simultaneously, including m1A, m5C, and Ψ.36 Beyond the expected m5C result, m1A was located via Dimroth rearrangement to m6A during bisulfite treatment that converted a cDNA synthesis stop to a bypass, and Ψ was found by a deletion signature following reverse transcription. The ongoing mystery with this method was why does bisulfite treatment of Ψ result in a deletion signature during cDNA synthesis?
In 1974, Singhal studied the bisulfite reaction with the Ψ nucleoside and identified two stable product peaks that retained the aromatic UV-vis absorbance at ~265 nm, similarly to Ψ, and the products formed a vicinal-diol borate complex supporting the conclusion that the ribose sugar was still present.16 Curiously, Singhal concluded bisulfite formed a stable adduct to the heterocycle (Scheme 1B), which would no longer be aromatic, a conclusion inconsistent with the UV-vis data. The only other report of this reaction comes from a Ph.D. dissertation from Shapiro’s laboratory (Donald W. Everett, Ph.D.; NYU, 1980).17 The bisulfite reaction on the Ψ nucleoside was found to yield two products that had the same UV-vis spectra as reported by Singhal. Further, the products had masses consistent with mono-bisulfite adducts, while the 1H-NMR for the two products were different with respect to the 1‵ proton signals, and the optical rotary dispersion spectra were mirror images of one another with subtle peak shape differences. Everett and Shapiro proposed that bisulfite was adducted to a ribose ring-opened nucleoside at the 1‵ carbon yielding a new carbon to sulfur covalent bond, and the two peaks represented diastereomeric products (Scheme 1B); however, the results were never published.
In the present report, we harnessed modern spectroscopic methods to discover that bisulfite reacts with the Ψ nucleoside to form two stable aromatic end products, both of which are mono-bisulfite adducts at C1’ and have a ring-opened ribose. However, instead of being a pair of diastereomers, these two products are constitutional isomers in which one is an O-adduct while the other is an S-adduct of bisulfite. Additionally, we found four isomeric unstable intermediates of bisulfite adducted to the heterocyclic ring of Ψ, and their decomposition pathways were determined. Lastly, the stable Ψ-bisulfite ribose adducts were synthesized in a model RNA template strand, and reverse transcription was found to generate a deletion at the adduct consistent with the previous RBS-Seq studies.36 Herein, we present the experiments that led us to unravel mysteries surrounding this unusual chemistry of high biological relevance.
Results
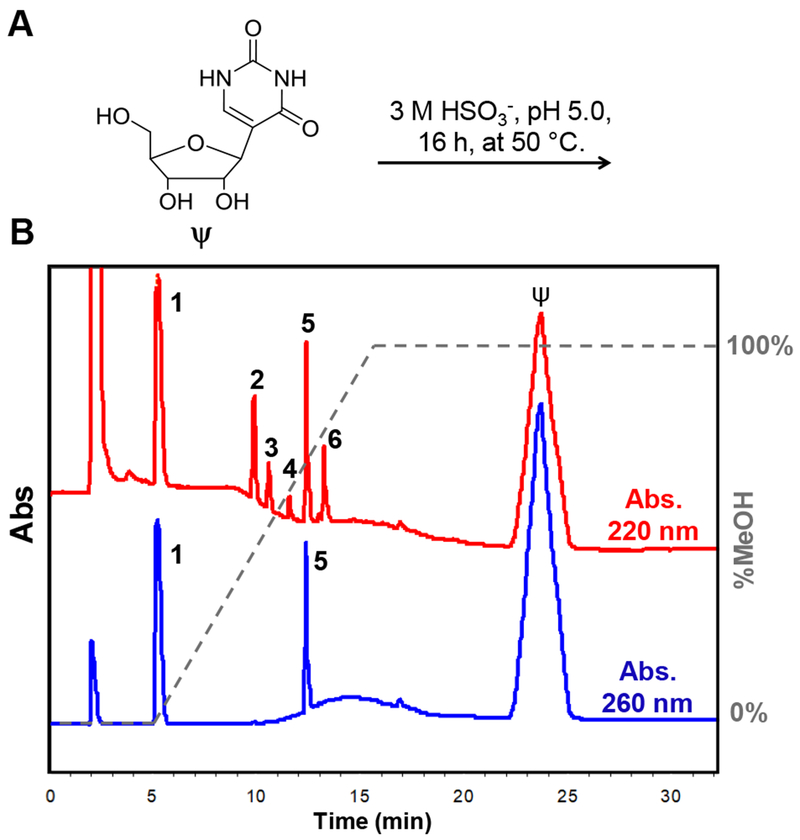
Interrogation of the reaction between Ψ and bisulfite was first studied in the nucleoside context. When Ψ was allowed to react with sodium bisulfite following the previous studies conditions (3 M NaHSO3, pH 5, 16 h, 50 °C; Figure 1A),36 Hypercarb™ HPLC analysis revealed six product peaks when the reaction was monitored at 220 nm (1-6; Figure 1B). When the reaction was analyzed at 260 nm, two peaks remained out of the six (1 and 5; Figure 1B). This initial analysis suggested products 1 and 5 have aromatic components and products 2, 3, 4, and 6 do not. Each peak was isolated and submitted for ESI−-MS analysis to discover that all six products have the same mass, consistent with a mono-bisulfite adduct (Figure S1). These initial data show that six isomeric products are formed when Ψ is allowed to reaction with bisulfite. The four products that are no longer aromatic (2, 3, 4, and 6) will be described in greater detail below because they were found to be intermediates leading to stable end products. Products 1 and 5 could be the diastereomers described by Everett and Shapiro;17 however, these products elute under very different mobile phase compositions (A = 20 mM NH4OAc (pH 7) and B = MeOH, 1 = 0% B and 5 = 65% B; Figure 1B), which is not typical of nucleoside diastereomers in our hands on this HPLC column.37
Figure 1.
The Ψ-bisulfite reaction yields six products. (A) The reaction conditions leading to (B) the HPLC analysis monitored at 220 nm or 260 nm to identify those peaks that are no longer aromatic or retain aromaticity, respectively. The Hypercarb™ HPLC mobile phase composition consisted of A = 20 mM NH4OAc (pH 7) and B = MeOH, in which 1 eluted with 0% B and 5 eluted with 65% B.
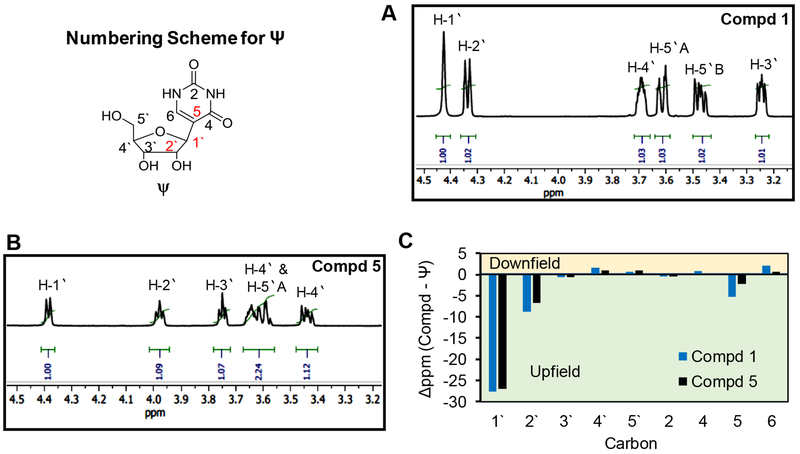
Prior to structural analysis of 1 and 5, purified samples of each were incubated under desulfonation conditions (pH 9, 70 °C, 2 h), as well as at pH 5, 7, or 9 at 70 °C for 24 h and both compounds were recovered without degradation indicating both to be stable bisulfite adducts. First, product 1 was analyzed by NMR for identification of proton and carbon chemical shifts to aid in structure determination. The 1H-NMR assignments described are supported by short range correlation 1H,1H-COSY and long-range correlation 1H,1H-TOCSY NMR analyses that were particularly helpful for assigning the ribose proton resonances (Figures S2–S4). In compound 1, one aromatic proton exists at the C6 carbon (7.8 ppm), and signals for each sugar proton were present. The key peak in the 1H-NMR for 1 was the anomeric proton resonance at 4.4 ppm (Figure 2A); however, it appears as a broad singlet. Comparison of the anomeric proton on 1 with Ψ provided a modest 0.1 ppm upfield shift. All other ribose protons in 1 were present and provided the anticipated splitting patterns. Noteworthy, the 1H-NMR for 1 was nearly identical to one of the spectra reported by Everett and Shapiro.17 The 13C-NMR spectrum for 1 was assigned using the previously determined 1H resonances and the 1H,13C-HSQC spectrum (Figures S5 and S6). In Figure 2C, a plot of the chemical shift changes in 13C signals in 1 relative to Ψ (Δppm = 1 - Ψ) identifies that the 1‵ carbon resonance was impacted the most, followed by the 2‵ carbon resonance of the sugar, and carbon 5 resonance of the heterocyclic ring. The MS and NMR data support that compound 1 is a mono-bisulfite adduct at the 1‵ carbon.
Figure 2.
NMR data for compounds 1 and 5. The 1H-NMR for (A) 1 and (B) 5 showing the sugar resonances. The assignments are interpreted with the assistance of 1H,1H-COSY and 1H,1H-TOCSY spectra. Panel C is a plot of the change in 13C signals for 1 and 5 relative to the parent nucleoside Ψ (Δppm = 1 - Ψ). The assignments were made with the aid of 1H,13C-HSQC spectra. The numbering scheme for Ψ is provided in the upper left-hand corner of the figure and all NMR spectra can be found in Figures S2–S11.
Secondly, product 5 was analyzed by the same NMR techniques as described for compound 1. The 1H-NMR spectrum for 5 showed one aromatic proton at carbon 6 and all ribose protons (Figures 2B and S7–S9). More interestingly, the 1‵ proton on 5 presented as an expected doublet that was modestly shifted upfield by 0.2 ppm relative to Ψ. The 13C-NMR peaks for 5 showed the greatest deviation from the parent nucleoside Ψ at the 1‵, 2‵, and 5 carbons (Figures 2C, S10, and S11). Additionally, the 1H-NMR spectrum for 5 is very similar to the other spectrum reported by Everett and Shapiro.17 The MS and NMR data also support compound 5 is a mono-bisulfite adduct at the 1‵ carbon.
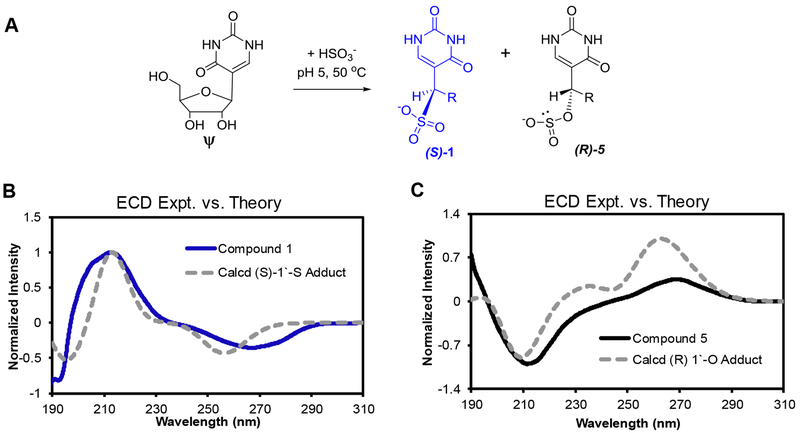
The data to this point might suggest that 1 and 5 are diastereotopic as was suggested by Everett in his dissertation;17 however, the vastly different HPLC retention profiles led us to question this assignment (Figure 1B). Attempts to crystallize the purified products failed to produce diffractable crystals in our hands. Analysis of the IR spectra and fragmentation mass spectrometry (ESI−-MS/MS) spectra for 1 and 5 (Figure S1) confirmed the results obtained from the NMR data, but added no new insights. Thus, Raman spectroscopy was conducted to interrogate the nature of the chemical bonds in 1 and 5. Compound 1 provided a Raman spectrum with a tall peak at 782 cm−1 consistent with a C-S stretch; a similar signal was not present in the spectrum for 5 (Figure S12). Additionally, a distinctive increase is noted in the S-O stretch at 1035 cm−1, which is characteristic of molecules with an R-SO3 configuration,38 and there is an increase in the S=O stretch consistent with the presence of the second S=O bond in compound 1.38 Finally, the strong mode at 980 cm−1 is likely due to S=O symmetric stretching typical of sulfonate compounds,39 and shows a large increase in intensity typical of the increased polarizability in compounds containing opposing S=O bonds. These data provide the basis for our assignments that 1 is a S-adduct of bisulfite at the 1‵ carbon in which the ribose is ring opened, and 5 is an O-adduct of bisulfite to the 1‵ carbon, also with a ring-opened sugar (Figure 3A).
Figure 3.
Stable ring-opened ribose adducts of bisulfite to Ψ are assigned as S (1) and O (5) covalent attachments. (A) Scheme illustrating the reaction of bisulfite with Ψ yielding the constitutional isomer adducts 1 and 5. (B and C) Comparisons of experimental versus theoretical ECD spectra for 1 and 5 to determine the stereochemistry about the 1‵ carbon in each adduct. The theoretical spectra were calculated using TD-DFT calculations implemented in the Gaussian09 program with the M06-2X functional with the 6-311++G(2d,2p) basis set while implicitly modeling water with the polarizable continuum model. The calculation for the (S) isomer of the S-adduct was red shifted by 14 nm and the (R) isomer of the O-adduct was red shifted by 22 nm to match the experimental spectra.
Confirmation of the connectivity assignments for 1 and 5 was then supported by a derivatization experiment. The O-adduct should be susceptible to oxidation of the bisulfite group to sulfate because of the lone pair on sulfur; such an oxidation would then create a good leaving group allowing reclosure of the ribose, yielding Ψ or a pyranose isomer depending on whether the 4‵ or 5‵ OH attacks (Figure 4A). In contrast, the S-adduct should not be prone to oxidation and should remain underivatized following the oxidation reaction (Figure 4A). The mild oxidant I2 was selected for this test because it readily oxidizes bisulfite but does not damage the heterocyclic ring complicating the secondary reaction. Treatment of 1 with I2 did not lead to formation of Ψ, while 5 did react with I2 to yield Ψ on the basis of HPLC analysis (Figure 4B). These derivatization reactions add further support for 1 as the S-adduct and 5 as an O-adduct.
Figure 4.
Oxidative derivatization supports 1 as an S-adduct and 5 as an O-adduct. (A) Scheme O vs. S bisulfite oxidation products. (B) Hypercarb™ HPLC analysis showing 5 is susceptible to oxidative derivatization.
We found it surprising that 1 and 5 eluted as single peaks from the Hypercarb™ HPLC column, which typically resolves nucleoside diastereomers,37 suggesting 1 and 5 are single stereoisomers about the 1‵ carbon. This claim is also supported by the NMR data, in which single peaks for each resonance were observed. Determination of the absolute configurations at the 1‵ carbons of 1 and 5 was achieved via their electronic circular dichroism (ECD) spectra and comparison of the experimental and theoretical spectra for each possible isomer of bisulfite adduction at the 1‵ carbon. The theoretical spectra were calculated using time-dependent density function theory (TD-DFT) calculations as implemented in the Gaussian09 program with the M02-6X functional and 6-311++G(2d,2p) basis set while implicitly defining the water solvent with the polarizable continuum model (PCM). The input structures for the TD-DFT calculations were geometrically optimized using the B3LYP functional with the same basis set as described for the TD-DFT calculations. We have experience applying this approach for stereochemical assignments of modified nucleosides.37 The best experimental versus theoretical overlap with 1 was the (S) isomer of the S-adduct (Figures 3B and S13), and the best overlap with 5 was the (R) isomer of the O-adduct (Figures 3C and S14). Beyond providing experimental evidence for the stereochemistry about the 1‵ carbon for 1 and 5, these calculations add another level of support for 1 being the S-adduct of bisulfite and 5 being the O-adduct of bisulfite to Ψ (Figure 3A). A final noteworthy point is that the ECD spectra for 1 and 5 are identical in the direction of light rotation to those reported by Everett and Shapiro for these two compounds using optical rotary dispersion.17 Comparison of the spectra identified a few subtle differences in the peak shapes that can be explained by different experimental approaches and instruments.
Products 2, 3, 4, and 6 that were only detectable at 220 nm have masses consistent with mono-bisulfite adducts as stated above. Experiments to determine their stabilities were then conducted. Each intermediate compound was HPLC purified and then subjected to desulfonation conditions (pH 9, 70 °C, 2 h) followed by HPLC reanalysis to identify the decomposition products. Compounds 2 and 6 eliminate bisulfite and revert back to Ψ in nearly quantitative yields, while 3 and 4 rearrange to compound 5 also in nearly quantitative yields (>95%; Figure S15). Mild incubation (pH 7, 22 °C, 24 h) of the compounds (2, 3, 4, and 6) also found decomposition or rearrangement to the same products in yields >75%. In the RBS-Seq report, a role for Mg2+ to drive the reaction to products that yield a deletion signature during cDNA synthesis was suggested. Thus, the intermediate degradation process was monitored in the nucleoside context with Mg2+ following the RBS-Seq protocol in which a Mg2+ dependency for intermediate degradation was not observed (Figure S15). The difference in Mg2+ dependency likely results from the difference in context that exists between the nucleoside studies reported here and the RNA studies in the previous work. These findings identify compounds 1 and 5 as the final products of the reaction.
Attempts to characterize compounds 2, 3, 4, and 6 by 1H- and 13C-NMR were pursued; however, as a result of the instability of these compounds after purification only 2 was successfully analyzed to yield spectra that were a composite of 2 and its degradation product Ψ (Figures S16 and S17). The known NMR peaks for Ψ40 enabled us to deconvolute the spectra to identify those peaks occurring from 2. The key diagnostic feature found for 2 was in the 13C-NMR spectra in which the aromatic carbons C5 and C6 were no longer present and observed at frequencies associated with sp3 hybridized carbon atoms (73.6 and 66.6 ppm; Figure S17). This supports the base adduct of bisulfite in 2. The lack of further characterization for 3, 4, and 6 limits our ability to assign their structures; however, on the basis of known chemistry of bisulfite with pyrimidines, 2,41 the rapid of rate of desulfonation of these products upon heating at pH 7 or greater (Figure S15), we propose these represent base adducts of bisulfite to Ψ. Consistent with the established mechanism,2,41 there should be four stereoisomers of bisulfite adducted to the heterocylic ring of Ψ that was observed with compounds 2, 3, 4, and 6. In conclusion, these results support bisulfite adduct formation at C6 of the heterocyclic base for all intermediates (Figures 1B, S1, and S16–S17).
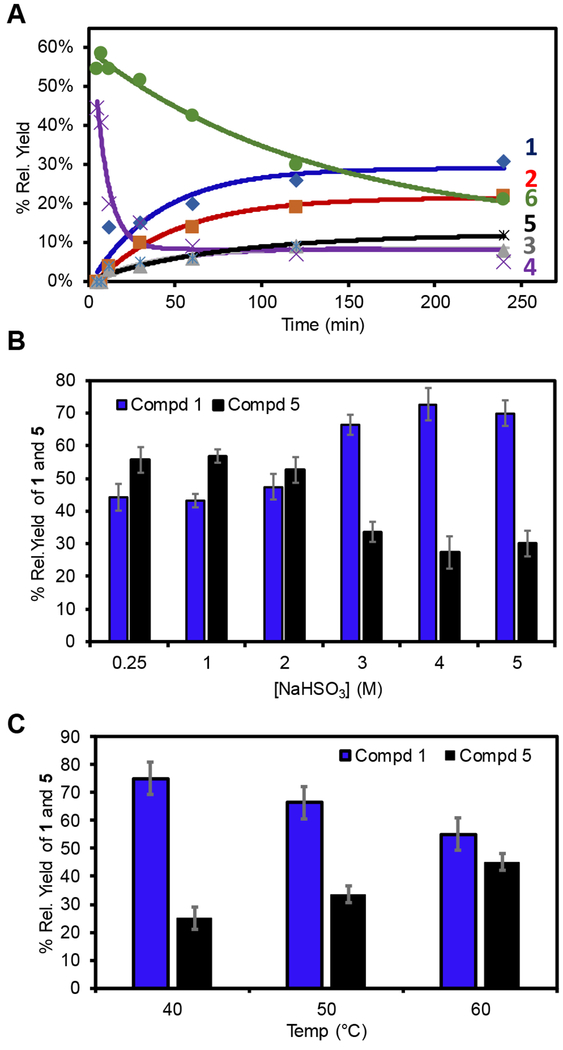
Understanding the structure of the products formed when Ψ was allowed to react with bisulfite, and how these products decompose and rearrange, led us to probe the reaction in more detail. First, a time-course analysis in product evolution from 0-240 min was conducted at pH 5 with 3 M NaHSO3 at 50 °C. Product formation was monitored by HPLC at 220 nm, in which it was assumed that all products have similar extinction coefficients; thus, the peak areas were directly compared. The time profile found intermediates 4 and 6 were initially formed in highest relative yield, and as the reaction progressed past 150 min, compound 1 was the major product observed and compound 5 was detected at approximately one-half the relative concentration of 1 (Figure 5A). The 2:1 relative yield for 1 and 5 was maintained even after a 16 h reaction. Next, the [NaHSO3] versus yields of 1 and 5 were monitored to find the following trend (Figure 5B). When the concentration of bisulfite was increased from 0.25 to 2 M the yields of 1 and 5 were nearly 1:1. In contrast, reactions conducted above 2 M bisulfite led to the approximate 2:1 yield of 1 to 5. Additionally, the formation of compound 5, the O-adduct, showed a temperature dependence with markedly higher yields at higher temperatures compared to compound 1 (Figure 5C). These observations provide useful insights as described in the discussion section below.
Figure 5.
(A) Time-dependent product yields for all products, (B) [NaHSO3]-dependent, (C) temperature-dependent yields for 1 (blue) and 5 (black). The yields were determined by Hypercarb™ HPLC analysis following the reaction at 220 nm, in which it was assumed that products 1-6 have the same extinction coefficients.
The ultimate goal of these studies was to understand how the reaction between Ψ and bisulfite in an RNA strand leads to a deletion signature during cDNA synthesis in RBS-Seq.36 A 24-mer RNA template strand based on one of the control sequences studied in RBS-Seq36 was prepared by solid-phase synthesis to contain Ψ at a known location for study by reverse transcription post bisulfite treatment. The RNA oligomer was treated with bisulfite under the established conditions at pH 5, after which the sample was subjected to desulfonation by incubating the sample at pH 9 for 2 h at 37 °C. The treated RNA was analyzed by anion-exchange HPLC separation to determine the conversion to product following the two-step reaction. The HPLC analysis found a new peak in >90% yield that was purified and verified by ESI−-MS to be a mono-bisulfite-adducted RNA strand (Figure S18). Studies were not pursued to determine the ratio of products 1 and 5 in the RNA template. Lastly, the RBS-Seq report highlighted an importance for Mg2+ in generation of the generation signature that appeared to occur during library preparation; however, the study did not decouple the absolute requirement of this divalent ion for polymerization and how Mg2+ can impact RNA secondary structures necessary for the deletion signature. In the present studies, Mg2+ was only added during reverse transcription because preparation of a sequencing library was not required for the present analysis.
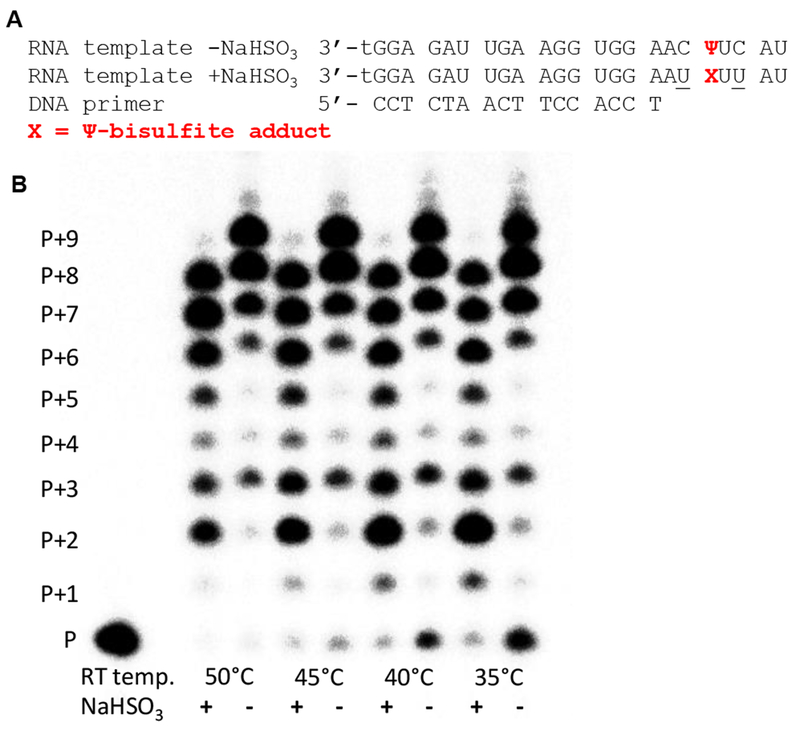
Next, running-start reverse transcription of the template RNA before and after bisulfite treatment was conducted using SuperScript™ III as the polymerase that we note is a small change from the SuperScript™ II that was used during RBS-Seq.36 On the basis of the manufacturer’s notes on the two SuperScript™ products, the key difference is an increase in thermal stability for III vs. II and for increased cDNA yields from structured RNA templates. The native RNA template was reverse transcribed to produce a full-length cDNA on the basis of denaturing polyacrylamide gel electrophoresis (PAGE); in contrast, the HPLC-purified bisulfite-adducted RNA produced a cDNA one nucleotide shorter (Figure 6). Note that SuperScript™ III often yields n+1 and n+2 oligomers due to a propensity for blunt-end addition of additional ATPs;42 thus, the bisulfite (−) lanes in Figure 6 show full length (P+7), n+1 (P+8), and n+2 (P+9) bands while the bisulfite (+) lanes reproduce this pattern exactly but at lengths one nucleotide shorter (P+6, P+7, and P+8). An additional feature observed in the bisulfite (+) lanes is significant pausing before the adducted Ψ (P+2) that was not observed in the bisulfite (−) lanes. This final observation confirms reverse transcription past the Ψ adduct results in a deletion mutation consistent with published RBS-Seq findings.36 A study of the temperature dependency of reverse transcription from 35-50 °C found the deletion mutation occurs in high yield with only modest impact of the temperature at which primer extension was conducted.
Figure 6.
Primer extension assay with an RNA template containing Ψ at a known location before and after bisulfite treatment. (A) The RNA template and DNA primer studied (t = thymidine; the 3` t increased RNA synthesis yields). (B) Reverse transcription followed by PAGE analysis at different temperatures (P = unextended primer, and P + number = number of dNTPs incorporated).
Discussion
Recently, we assisted the Cairns laboratory to discover RBS-Seq, a bisulfite-based reaction on RNA that permits sequencing of three RNA modifications simultaneously, including m1A, m5C, and Ψ.36 The novel observation was that bisulfite treatment of Ψ led to a clear deletion mutation during cDNA synthesis that provided a sequencing signature to locate this epitranscriptomic modification. Pseudouridine has long been known to play a role in tRNA and rRNA tertiary structure,24,43 and its importance is underscored by the fact that 13 pseudouridine synthetases have been found in humans, four of which are established to operate on mRNA.44 The recent findings that levels of Ψ vary with cellular stress and that high levels of Ψ are formed in viral RNA have reinvigorated interest in this RNA modification.34,35,45,46
Sequencing for modifications is best accomplished when two criteria are met: (1) the intact nucleic acid can be treated with a reagent that reacts quantitatively with a single type of base to convert it to a different base that can be amplified in an unbiased manner for high-throughput sequencing. C vs. m5C represents such a case in which the reaction of the bisulfite nucleophile is at least 50-fold greater with C compared to m5C.4 (2) Ideally, the reagent should react with the modified base, present in lower abundance, rather than the unmodified base. In this way, a positive signal is observed rather than seeking the absence of a reaction, which could be due to any number of factors. C vs.m5C does not meet this criterion, but the reaction remains useful, particularly for DNA where the strong sequence bias for 5’-CpG sites serves as a double-check of the presence of m5C. In contrast, the RBS-Seq method positively identifies Ψ compared to U, because only Ψ provides the deletion signature, and it does so in high yields.36
The goal of the present study was to understand the bisulfite chemistry with Ψ that results in the deletion signature. We initially addressed this by subjecting the Ψ nucleoside to the established bisulfite treatment and identified six new products that all had masses consistent with mono-bisulfite adducts (Figures 1 and S1). Products 1 and 5 retain aromaticity of the heterocycle giving a strong 260-nm absorbance, while 2, 3, 4, and 6 were only detectable at 220 nm. Stability studies found 1 and 5 to be the stable end products of the reaction that were formed in a 2:1 ratio, respectively, under the established RBS-Seq protocol (Figure S15).36
The structures for 1 and 5 were proposed on the basis of complementary 1D- and 2D-NMR analysis to identify both products were bisulfite adducts at the 1‵ carbon (Figures 2 and S2–S11). Raman spectroscopy of 1 and 5 provided a key peak for compound 1 at 782 cm−1 that supports an S-adduct of bisulfite at the 1‵ carbon resulting in a ring-opened sugar. A similar peak was not observed in the Raman spectrum for 5 leading us to propose an O-adduct of bisulfite at the 1‵ carbon also resulting in a ring-opened ribose (Figure S12). The O- vs. S-adducts were further confirmed by a derivatization reaction (i.e., mild oxidation to the sulfate analog) that yields a positive result for an O-adduct and a negative result for an S-adduct (Figure 4).
A surprising finding was the observation of a single stereoisomer of each adduct type at the 1‵ carbon, in which the configurations were assigned by ECD interpreted via TD-DFT calculations (Figure 3B). Thus, 1 is the (S) isomer of the S-adduct between bisulfite and Ψ with a ring-opened ribose and 5 is the (R) isomer of the O-adduct between bisulfite and Ψ with a ring-opened ribose (Figure 3A); both adducts are to the ribose C1’, on the basis of 13C NMR assignments (Figure 2C). The unusual lack of splitting to the C1’-H of adduct 1 must be attributable to a rare, but possible, preferred conformation at which the Karplus curve predicts a coupling constant of 0 Hz (Figure 2A). The proposed structures also provide an explanation for the different HPLC elution profiles for 1 and 5 that led us to dig deeper in the first place and to identify the different covalent attachments leading to 1 and 5. Compound 1 is a sulfonic acid derivative expected to have a low pKa value (< 0), while compound 5 is a sulfinic acid derivative with a much higher pKa (2-7), and these vastly different acidities likely lead to the different HPLC retention times observed.
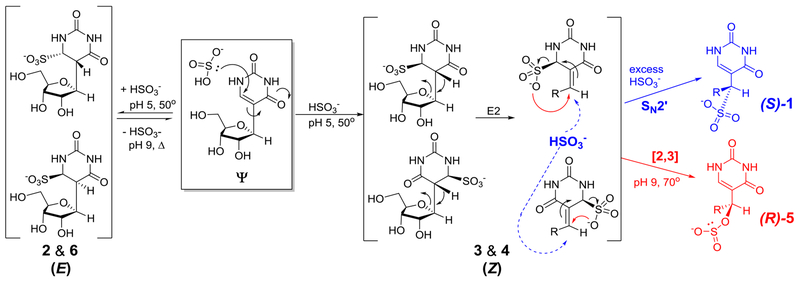
On the basis of the structures and the [NaHSO3] dependency in the yields of 1 and 5, a mechanism is proposed for their formation that addresses the final stereochemistry at the 1‵ carbon of each adduct (Figures 3A and 5B, Scheme 2). Conjugate addition of the nucleophilic sulfur of bisulfite to C6 of Ψ at pH 5 generates four diastereomeric base adducts, represented by peaks 2, 3, 4 and 6 in the HPLC chromatogram (Figure 1B); two of the isomers are E and two are Z. Isomers 2 and 6 must have a trans orientation of C6-sulfite and C5-H, because heating at pH 9 leads to elimination of HSO3− to return to starting material Ψ, mostly likely through an E2 mechanism. Overall, this chemistry is essentially identical to the bisulfite sequencing reactions of C vs. m5C for which pH-reversible conjugate addition to C6 is sterically encumbered for m5C, and therefore much slower. Hayatsu has previously found anti elimination for bisulfite adducted T to proceed efficiently, while syn elimination from T required increased heat and alkaline conditions.41
Scheme 2.
Proposed mechanism of formation of (S)-1 and (R)-5.
The outcome of isomers 3 and 4 is more complex; we propose they have a cis relationship of the C6-SO3− and C5-H groups, and therefore the standard antiperiplanar geometry for an E2 elimination of bisulfite (H + SO3−) cannot be attained. Instead each of these isomers can adopt a conformation in which the ribose O4’ is antiperiplanar to the acidic hydrogen at C6. Thus, an E2 elimination leads to ribose ring opening of defined stereochemistry (Scheme 2; note, individual assignments of 2 vs. 6 and 3 vs. 4 could not be made). Peaks 3 and 4 are the smallest components of the HPLC chromatogram because these products readily undergo further chemistry, even at pH 5, to produce 1 and 5. We are uncertain at this stage whether peaks 3 and 4 are the initial bisulfite adducts (two E isomers) or their ring-opened isomers. Support for the ring-opened structures is derived from the UV-vis spectra for 3 and 4 that have greater conjugation than the ring-closed structures 2 and 6 leading to a small but significant absorption at 260 nm (Figure S19). In any case, both 3 and 4 can lead to both (S)-1 and to (R)-5 in the following ways: (1) high concentrations of bisulfite increase the yield of 1 (Figure 5B) that can be formed from either of the ring-opened 3 or 4 structures by an SN2’ reaction with anti stereochemistry (Scheme 2, blue). The stereochemistry of SN2’ reactions was debated decades ago;47,48 apparently the reaction surface is sufficiently plastic that the nature of the nucleophile, the leaving group and the planarity of the intermediate carbon play roles in determining the syn/anti preference. Here we propose an anti orientation of the attacking HSO3− and the leaving HSO3− because these groups are very bulky, hence disfavoring the syn stereochemistry often observed for SN2’ reactions. (2) Higher temperatures favor the conversion of both 3 and 4 to compound 5 (Figure 5C), and both lead to the (R) isomer (Scheme 2, red). This reaction must be a [2,3] sigmatropic shift because of the carbon-oxygen bond formation at C1’. Only intramolecular attack of an oxygen could create the C-O bond; any bimolecular nucleophilic attack would create a C-S bond due to the higher nucleophilicity of sulfur. Finally, we point to the fact that U does not readily undergo reactions parallel to this, i.e., to ring-opened adducts, because bisulfite adducts to the 5,6-double bond of U do not place a hydrogen at an appropriate site for E2 ring-opening akin to 3 and 4.
Several features of the mechanistic scheme are noteworthy. First, after the stereorandom conjugate addition of bisulfite to C6 of Ψ producing four diastereomeric adducts, all subsequent reactions—E2, SN2’ and the [2,3]-sigmatropic shifts—are stereospecific. The enantiopure outcomes of (S)-1 and to (R)-5 are directed by the absolute configuration at C-1’ in Ψ. Secondly, while bisulfite addition/elimination reactions have been well studied with cytidine, the SN2’ reaction in Scheme 2 is unprecedented. The driving force for this reaction in the forward direction must be the rearomatization of the uracil ring. Thirdly, we are unaware of any reports of [2,3] sigmatropic shifts involving a sulfonate.
The final step of our studies was to treat a synthetic RNA strand containing a single Ψ at a known site with bisulfite followed by desulfonation to generate the adducts 1 and 5. The HPLC-purified, adducted strands were used as templates for reverse transcription to discover a deletion signature when the adduct was present (Figure 6). A prior study by the Grollman laboratory found that when a DNA polymerase traverses a ring-opened sugar in a DNA template strand, the damaged site is extruded, and polymerization continues yielding a deletion mutation.49 Apparently, the ribose ring-opened adducts of bisulfite to Ψ (1 and 5) in an RNA template also result in extrusion during reverse transcription leading to a deletion signature, on the basis of the present findings. The deletion signature yield observed during reverse transcription was nearly quantitative, while the deletion signature found during DNA polymerization was modest suggesting that both DNA vs. RNA context and the nature of the polymerase have an impact on the yield. The critical details differentiating the deletion yield between the two studies is not known at present; although, reverse transcriptases bypass modified nucleotides with higher efficiency albeit with lower fidelity.50 A final study to ensure that the ribose adducts did not simply change the acid-base properties or hydrogen bonding potential of Ψ to cause the deletion signature was ruled out via an acid-base titration study of 1 and 5. The titrations were followed by UV-vis to find that the pKa values for the heterocyclic rings of 1 and 5 were within error of the measured pKa for Ψ at 9.5 (Figure S20). This final study aids in the claim that opening the furanose ring of Ψ via bisulfite adduction leads to the deletion signature during cDNA synthesis.
These deletions are conveniently identified during next-generation sequencing during the RBS-Seq procedure for identification of sites in the transcriptome in which a U was isomerized to Ψ.36 The key advantage provided by read–through of the adducted Ψ is that more than one modification per template strand can be mapped. This approach thus demonstrates a significant advantage over other methods for locating Ψ by, for example, introduction of polymerase stops using CMC prior to cDNA synthesis.31–35 Identification of RNA modifications by a stop in cDNA synthesis is notorious for yielding false positives from reverse transcription stops resulting from other features of the RNA. Additionally, only one Ψ per strand can be detected by the CMC approach.
Conclusions
The discovery that the epitranscriptomic modifications m1A, m5C, and Ψ can be mapped by bisulfite treatment via RBS-Seq provides a significant advance in sequencing for these critical modifications.36 Renewed interest in Ψ, due to better experimental approaches to its detection, is leading to new insights into the role of this modification as a modulator of RNA function and as a response to cellular stress.34,35,45,46 During RBS-Seq, Ψ is located via a deletion signature that occurs after bisulfite treatment. Herein, we applied the bisulfite reaction to Ψ, first in the nucleoside context to monitor the reaction in which six isomeric mono-bisulfite products were formed. Two of these products, 1 and 5, are stable end products with bisulfite adducted at the 1‵ carbon, resulting in opening of the sugar. Prior work reported in a Ph.D. dissertation from the Shapiro laboratory studied the bisulfite addition to Ψ and proposed a pair of diastereomers being formed with S-adduction at the 1‵ carbon;17 however, puzzling data, such as the unsplit C1’ proton of compound 1 must have discouraged them from publication of the results. Using modern spectroscopic techniques coupled with classical product derivatization, we found that the two products formed are constitutional isomers in which 1 is the (S) isomer of the S-adduct of bisulfite at the 1‵ carbon and 5 is the (R) isomer of the O-adduct at the same carbon (Figure 3A). These products are highly stable and formed in a ratio of 2:1 for 1 and 5, respectively. Several noteworthy aspects of the reaction mechanism include the series of stereoselective reactions—E2, SN2’ and a [2,3]-sigmatropic shift—with the latter two being unprecedented in the bisulfite literature (Scheme 2).
In a second part of the study, we treated a synthetic RNA template with bisulfite to generate 1 and 5 at a known location to then find that a deletion occurs after reverse transcription. This final observation in tandem with all the other studies identifies the source of the deletion signature at Ψ that occurs when RBS-Seq is applied to the transcriptome.36 The ability to locate the epitransciptomic modification Ψ by a deletion signature during cDNA synthesis permits location of more than one modification per template strand that is a significant advance over prior methods.31–35 The initial observation of bisulfite treatment of dC/C to effect deamination to dU/U, while the same reaction of 5mC/m5C proceeds with 50x slower kinetics has been harnessed by researchers to sequence these epigenomic/epitranscriptomic modifications via high-throughput sequencing. Application of this technique has shed light on many aspects of chemical modification of DNA and RNA to impact cellular phenotype. This classical reaction, once harnessed to facilitate recrystallization of aldehydes and ketones, has revolutionized our understanding of biology and now deserves reintroduction to students of organic chemistry.
Methods
Nucleoside reaction and product characterization.
The Ψ nucleoside is commercially available, as well as all other reagents that were used without further purification. The Ψ nucleoside reactions with bisulfite were conducted following the RBS-Seq protocol.36 In a reaction volume of 1 mL was added 1 mM Ψ and 3 M freshly prepared NaHSO3 at pH 5.0. The reaction mixture was placed in a 50 °C heat block to react for 16 h, after which the solution was removed from the heat and directly analyzed by HPLC using a Hypercarb™ column running A = 20 mM NH4OAc (pH 7) and B = MeOH. The HPLC method consisted of a flow rate of 1 mL/min holding an isocratic gradient of 0% B for the first 5 min followed by a linear increase in B to 100% over 10 min that was held for 15 min. The HPLC analysis indicated six product peaks of which 1 and 5 were the stable end products and were fully characterized and the others were unstable intermediates characterized by mass spectrometry. 1. tR = 5 min. HRMS (ESI−-TOF) calcd for C9H13N2O9S [M-H]− = 325.0342; found 325.0349. ESI−-TOF-MS/MS parent = 325.0349, daughters = 204.9920, 183.0407, 161.9861, 153.0299, 141.0299, 140.0345, 123.0196, 110.0243, 98.0244, 80.9651, and 79.9573. 1H-NMR (500 MHz, D2O) δ 7.76 (s, 1 H), 4.40 (s, J = 0 Hz, 1 H), 4.31 (dd, J = 8.32 Hz, 1 H), 3.67 (m, J = 2.45 Hz, 1 H), 3.61 (d, J = 2.17 Hz, 1 H), 3.45 (d, J = 6.85 Hz, 1 H), 3.22 (m, J = 4.89 Hz, 1 H). 13C-NMR (125 MHz, D2O) δ 168.6, 155.0, 146.1, 107.7, 75.1, 74.9, 73.0, 64.5, 58.1. UV-vis in ddH2O λmax = 265 nm (ε = 6,000 L•mol−1•cm−1); ECD in ddH2O λ nm (c = 2 × 10−4 M in ddH2O) 266 (Δε = +14.4) and 212 (Δε = −40.3). Raman shift cm−1 (ddH2O) 440, 550, 650, 775, 900, 950, 1050, 1400, 1625, 2900. 5. tR = 12.5 min. HRMS (ESI−-TOF) calcd for C9H14N2O9S [M - H]− = 325.0342; found 325.0341. ESI−-TOF-MS/MS parent = 325.0341, daughters = 204.9917, 183.0396, 161.9855, 153.0298, 141.0296, 140.0344, 123.0192, 110.0240, 98.0242, and 80.9652. Intermediates. 1H-NMR (500 MHz, D2O) δ 7.61 (s, 1 H), 4.39 (d, J = 6.85 Hz, 1 H), 3.98 (dd, J = 6.85 Hz, 1 H), 3.75 (dd, J = 5.87 Hz, 1 H), 3.65 (m, J = 3.41 Hz, 1 H), 3.58 (d, J = 3.22 Hz, 1 H), 3.44 (d, J = 6.85 Hz, 1 H). 13C-NMR (125 MHz, D2O) δ 184.0, 167.9, 155.0, 144.7, 110.8, 75.0, 74.2, 64.9, 58.8. UV-vis in ddH2O λmax = 266 nm (ε = 6,200 L•mol−1•cm−1); ECD in ddH2O λ nm (c = 1 × 10−4 M in ddH2O) 268 (Δε = −8) and 211 (Δε = +22). Raman shift cm−1 (ddH2O) 550, 775, 900, 950, 1050, 1625, 2900. 2. tR = 10 min (ESI−-MS) calcd [M-H]− = 325.27; found 325.00. 1H-NMR (500 MHz, D2O) δ 4.43 (s, 1 H), 3.97 (m, J = 4.89 Hz, 1 H), 3.89 (m, J = 5.87 Hz, 1 H), 3.79 (m, J = 4.89 Hz, 1 H), 3.46 (dd, J = 5.38 Hz, 1 H). 13C-NMR (125 MHz, D2O) δ 173.1, 143.9, 86.7, 82.6, 75.0, 73.6, 73.2, 66.6, 64.1. 3. tR = 10.5 min (ESI−-MS) calcd [M-H]− = 325.27; found 325.07. 4. tR = 11.5 min (ESI−-MS) calcd [M-H]− = 325.27; found 325.07; 6. tR = 13 min (ESI−-MS) calcd [M-H]− = 325.27; found 325.18.
Density functional theory calculations.
All calculations were performed using DFT methods as implemented in the Gaussian09 package, and visualized using GaussView (v5).52 The 6-311++G(2d,2p) basis set,53,54 B3LYP55,56 or M06-2X functional,57 and the polarizable continuum model (PCM)58,59 for implicit definition of water were used for the calculations. The computed ECD spectra were completed using TD-DFT theory to calculate 25 excitations.60–62 Complete details for plotting the theoretical spectra has previously been reported by our laboratory.37
Polymerase bypass assay.
The RNA template and DNA primer were synthesized by solid-phase synthesis following established protocols using commercially available phosphoramidites (RNA = 5‵-UA CUΨ CAA GGU GGA AGU UAG AGG t-3‵; and DNA = 5‵-CCT CTA ACT TCC ACC T-3‵). The RNA strand was synthesized with a 3‵ thymidine (t) to increase the synthesis yield, and it does not impact the reverse transcription reaction. The Ψ-containing RNA strand was subjected to the bisulfite reaction as described for the nucleoside, with one change. After the 16 h reaction, the NaHSO3 was removed via a NAP-5 column, after which the RNA was desulfonated by heating at 37 °C in 1 M Tris buffer (pH 9) for 2 h. Upon desulfonation, the RNA was immediately purified by anion-exchange HPLC using the mobile phases A = 1:9 MeCN:ddH2O and B = 1.5 M NaOAc (pH 7) in 1:9 MeCN:ddH2O. The HPLC method was initiated at 25% B and increased linearly to 100% B over 30 min with a 1 mL/min flow rate while monitoring the elution via absorbance at 260 nm. The HPLC-purified adducted RNA strands were characterized by ESI−-MS: calcd mass = 7854.7; found 7854.1. In brief, the reverse transcription experiments were conducted by first placing a 5‵ 32P on the end of the DNA for visualization of the polymerization reaction by denaturing PAGE. Next, a reaction with 10-μL volume containing 100 nM pre-annealed RNA template and DNA primer duplex, 3 mM MgCl2, 10 mM DTT, and 500 μM of each dNTP was prepared. The extension reaction was thermally equilibrated at the desired reaction temperature for 15 min prior to addition of 10 U of SuperScript™ III. The reaction was allowed to progress for 40 min followed by heat denaturation to quench the reaction prior to analysis by PAGE. Complete experimental details are provided in the Supporting Information.
Supplementary Material
Acknowledgments
Financial support of this research was provided by the NIH via grant no. R01 GM093099 to CJB and the NSF via grant no. CHE 1904424 to JMH. The DNA and RNA strands were synthesized by the University of Utah Health Sciences Core facilities that are supported in part by a National Cancer Center Support grant (P30 CA042014). The support and resources from the Center for High Performance Computing at the University of Utah are gratefully acknowledged. We are grateful to Prof. Jon Rainier (Univ. of Utah) for helpful discussions. The work presented herein was inspired by the astute observations of Dr. Vahid Khoddami and Prof. Bradley Cairns at the University of Utah while developing RBS-Seq.
Footnotes
Supporting Information
Supporting Information is available.
Complete experimental details, MS, NMR, HPLC, IR, Raman spectra, UV-vis, and DFT optimized geometries and energies.
Conflict of Interest
The authors are not in conflict of interest in this work.
References
- (1).Morrison RT; Boyd RN: Organic Chemistry; Allyn and Bacon, Inc.: Boston, MA, USA, 1973; Vol. 3rd. [Google Scholar]
- (2).Shapiro R; Servis RE; Welcher M Reactions of uracil and cytosine derivatives with sodium bisulfite. J. Am. Chem. Soc . 1970, 92, 422–424. [Google Scholar]
- (3).Hayatsu H; Wataya Y; Kai K; Iida S Reaction of sodium bisulfite with uracil, cytosine, and their derivatives. Biochemistry 1970, 9, 2858–2865. [DOI] [PubMed] [Google Scholar]
- (4).Wang RY; Gehrke CW; Ehrlich M Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Res . 1980, 8, 4777–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Frommer M; McDonald LE; Millar DS; Collis CM; Watt F; Grigg GW; Molloy PL; Paul CL A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U.S.A . 1992, 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Booth MJ; Raiber E-A; Balasubramanian S Chemical methods for decoding cytosine modifications in DNA. Chem. Rev . 2015, 115, 2240–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Song CX; Yi C; He C Mapping recently identified nucleotide variants in the genome and transcriptome. Nat. Biotechnol . 2012, 30, 1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Booth MJ; Branco MR; Ficz G; Oxley D; Krueger F; Reik W; Balasubramanian S Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 2012, 336, 934–937. [DOI] [PubMed] [Google Scholar]
- (9).Yu M; Hon GC; Szulwach KE; Song CX; Zhang L; Kim A; Li X; Dai Q; Shen Y; Park B; Min JH; Jin P; Ren B; He C Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 2012, 149, 1368–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Song CX; Szulwach KE; Dai Q; Fu Y; Mao SQ; Lin L; Street C; Li Y; Poidevin M; Wu H; Gao J; Liu P; Li L; Xu GL; Jin P; He C Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 2013, 153, 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Booth MJ; Marsico G; Bachman M; Beraldi D; Balasubramanian S Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution. Nat. Chem . 2014, 6, 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lu X; Song CX; Szulwach K; Wang Z; Weidenbacher P; Jin P; He C Chemical modification-assisted bisulfite sequencing (CAB-Seq) for 5-carboxylcytosine detection in DNA. J. Am. Chem. Soc . 2013, 135, 9315–9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wang Y; Zhang X; Zou G; Peng S; Liu C; Zhou X Detection and application of 5-formylcytosine and 5-formyluracil in DNA. Acc. Chem. Res . 2019, 52, 1016–1024. [DOI] [PubMed] [Google Scholar]
- (14).Dietzsch J; Feineis D; Hobartner C Chemoselective labeling and site-specific mapping of 5-formylcytosine as a cellular nucleic acid modification. FEBS Lett . 2018, 592, 2032–2047. [DOI] [PubMed] [Google Scholar]
- (15).Carell T; Kurz MQ; Muller M; Rossa M; Spada F Non-canonical bases in the genome: The regulatory information layer in DNA. Angew. Chem. Int. Ed. Engl . 2018, 57, 4296–4312. [DOI] [PubMed] [Google Scholar]
- (16).Singhal RP Chemical probe of structure and function of transfer ribonucleic acids. Biochemistry 1974, 13, 2924–2932. [DOI] [PubMed] [Google Scholar]
- (17).Everett DW Part i: reaction of pseudouridine with bisulfite. part ii: reaction of glyoxal with guanine derivatives: a spectrophotometric probe of molecular structure. Ph.D. Dissertation, New York University, 1980. [Google Scholar]
- (18).Nachtergaele S; He C Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet . 2018, 52, 349–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Meyer KD; Jaffrey SR Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol . 2017, 33, 319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Huber SM; Leonardi A; Dedon PC; Begley TJ The versatile roles of the tRNA epitranscriptome during cellular responses to toxic exposures and environmental stress. Toxics 2019, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Martinez NM; Gilbert WV Pre-mRNA modifications and their role in nuclear processing. Quant. Biol . 2018, 6, 210–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Roundtree IA; Evans ME; Pan T; He C Dynamic RNA modifications in gene expression regulation. Cell 2017, 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Song J; Yi C Chemical modifications to RNA: A new layer of gene expression regulation. ACS Chem. Biol . 2017, 12, 316–325. [DOI] [PubMed] [Google Scholar]
- (24).Jiang J; Seo H; Chow CS Post-transcriptional modifications modulate rRNA structure and ligand interactions. Acc. Chem. Res . 2016, 49, 893–901. [DOI] [PubMed] [Google Scholar]
- (25).Fisher AJ; Beal PA Structural basis for eukaryotic mRNA modification. Curr. Opin. Struct. Biol . 2018, 53, 59–68. [DOI] [PubMed] [Google Scholar]
- (26).Harcourt EM; Kietrys AM; Kool ET Chemical and structural effects of base modifications in messenger RNA. Nature 2017, 541, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Grozhik AV; Jaffrey SR Distinguishing RNA modifications from noise in epitranscriptome maps. Nat. Chem. Biol . 2018, 14, 215–225. [DOI] [PubMed] [Google Scholar]
- (28).Morse DP; Bass BL Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry 1997, 36, 8429–8434. [DOI] [PubMed] [Google Scholar]
- (29).Hafner M; Landthaler M; Burger L; Khorshid M; Hausser J; Berninger P; Rothballer A; Ascano M; Jungkamp A-C; Munschauer M; Ulrich A; Wardle GS; Dewell S; Zavolan M; Tuschl T Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Squires JE; Patel HR; Nousch M; Sibbritt T; Humphreys DT; Parker BJ; Suter CM; Preiss T Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res . 2012, 40, 5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Carlile TM; Rojas-Duran MF; Zinshteyn B; Shin H; Bartoli KM; Gilbert WV Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Li X; Zhu P; Ma S; Song J; Bai J; Sun F; Yi C Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol . 2015, 11, 592–597. [DOI] [PubMed] [Google Scholar]
- (33).Lei Z; Yi C A radiolabeling-free, qPCR-based method for locus-specific pseudouridine detection. Angew. Chem. Int. Ed. Engl . 2017, 56, 14878–14882. [DOI] [PubMed] [Google Scholar]
- (34).Schwartz S; Bernstein DA; Mumbach MR; Jovanovic M; Herbst RH; Leon-Ricardo BX; Engreitz JM; Guttman M; Satija R; Lander ES; Fink G; Regev A Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Lovejoy AF; Riordan DP; Brown PO Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PloS One 2014, 9, e110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Khoddami V; Yerra A; Mosbruger TL; Fleming AM; Burrows CJ; Cairns BR Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc. Nat. Acad. Sci. U.S.A . 2019, 116, 6784–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Fleming AM; Orendt AM; He Y; Zhu J; Dukor RK; Burrows CJ Reconciliation of chemical, enzymatic, spectroscopic and computational data to assign the absolute configuration of the DNA base lesion spiroiminodihydantoin. J. Am. Chem. Soc . 2013, 135, 18191–18204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Houlton HG; Tartar HV Raman spectra of sodium alkyl sulfonates and sulfinates. J. Am. Chem. Soc . 1938, 60, 544–548. [Google Scholar]
- (39).Ben Mabrouk K; Kauffmann TH; Aroui H; Fontana MD Raman study of cation effect on sulfate vibration modes in solid state and in aqueous solutions. J. Raman Spectrosc . 2013, 44, 1603–1608. [Google Scholar]
- (40).Wishart DS; Knox C; Guo AC; Eisner R; Young N; Gautam B; Hau DD; Psychogios N; Dong E; Bouatra S; Mandal R; Sinelnikov I; Xia J; Jia L; Cruz JA; Lim E; Sobsey CA; Shrivastava S; Huang P; Liu P; Fang L; Peng J; Fradette R; Cheng D; Tzur D; Clements M; Lewis A; De Souza A; Zuniga A; Dawe M; Xiong Y; Clive D; Greiner R; Nazyrova A; Shaykhutdinov R; Li L; Vogel HJ; Forsythe I HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res . 2009, 37, D603–D610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Hayatsu H Discovery of bisulfite-mediated cytosine conversion to uracil, the key reaction for DNA methylation analysis--a personal account. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci . 2008, 84, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Ohtsubo Y; Nagata Y; Tsuda M Efficient N-tailing of blunt DNA ends by Moloney murine leukemia virus reverse transcriptase. Sci. Rep . 2017, 7, 41769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Motorin Y; Helm M tRNA stabilization by modified nucleotides. Biochemistry 2010, 49, 4934–4944. [DOI] [PubMed] [Google Scholar]
- (44).Li X; Ma S; Yi C Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr. Opin. Chem. Biol . 2016, 33, 108–116. [DOI] [PubMed] [Google Scholar]
- (45).Jaffrey SR An expanding universe of mRNA modifications. Nat. Struct. Mol. Biol . 2014, 21, 945–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).McIntyre W; Netzband R; Bonenfant G; Biegel JM; Miller C; Fuchs G; Henderson E; Arra M; Canki M; Fabris D; Pager CT Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res . 2018, 46, 5776–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Stohrer W-D On the stereochemistry of the SN2` reaction. Angew. Chem. Int. Ed. Engl . 1983, 22, 613–614. [Google Scholar]
- (48).Stork G; Kreft AF Stereochemistry of the SN2’ reaction. “Concerted” allylic displacement in an acyclic system: anti displacement with thiolate anion. J. Am. Chem. Soc . 1977, 99, 3851–3853. [Google Scholar]
- (49).Shibutani S; Takeshita M; Grollman AP Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the “A rule”. J. Biol. Chem . 1997, 272, 13916–13922. [DOI] [PubMed] [Google Scholar]
- (50).Potapov V; Fu X; Dai N; Corrêa IR Jr.; Tanner NA; Ong JL Base modifications affecting RNA polymerase and reverse transcriptase fidelity. Nucleic Acids Res . 2018, 46, 5753–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Frisch MJ; Trucks GW; Schlegel HB; Scuseria GE; Robb MA; Cheeseman JR; Scalmani G; Barone V; Petersson GA; Nakatsuji H; Li X; Caricato M; Marenich AV; Bloino J; Janesko BG; Gomperts R; Mennucci B; Hratchian HP; Ortiz JV; Izmaylov AF; Sonnenberg JL; Williams.Ding F; Lipparini F; Egidi F; Goings J; Peng B; Petrone A; Henderson T; Ranasinghe D; Zakrzewski VG; Gao J; Rega N; Zheng G; Liang W; Hada M; Ehara M; Toyota K; Fukuda R; Hasegawa J; Ishida M; Nakajima T; Honda Y; Kitao O; Nakai H; Vreven T; Throssell K; Montgomery JA Jr.; Peralta JE; Ogliaro F; Bearpark MJ; Heyd JJ; Brothers EN; Kudin KN; Staroverov VN; Keith TA; Kobayashi R; Normand J; Raghavachari K; Rendell AP; Burant JC; Iyengar SS; Tomasi J; Cossi M; Millam JM; Klene M; Adamo C; Cammi R; Ochterski JW; Martin RL; Morokuma K; Farkas O; Foresman JB; Fox DJ: Gaussian 16 Rev. C.01. Wallingford, CT, 2016. [Google Scholar]
- (52).Dennington R; Keith T; Millam J, GaussView, Version 5; Semichem, INc: Shawnedd Mission, KS, 2009. [Google Scholar]
- (53).Krishnan R; Binkley JS; Seeger R; Pople JA Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J. Chem. Phys . 1980, 72, 650–654. [Google Scholar]
- (54).Frisch MJ; Pople JA; Binkley JS Self-consistent molecular orbital methods 25. Supplementary functions for Gaussian basis sets. J. Chem. Phys . 1984, 80, 3265–3269. [Google Scholar]
- (55).Becke AD Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys . 1993, 98, 5648–5652. [Google Scholar]
- (56).Lee C; Yang W; Parr RG Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [DOI] [PubMed] [Google Scholar]
- (57).Zhao Y; Truhlar D The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc . 2008, 120, 215–241. [Google Scholar]
- (58).Scalmani G; Frisch MJ Continuous surface charge polarizable continuum models of solvation. I. General formalism. J. Chem. Phys . 2010, 132, 114110. [DOI] [PubMed] [Google Scholar]
- (59).Tomasi J; Mennucci B; Cammi R Quantum mechanical continuum solvation models. Chem. Rev . 2005, 105, 2999–3093. [DOI] [PubMed] [Google Scholar]
- (60).Autschbach J; Ziegler T; van Gisbergen SJA; Baerends EJ Chiroptical properties from time-dependent density functional theory. I. Circular dichroism spectra of organic molecules. J. Chem. Phys . 2002, 116, 6930–6940. [Google Scholar]
- (61).Bauernschmitt R; Ahlrichs R Treatment of electronic excitations within the adiabatic approximation of time dependent density functional theory. Chem. Phys. Lett . 1996, 256, 454–464. [Google Scholar]
- (62).Stratmann RE; Scuseria GE; Frisch MJ An efficient implementation of time-dependent density-functional theory for the calculation of excitation energies of large molecules. J. Chem. Phys . 1998, 109, 8218–8224. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.