Abstract
In fields ranging from environmental toxicology to drug discovery, it is critical to identify how multiple chemical compounds interact to perturb biological systems. Isolation-based approaches fail to incorporate multi-constituent interactions, such as synergy. We have developed an approach called “Simplify”, which identifies mixture constituents that interact to achieve biological effects. Simplify combines biological and mass spectrometric datasets and uses an ‘activity index’ to predict mixture interactions. Using the plant Salvia miltiorrhiza as a case study, we employed Simplify to identify four individual constituents that contribute to antimicrobial activity, three additives and one synergist. Our study is the first to enable identification of unknown synergists prior to isolating them, demonstrating the ability of the Simplify workflow to predict key contributors to the biological effect of a complex mixture. While utilized for natural products discovery in this study, this approach is expected to prove useful across multiple disciplines that rely on mixture analysis.
Keywords: Synergy, complexity, bioinformatics, metabolomics, natural products, potentiation
Graphical Abstract
Mixture analysis plays a key role in studying biological processes and interactions. A complex mixture under study, whether it be an environmental pollutant such as cigarette smoke, a microbial community collected from the deep ocean, or a botanical extract, is frequently reduced to the contributions of individual constituents.1-4 Increasingly, studies demonstrate that individual constituents behave very differently alone than they do within a complex mixture.1,5-8 For example, environmental exposures occur as complex mixtures which may include additive and synergistic effects5,9-11 while bacterial symbionts produce complex mixtures of chemical cues that can activate, synergize, and inhibit developmental processes in multicellular hosts.12. Similarly, many natural products are employed therapeutically as mixtures; thus, it is of interest to know how such mixtures act in their complex form. Although the presence of combination effects within complex mixtures has been well established, most drug discovery efforts,13 toxicological evaluations,10,14 and environmental regulatory protocols15,16 are geared towards identifying and monitoring single constituents that are responsible for the observed biological effect. This is often achieved through the utilization of bioassay-guided fractionation13 or effect-directed analysis,15 in which biological testing results guide the fractionation procedure. While these approaches have been successfully applied in multiple fields, combination effects are often overlooked with isolation-based workflows.
Recently, effective bioinformatics tools have been developed that can integrate biological and chemical datasets to predict which mixture constituents possess bioactivity.1,16,17 Several reports illustrate the applications of biologically-guided metabolomics studies (“biochemometrics” approaches) to identify putative active compounds from natural product mixtures.1,17-19 Non-targeted metabolomics analyses have also been utilized to identify bacterial biotransformation products of common environmental contaminants that contribute to the genotoxicity of bioremediated soils.16 Until this point, these approaches cannot predict whether mixture components are interacting synergistically, additively, or antagonistically without purifying and testing compounds in isolation. Existing assays capable of disentangling synergy from additivity are time consuming and require large quantities of material.1,3 High throughput synergy screening assays have been developed that utilize less time and material and applied to identify additives and synergists from complex extracts.19,20 However, these assays can only identify the presence of specific combination effects within an extract rather than the specific chemical components contributing to this activity. For example, it has been estimated that the metabolome of a single biological organism may contain more than 1,000 unique detectable metabolites.22 Isolating each of these metabolites would be infeasible, and even if isolation were possible, testing them in combination, even two-by-two at a single concentration, would require 499,500 assays.23 Thus, new tools are needed to enable the efficient identification of constituents that interact to achieve biological activity without relying on isolation and random re-combination.
The goal of this work was to develop a predictive approach to prioritize the isolation of mixture constituents that interact synergistically, additively, or antagonistically. As a case study, we utilized the plant Salvia miltiorrhiza Bunge (Lamiaceae) (Chinese red sage or danshen). Used for over 2000 years in China,24-29 S. miltiorrhiza remains one of today’s most popular traditional medicines.24,27,28 Over 70 constituents have been identified in S. miltiorrhiza, making it a well-characterized model organism. Herein, we demonstrate the effectiveness of Simplify, an approach combining bioactivity studies with metabolomics models towards the identification of synergists and additives that exert combined effects. Importantly, Simplify represents the first approach capable of distinguishing synergists from additives prior to compound isolation. The methodology described herein is relevant beyond the field of natural products and could prove useful to researchers investigating the activity of mixtures in fields ranging from toxicology to pharmacology to drug discovery.
EXPERIMENTAL SECTION
General Experimental Procedures.
UPLC-MS analysis was conducted with a Thermo-Fisher Q-Exactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific, MA, USA) coupled to an Acquity UPLC system (Waters Corporation, Milford, MA, USA) using a reversed-phase column (BEH C18, 1.7 μm, 2.1 × 50 mm, Waters Corporation, Milford, MA, USA). Each sample was analyzed in triplicate at 0.1 mg mL−1 in methanol with a 3μL injection. A binary gradient of water with 0.1% formic acid (A) and acetonitrile with 0.1% formic acid (B) began at 10% B for 0.5 min and increased 100% B from 0.5–8.0 min, then held for 30 s. From 8.5–9.0 min, starting conditions were re-established, and the composition remained at 10% B until 10.0 min. Analysis was conducted in positive and negative ion modes with 150–1500 m/z, scan time 200 ms, capillary temperature 256°C, S-lens RF 50.00, spray voltage 3.50 kV, sheath gas flow 47.50, and auxiliary gas flow 11.25. A data-dependent method that fragmented the top four ions with HCD of 35.0 was employed.
Plant Material and Extraction.
Fresh roots of S. miltiorrhiza Bunge (Lamiaceae) were collected on November 8, 2016 (Batch #CRS1016F1, 41W501 Keslinger Rd, Elburn, IL, USA). Plants were identified by Richard Cech at Strictly Medicinal Seeds, and a voucher specimen grown from the same seed line was collected September 3, 2017 and deposited at the herbarium of the University of North Carolina at Chapel Hill (NCU652634). Fresh roots were dug, washed, chopped, and air dried, yielding 500 g. Roots were ground with a Wiley Mill Standard Model No. 3 (Arthur Thomas Co.), extracted in MeOH at 160 g L−1 for 24 h, and filtered. This process was repeated with the same material every 24 h for 72 h. The final extract was concentrated in vacuo and partitioned with 10% aqueous MeOH and hexane (1:1) for defatting. The MeOH layer was partitioned in EtOAc:MeOH:H2O (4:5:1). The EtOAc layer was washed with a 1% NaCl solution (1:1) to remove hydrosoluble tannins and the EtOAc extract was dried under nitrogen yielding 18.32 g (Figure S1).
Chromatographic Separation and Isolation.
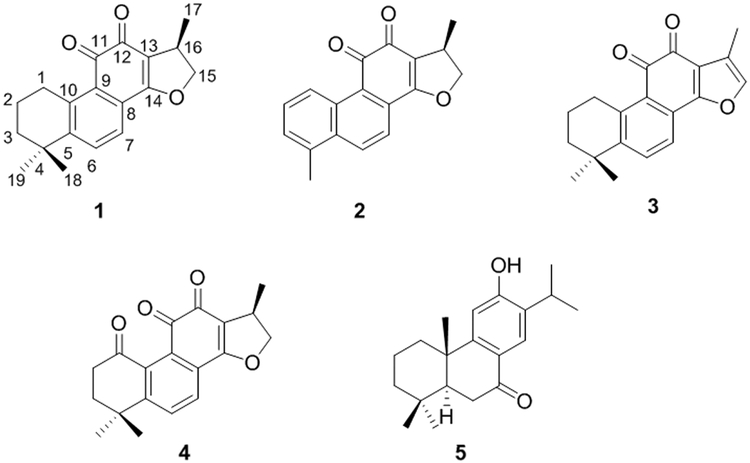
Structures of compounds isolated in this study are shown in Figure 1. See Supporting Information for details on chromatographic separation of complex fractions, and isolation of compounds 1, 4, and 5 (Figure S1). Compounds 2 and 3 were identified by matching fragmentation spectra to those of standard compounds (Figures S2 and S3).
Figure 1. S. miltiorrhiza compounds identified during this study.
Compounds 1-5 correspond to cryptotanshinone, dihydrotanshinone I, tanshinone IIA, 1-oxocryptotanshinone, and sugiol, respectively.
Sugiol (5): white amphorphous powder; HRESIMS m/z 301.2159 [M+H]+ (calculated for C20H29O2+, 301.2167, −2.6 ppm). MS/MS data of the isolated compound match fragmentation patterns of the compound found in S. miltiorrhiza extract (Figure S4). NMR data from the literature are inconsistent and incomplete,30-36 and improved spectra are provided (Table S1, Figures S5-S9). A previous report was conducted in DMSO-d6,34 and for further confirmation on the identity of this compound, we ran an additional 1H NMR (500 MHz, DMSO-d6), which matched literature values (Figure S9).34
Cryptotanshinone (1): red crystalline solid; HRESIMS m/z 297.1487 [M+H]+ (calculated for C19H21O3+, 297.1490, 1.01 ppm); 1H NMR (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) are consistent with previous reports,37 provided as Supporting Information (Figures S10-S11). Experimental MS/MS data match the purchased standard (Figure S12).
1-oxo-cryptotanshinone (4): orange amorphous powder; HRESIMS m/z 311.1277 [M+H]+ (calculated for C19H19O4+, 311.1283, 1.93 ppm); 1H NMR (500 MHz, CDCl3) are consistent with previous reports,38 and are provided as Supporting Information (Figure S13). Compound degradation occurred before additional spectra could be obtained.
Antimicrobial Assays.
To evaluate antimicrobial activity, broth microdilution assays were conducted using a clinically relevant strain of methicillin-resistant Staphylococcus aureus (MRSA, strain USA300 LAC AH1263)39 using CLSI guidelines40 as described previously.1,8,17,18 Stock solutions were prepared in DMSO and diluted with Mueller-Hinton Broth (MHB) for a final concentration of 2% DMSO and a sample concentration ranging from 0–100 μg mL−1. Full dose-response curves of the known antimicrobial cryptotanshinone (compound 1) were conducted during each screening, and served as positive control.24,25,28 A minimum inhibitory concentration (MIC), defined as the concentration where no statistically significant changes in OD600 were found between treated wells and wells with samples but no bacteria, was calculated for each compound. MIC curves were fit with a four-parameter logistic model in GraphPad Prisom 8.2.0 (Graphpad Software, La Jolla, CA, USA).
Activity Prediction and the Activity Index.
To identify a subset of S. miltiorrhiza fractions for synergy testing, the Simplify workflow was employed (Figure 2). The S. miltiorrhiza extract and chromatographic fractions were analyzed with UPLC-MS to obtain metabolite profiles. Cryptotanshinone concentrations were calculated for each fraction (Figure S1) using a calibration curve (Figure S14). After two rounds of chromatographic separation, synergists had been separated from cryptotanshinone. To avoid misidentifying these fractions as inactive, a sub-inhibitory concentration of cryptotanshinone (3 μg mL−1) was spiked into each sample prior to antimicrobial testing and LC-MS. Dose-response curves of cryptotanshinone were used to predict activity of fractions based on their concentration of cryptotanshinone. Predicted and observed bioactivities of fractions were compared, and activity indices calculated (equation 1):
| (equation 1) |
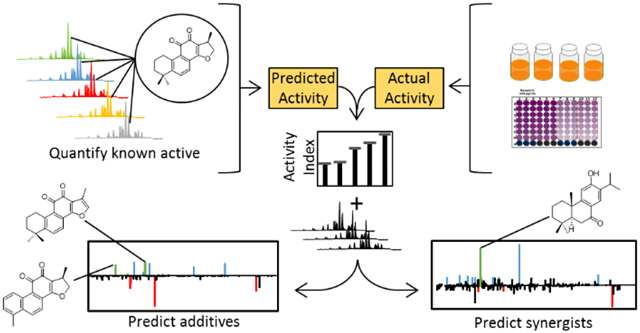
Figure 2. Workflow for the Simplify Approach.
A known active compound is quantified in each complex mixture. An activity index (equation 1) is calculated by comparing the predicted and actual activity for each fraction. Fractions with activity indices greater than 110 are prioritized for follow up testing. Activity indices are used in SR modeling to predict constituents act additively or synergistically, which are subsequently isolated to confirm activity.
The activity index represents the percent enhancement when actual activity is ratioed to predicted activity; thus, fractions with an activity index >100 potentially contain synergists or additives, while fractions with an activity index <100 may contain antagonists.
Synergy Assessment.
Broth microdilution checkerboard assays41 were utilized to pinpoint the type of combination effects between S. miltiorrhiza fractions (Figure S1) and cryptotanshinone. Combination effects can occur over a broad range of sample concentrations. As such, the checkerboard assay, with its ability to acquire and compare dose-response curves using different dose ratios of samples under study, was chosen as the biological test for evaluating synergy.42 A subset of S. miltiorrhiza fractions where chosen for testing that demonstrated activity indices greater than 110 or less than 90 and were present in sufficient concentration. Pure compounds 2, 3, and 5 were also tested. Cryptotanshinone concentrations ranged from from 0–25 μg mL−1 while other pure compounds and fraction concentrations ranged from 0–100 μg mL−1. The vehicle control was 2% DMSO in MHB. Fractional inhibitory concentration indices (ΣFICs) were then calculated (equation 2), where A and B are the samples tested, IC50A represents the IC50 of A alone, IC50B represents the IC50 of B alone, [A] is the IC50 of A in the presence of B, and [B] is the IC50 of B in the presence of A.41
| (equation 2) |
IC50 values for A and B in combination were defined as the lowest tested concentrations that led to ≥ 50% growth inhibition. Conservative values were used to assign combination effects, as recommended recently.42 Synergistic effects have ΣFICs ≤ 0.5, additive effects range from 0.5 to 1.0, non-interactive effects range from 1.0 to 4.0, and antagonistic effects have an ΣFIC ≥ 4.0.
Metabolomics Data Analysis.
Baseline Correction/MZMine Parameters.
Datasets acquired in positive and negative modes for triplicate injections of each sample were analyzed, aligned, filtered, and peak picked using MZMine 2.21.2 (http://mzmine.sourceforge.net/).43 Chromatograms were built for all m/z values that were detected for longer than 0.1 min. Modeling was completed with the third set of fractions (SM-3–2-1 through SM-3–2-9 and SM-3–4-1 through SM-3–4-5, Figure S1) using a noise level (absolute value) of 7.0 × 106 for negative mode and 2.0 × 106 for positive mode. The m/z tolerance was 5ppm, and the intensity variation tolerance was 20%. Peaks were aligned into a single peak if they eluted within 0.2 min from one another and had less than 5 ppm difference in m/z values. Retention time, m/z, and peak area was imported into Excel (Microsoft, Redmond, WA, USA) and positive and negative ion data were combined as a single peak list. Data reduction included removing all m/z- retention time pairs with peak areas >1.0 × 107 in the MeOH blank and those with an m/z ratio below 200 or above 900 or eluting before 1.5 or after 9 min. Biological activity data were added in the form of activity indices calculated with percent inhibition data at 100 μg mL−1. Checkerboard assay results of the prioritized subset of fractions were used to divide samples into two groups, one with high activity indices due to synergy and one that had high activity indices due to additivity. Non-interactive fractions were included in both models.
Selectivity Ratio (SR) Analysis.
Sirius version 10.0 statistical software (Pattern Recognition Systems AS, Bergen, Norway)44 was used for statistical modeling. To remove interference, triplicate injections of samples were subjected to hierarchical cluster analysis and filtering of interferents as described in a recent publication.45 If triplicate injections did not cluster, variables illustrating high peak area variability (above 1.5 × 108) within triplicate injections were removed, as were their associated in-source fragments, mass spectral adducts, and isotopologues (Table S2). Two SR models were produced, one using a subset of synergistic and indifferent fractions (SM-3–2-1 through SM-3–2-9, Figure S1), and one using a subset of additive and indifferent fractions (SM-3–4-1 through SM-3–4-5, Figure S1). Each subset underwent an internally cross-validated PLS analysis using 100 iterations and a significance level of 0.05. Activity indices at the 100 μg mL−1 level were used as the dependent variable guiding separation between groups. Because fractions with high activity indices contained additive or synergistic properties, ions with high selectivity ratios are likely to be synergists and additives. Should checkerboard assays reveal the presence of antagonism, the reciprocal of the activity index should be used as the dependent variable guiding separation of a subset of antagonistic and indifferent fractions, as follows:
| (equation 3) |
By using the reciprocal activity index, antagonists that contribute to a decrease in the observed activity will increase the reciprocal activity index and can be identified with high selectivity ratios. To simplify interpretation and remove correlated noise from, a final filtering step, in which variables showing less than 10% peak area variance across samples were given a SR of 0, was included.8
RESULTS AND DISCUSSION
Simplify Reveals a Mismatch Between Predicted and Observed Activity and Enables Prediction of Mixture Constituents Responsible for Combination Effects.
With the Simplify approach (Figure 2), the mixture is chromatograph-ically separated, and the activity of the resulting fractions is predicted based on the quantity of a known active constituent, which is either naturally present or spiked into the mixture. If no active mixture constituents are known, an active component that is not naturally present can be added to the mixture to identify mixture components that interact with the added compound. An activity index (equation 1) is then calculated for each fraction. To distinguish between additivity and synergy, fractions with sufficient material and activity indices > 110 undergo checkerboard assays and ΣFIC indices are calculated (equation 2).38 Similarly, fractions with activity indices < 90 also undergo checkerboard assays and ΣFIC calculations to determine if antagonistic interactions are present.
Several reviews have been published on the interpretation of isobolograms and ΣFICs for evaluating combination effects.42,47-50 In short, an isobologram is a plot where each x,y data pair corresponds to a dose combination at which a desired activity is achieved (e.g. IC50). The shape of the isobologram defines the nature of the interaction and the ΣFIC index is a quantitative measure calculated using the same data.41 ΣFIC values are used to sort fractions by the type of combination effect they exhibit, and activity indices are used to guide SR analyses. The application of the activity index is what makes the Simplify process unique as compared to other approaches. Biochemometric analysis of these data enables identification (based on detected m/z and retention time) of specific mixture constituents that are expected to exhibit interactions. These compounds can then be prioritized for isolation and structure elucidation and tested to confirm predictions.
The Activity of Cryptotanshinone is Enhanced by Both Additives and Synergists in S. miltiorrhiza.
Cryptotanshinone from S. miltiorrhia has antibacterial activity against a broad range of bacteria, both alone24,25,28 and in combination with existing antibiotics.24,25 The first four steps of the Simplify approach (Figure 2) were employed at each stage of the fractionation process. S. miltiorrhiza was extracted and fractionated, and cryptotanshinone (1) was quantified in each fraction. The predicted activity, based on the activity of pure cryptotanshinone, was compared to the observed activity to identify fractions for which activity could not be explained by cryptotanshinone alone. These fractions were prioritized for follow up testing to identify specific combination effects. The first round of fractionation yielded eight fractions (SM-1 through SM-8, Figure S1), three of which possessed enhanced antimicrobial activity that was not explained by cryptotanshinone (fractions SM-1, SM-3, and SM-5, Figure 3A). To determine whether this enhancement in activity was synergistic or additive, checkerboard assays41,51 were conducted, in which a series of cryptotanshinone dose-response curves were collected in the presence of varying concentrations of the fraction under study. Using these data, isobolograms were plotted and ΣFICs were calculated (Figure 3B-3D). SM-1 and SM-3 had ΣFIC values < 0.5, indicating that synergists were present in these mixtures. SM-5 had an ΣFIC of 0.75, illustrating that additives rather than synergists were present in this sample.
Figure 3.
Comparison of predicted to actual activity (A), black bars denote the activity due to cryptotanshinone and gray bars represent the observed fraction activity at 10 μg mL−1. Cryptotanshinone’s MIC matches previous reports.46 Fractions SM-1, SM-3, and SM-5 were prioritized for synergy testing. SM-1 showed synergy with an ΣFIC of 0.38 (B) as did SM-3 with an ΣFIC of 0.19 (C). SM-5 is additive with an ΣFIC of 0.75 (D). Equation 2 was used to calculate ΣFICs.
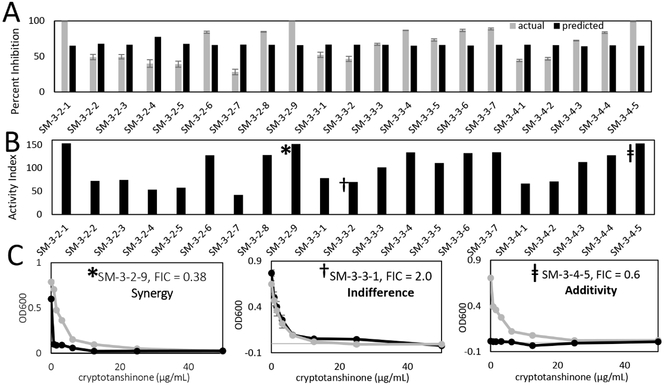
Of the fractions tested, SM-3 inhibited bacterial growth most strongly, and was prioritized for fractionation, yielding 4 simplified fractions (SM-3–1 through SM-3–4, Figure S1). The first fraction, SM-3–1, possessed antimicrobial activity, while the other fractions did not (Figure S15A). We expected that synergists had been separated from cryptotanshinone during the chromatographic separation process and tested the inactive fractions (SM-3–2 through SM-3–4) for synergy. Isobolograms and ΣFIC values for each of these fractions (Figure S15B-15D) revealed synergistic activity for all three, with ΣFIC values ranging from 0.14–0.40. Fractions SM-3–2, SM-3–3, and SM-3–4 were chromatographically separated into 21 simplified fractions. Because cryptotanshinone was no longer present at biologically relevant concentrations, it was spiked at sub-antimicrobial concentrations (3 μg mL−1) into samples for biological testing (Figure 2). Several fractions possessed greater than predicted activity (Figure 4A), a subset of which were prioritized for synergy testing (Table 1).
Figure 4.
A. Predicted and actual activities of third stage fractions resulting from chromatographic separation of S. miltiorrhiza fractions SM-3-2, SM-3-3, and SM-3-4 (see fractionation scheme in Figure S1) where black bars represent the predicted antimicrobial activity of each fraction due to cryptotanshinone and gray bars represent the actual activity of the fraction at 100 μg mL−1. Cryptotanshinone served as a positive control, consistent with previous reports.46 B. Activity indices (equation 1) of fractions SM-3-2-1 through SM-3-4-5, where bars represent the extent to which each fraction enhances or suppresses the activity of cryptotanshinone. C. Selected dose response curves of cryptotanshinone with (black) and without (gray) 100 μg mL−1 of synergistic (left), indifferent (middle), and additive fractions, corresponding to symbols in panel B.
Table 1. IC50, MIC, ΣFICs, and Activity Indices (AI) of S. miltiorrhiza extracts in combination with cryptotanshinone.
IC50 and MICs represent concentrations of individual extracts required to inhibit bacterial growth (USA300 LAC AH1263)39 by 50 or 100%, and ΣFICs indicate the interaction between extracts and cryptotanshinone.
| IC50
(μg mL−1) |
MIC (μg mL−1) |
ΣFIC † | AI ‡ | |
|---|---|---|---|---|
| SM-1 | 12.9 ± 1.7 | ≤ 25 | 0.38, synergy | -- |
| SM-3 | 9.8 ± 1.5 | ≤ 25 | 0.19, synergy | -- |
| SM-5 | 11.4 ± 1.5 | ≤ 25 | 0.75, additivity | -- |
| SM-3-2 | > 100 | > 100 | 0.26, synergy | -- |
| SM-3-3 | > 100 | > 100 | 0.40, synergy | -- |
| SM-3-4 | 46.0 ± 7.2 | ≤ 100 | 0.14, synergy | -- |
| SM-3-2-1 | > 100 | > 100 | 0.31, synergy | 153 |
| SM-3-2-7 | > 100 | > 100 | 1.25, indifference | 42 |
| SM-3-2-8 | > 100 | > 100 | 0.38, synergy | 128 |
| SM-3-2-9 | > 100 | > 100 | 0.38, synergy | 151 |
| SM-3-3-2 | > 100 | > 100 | 2.0, indifference | 70 |
| SM-3-4-1 | > 100 | > 100 | 2.0, indifference | 67 |
| SM-3-4-2 | > 100 | > 100 | 2.0, indifference | 71 |
| SM-3-4-3 | > 100 | > 100 | 0.75, additivity | 113 |
| SM-3-4-4 | > 100 | > 100 | < 1.0, additivity § | 127 |
| SM-3-4-5 | 12.3 ± 6.3 | ≤ 25 | 0.60, additivity | 153 |
± standard error
ΣFICs were calculated using equation 2.
Activity indices were calculated for third stage fractions only.
the highest concentration tested was 100 μg mL−1, which did not reach IC50. To conservatively estimate activity, 100 μg mL−1 was chosen as the IC50 of SM-3-4-4 to calculate the ΣFIC and yielded a result of 1.0. However, since the actual IC50 of SM-3-4-4 is higher than 100, the ΣFIC is lower than 1.0, categorized as additive.
In the third round of chromatographic separation, several fractions with sufficient material for further testing had activity indices at or below 71 (fractions SM-3–2-7, SM-3–3-2, SM-3–4-1, and SM-3–4-2) (Figure 4A). However, when they were tested for antagonism in a checkerboard assay,41 they had ΣFIC values of 1.25 (SM-3–2-7) or 2.0 (SM-3–3-2, SM-3–4-1, and SM-3–4-2) which we have classified as “noninteractive” (Table 1). Several researchers have considered ΣFIC indices ≥ 2.0 to be indicative of antagonism.52-54 Here we adopted a more conservative approach, as recommended previously,42,55 defining antagonism as ΣFIC >4.0 to enable better interpretation of pharmacological interactions and avoid reproducibility errors.
Selectivity Ratio Analysis Predicts Active Compounds and Characterizes the Nature of their Interactions.
The first steps of the Simplify approach enabled the prioritization of a subset of S. miltiorrhiza fractions whose activity was not explained by the presence of cryptotanshinone alone. To identify active constituents in these fractions, partial least squares (PLS) analysis was conducted on the mass spectral and biological activity data from a subset of fractions and visualized using SR analysis. SR analysis enables quantification and ranking of each variable’s impact on the biological response in question, independent of the abundance of the ion in the sampes; as such, it is not biased towards highly abundant mixture constituents. Each variable’s explained variance is then divided by its residual variance to produce a selectivity ratio, which enables the prediction of each variable’s contribution to biological activity. Rather than use raw activity data to guide analyses as has been done in previous studies,1,17,18 we used activity indices to measure the extent to which fractions enhanced or suppressed cryptotanshinone’s activity.
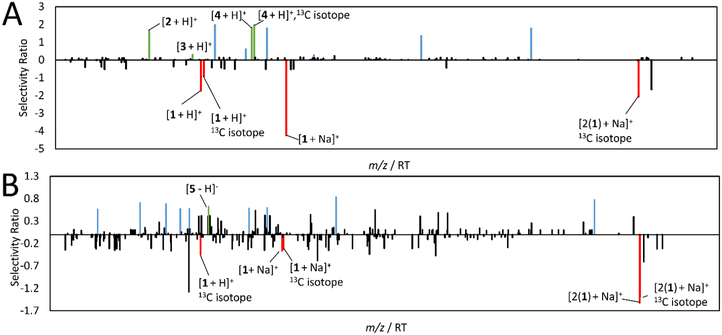
Using activity indices to predict putative active compounds, two PLS models were produced and visualized with SR plots. In these plots, each unique m/z – retention time pair is plotted on the x-axis, and the SR is plotted on y-axis. The SR, a ratio of explained to residual variance,56 represents each variable’s association with biological activity. Because fractions with activity indices > 110 possessed additive or synergistic activity (Figure 4, Table 1), variables with high SRs are most likely synergists and additives. The first SR model was built with mass spectral data and activity indices from fractions SM-3–4-1 through SM-3–4-5 (Figure S1). This internally cross-validated model generated 3 components that accounted for 97.1% of the independent (mass spectral) and 89.9% of the dependent (activity index) variation. The first SR plot was generated to predict additive compounds (Figure 5A). Of the 1263 ions included in the model, only 117 were assigned a SR above 0. The top ten predicted additives are listed in Table 2. The second SR model was built with fractions 3–2-1 through SM-3–2-9 (Figure S1) using activity indices and peak area data of 1263 individually detected ions. An internally cross-validated model was produced to predict synergistic compounds. This 3-component model used 51.8% of the mass spectral data to explain 91.4% of activity index variation. 127 ions were assigned an SR above 0 and the top ten predicted synergists were identified (Figure 5B, Table 2).
Figure 5. SR models used to predict additives and synergists.
High SRs correspond to m/z-retention time pairs that most likely possess activity. Each compound may be represented by more than one variable (i.e. isotopes and adducts). Compounds confirmed by NMR or MS-MS fragmentation are marked green, cryptotanshinone ions (compound 1) have been marked red, and unidentified variables have been marked blue. Compound 1 is not correlated with activity because it was spiked equally to all fractions. A. Additive SR plot using data from fractions SM-3-4-1 through SM-3-4-5. Dihydrotanshinone 1, tanshinone IIA, and 1-oxocryptotanshinone (compounds 2-4, respectively) were identified among the top ten additives. B. Synergistic SR plot using data from fractions SM-3-2-1 through SM-3-2-9. Sugiol (compound 5) was identified as a putative synergist
Table 2. Top ten ions predicted from additive and synergistic models.
Many were not abundant enough to isolate and confirm identities.
| m/z | RT | Polarity | Identity | SR | Activity |
|---|---|---|---|---|---|
| 245.117 | 4.586 | + | 0.561 | Synergistic | |
| 275.129 | 5.438 | + | 0.71 | Synergistic | |
| 279.103 | 5.217 | + | Compound 2* | 1.66 | Additive † |
| 287.164 | 6.102 | + | 0.686 | Synergistic | |
| 292.155 | 5.456 | + | 0.574 | Synergistic | |
| 295.135 | 6.529 | + | Compound 3* | 0.300 | Additive † |
| 295.136 | 5.523 | + | 0.573 | Synergistic | |
| 299.201 | 5.828 | − | Compound 5‡ | 0.627 | Synergistic † |
| 301.085 | 5.239 | + | 1.97 | Additive | |
| 309.113 | 5.172 | + | 0.607 | Additive | |
| 311.129 | 4.484 | + | Compound 4‡ | 1.82 | Additive |
| 311.129 | 4.885 | − | 0.584 | Synergistic | |
| 312.131§ | 4.475 | + | Compound 4‡ | 1.97 | Additive |
| 315.159 | 3.761 | + | 1.79 | Additive | |
| 315.196 | 5.625 | + | 0.598 | Synergistic | |
| 327.160 | 5.314 | + | 0.268 | Additive | |
| 332.185 | 5.621 | + | 0.834 | Synergistic | |
| 355.152 | 3.793 | + | 1.35 | Additive | |
| 393.287 | 4.809 | + | 1.78 | Additive | |
| 460.196 | 5.459 | + | 0.776 | Synergistic |
identity confirmed by comparing MS-MS patterns to a pure standard
activity confirmed by running full checkerboard assays
compound identity confirmed by NMR
m/z value corresponds to the 13C isotope of compound 4
Model Predictions Identified Additive and Synergistic Compounds.
Using SRs of individual constituents, we identified a subset of 20 putatively active compounds from the 1263 ions detected (Figure 5, Table 2). Two of the top ten predicted additives, dihydrotanshinone I (compound 2) and tanshinone IIA (compound 3) were identified by comparing fragmentation patterns of standard compounds to compounds detected in S. miltiorrhiza fractions (Figure S2 and S3). Compounds 2 and 3 were tested in combination with cryptotanshinone as previously described41,51 to confirm predictions of additivity, and demonstrated ΣFIC values of 0.68 and 0.61, respectively, confirming the predictions from SR analysis (Table 3). These compounds also demonstrated antimicrobial activity in isolation (MICs ≤ 6.25 and 25 and IC50 values of 2.2 ± 0.4 and 15.0 ± 8.4 μg mL−1 for 2 and 3, respectively) (Figure S16). 1-oxocryptotanshinone (compound 4), the top predicted additive, was isolated from S. miltiorrhiza for the first time. The yield was too low to confirm activity, but 4 is likely to be antimicrobial given its structural similarity to 1.
Table 3.
IC50, MIC, and ΣFICs of pure compounds combined with cryptotanshinone against S. aureus (USA300 LAC AH1263)39
| IC50
(μg mL−1) * |
MIC (μg mL−1) |
ΣFIC † | |
|---|---|---|---|
| Cryptotanshinone | 5.9 ± 2.2 | ≤ 25 | -- |
| Dihydrotanshinone I | 2.2 ± 0.4 | ≤ 6.25 | 0.68, additivity |
| Tanshinone IIA | 15.0 ± 8.4 | ≤ 25 | 0.61, additivity |
| Sugiol | > 100 | > 100 | 0.28, synergy |
± standard error
ΣFICs were calculated using equation 2.
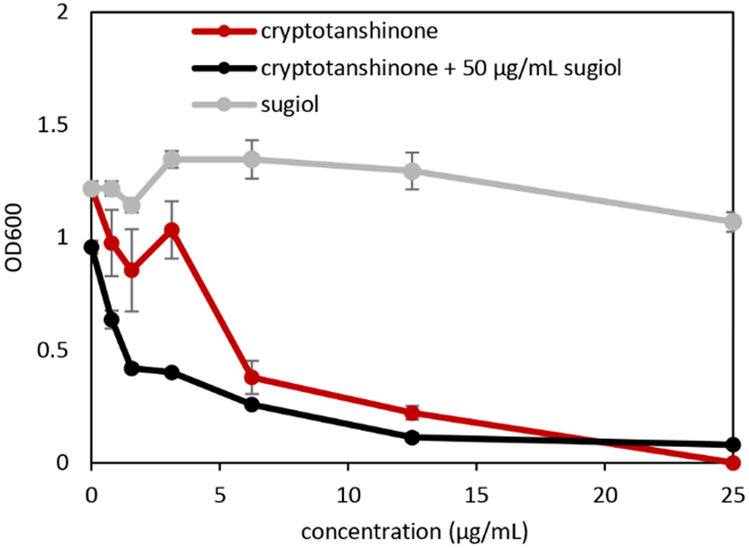
Several putative synergists were identified in S. miltiorrhiza. One of these, sugiol (compound 5), was isolated and its structure confirmed by MS-MS and NMR (Figures S4-S9, Table S1).34 Sugiol was identified as the 5th top contributor to synergy according to model predictions (Figure 5). In combination with cryptotanshinone, sugiol was synergistic (ΣFIC value of 0.28). Sugiol was inactive in isolation (IC50 and MICs >100 μg mL−1), but when sugiol was combined with cryptotanshinone, the IC50 of cryptotanshinone fell from 5.89 to 1.56 μg mL−1 (Figure 6).
Figure 6. Dose-response curves (± standard error) of cryptotanshinone, sugiol, and their combination.
Combined with cryptotanshinone, sugiol (inactive in isolation) causes a four-fold drop in cryptotanshinone’s IC50 (5.89 to 1.56 μg mL−1) illustrating synergy.
Simplify represents the first example of a bioinformatics approach in which compounds contributing to bioactivity were identified and the nature of their interactions characterized prior to their isolation. Using S. miltiorrhiza as a model organism, we identified three additive compounds and one synergistic compound. Using the SR plot designed to find synergists (Figure 5B), sugiol (compound 5) was correctly predicted to synergize with cryptotanshinone. Although this compound has been identified from S. miltiorrhiza previously,57 this is the first report of its synergistic activity. Using bioassay-guided fractionation, sugiol would have been missed because it is inactive in isolation.
Interestingly, the fractions used to produce the SR plot for predicting additives (Figure 5A), which all possessed additive rather than synergistic activity, came from the separation of the synergistic fraction SM-3–4. It is possible that synergists were lost during the chromatographic separation process due to irreversible binding to the column, or that multiple constituents were required for the synergistic effect of SM-3–4. Thus, while the obtained results provide a more comprehensive picture of constituents that contribute to the activity of S. miltiorrhiza than was possible using other approaches, it is still only part of the story. While it is possible to identify multiple active constituents, material limitations remain a central challenge in complex mixture characterization efforts. From over 1200 ions detected, not all of which represent a single compound, Simplify enabled isolation efforts to be geared towards the compounds most likely to be active. Several predictions generated with this approach were validated; all tested compounds possessed the expected additive or synergistic activity.
Simplify is compatible with existing bioassay- and effect-directed fractionation workflows and minimizes the time between discovery of combination effects and identification of synergists and additives. While this approach required three rounds of fractionation in order to produce selectivity ratio models, bioactivity-directed fractionation was not needed to identify active components. Using model predictions, we were able to return to untested fractions (SM-1–5 and SM-12) to isolate the predicted synergistic and additive compounds. Additionally, this approach is applicable to any complex mixture without the need for existing knowledge of its chemical composition. With this application, we sought to characterize the nature of the interactions within a well-characterized natural product mixture. However, researchers may still use this approach on a mixture of truly unknown composition to identify combination effects that occur between mixture constituents and a known, but not naturally present, active molecule. Although we did not identify antagonism in this study, this approach could also be utilized to identify constituents that act as antagonists.
CONCLUSIONS
Biological systems rely on diverse chemical interactions; not only do organisms themselves represent complex mixtures, but they interact with an array of organic and inorganic chemicals for survival. Currently, there is a gap in the way that we understand complex mixtures, whether they be natural products, environmental contaminants, or human microbiota, because these mixtures do not exert biological effects equal to the sum of their individual constituents. With this work, we illustrate the ability of Simplify to provide a more comprehensive picture of combination effects that occur in complex mixtures than is possible with reductionist approaches. Importantly, this approach can be modified to incorporate any biological activity data, combination effect model, or analytical platform. As such, we expect that this tool will be applicable in many fields for studying interactions in mixtures.
Supplementary Material
ACKNOWLEDGEMENTS
This work was done with NCCIH funds 5T32 AT008938 and 1R15 AT010191. Thanks to Richard Cech for plant material, Dr. Alexander Horswill for MRSA strains, and Dr. Olav Kvalheim for statistical expertise. Dr. Nick Oberlies, Dr. Ron Venters, and Sonja Knowles aided with NMR. MS analyses were conducted at UNCG’s Triad Mass Spectrometry Facility.
Footnotes
Supporting Information.
The chromatographic separation procedure, fractionation scheme, activity models, dose response curves, calibration curve, fragmentation and NMR spectra, and chemical contaminants are provided as Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website.
The authors declare that there are no conflicts of interest.
REFERENCES
- (1).Britton ER; Kellogg JJ; Kvalheim OM; Cech NB J. Nat. Prod 2018, 81, 484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Health Effects Institute. In Theoretical Approaches to Analyzing Complex Mixtures; Heath Effects Institute: Cambridge, MA, 1996. [Google Scholar]
- (3).Junio HA; Sy-Cordero AA; Ettefagh KA; Burns JT; Micko KT; Graf TN; Richter SJ; Cannon RE; Oberlies NH; Cech NB J. Nat. Prod 2011, 74, 1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Abreu NA; Taga ME FEMS Microbiol. Rev 2016, 40, 648–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Snyder SA J. Am. Water Works 2014, 106, 38–52. [Google Scholar]
- (6).Delfosse V; Dendele B; Huet T; Grimaldi M; Boulahtouf A; Gerbal-Chaloin S; Beucher B; Roecklin D; Muller C; Rahmani R Nat. Comm 2015, 6, 8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kortenkamp A Curr. Opin. Pharmacol 2014, 19, 105–111. [DOI] [PubMed] [Google Scholar]
- (8).Caesar LK; Kellogg JJ; Kvalheim OM; Cech NB J. Nat. Prod 2018, 82, 469–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Cedergreen N PLoS One 2014, 9, e96580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hayes AW; Li R; Hoeng J; Iskandar A; C Peistch M; Dourson M Toxicol. Res. Appl 2019, 3, 1–10. [Google Scholar]
- (11).Laetz CA; Baldwin DH; Collier TK; Hebert V; Stark JD; Scholz NL Environ. Heath Perspect. 2009, 117, 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Woznica A; Cantley AM; Beemelmanns C; Freinkman E; Clardy J; King N PNAS. 2016, 113, 7894–7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kinghorn AD Curr. Org. Chem 1998, 2, 597–612. [Google Scholar]
- (14).Iskandar AR; Gonzalez-Suarez I; Majeed S; Marescotti D; Sewer A; Xiang Y; Leroy P; Guedj E; Mathis C; Schaller JP; Vanscheeuwijck P; Frentzel S; Martin F; Ivanov NV; Peitsch MC; Hoeng J Toxicol. Mech. Methods 2016, 26, 389–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Brack W Anal. Bioanal. Chem 2003, 377, 397–407. [DOI] [PubMed] [Google Scholar]
- (16).Tian Z; Gold A; Nakamura J; Zhang Z; Vila J; Singleton DR; Collins LB; Aitken MD Environ. Sci. Technol 2017, 51, 7091–7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kellogg JJ; Todd DA; Egan JM; Raja HA; Oberlies NH; Kvalheim OM; Cech NB J. Nat. Prod 2016, 79, 376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Caesar LK; Kellogg JJ; Kvalheim OM; Cech RA; Cech NB Planta Med. 2018. 84, 9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Inui T; Wang Y; Pro SM; Franzblau SG; Pauli GF Fitoterapia 2012, 83, 1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Rodruiguez MV; Sortino MA; Ivancovich JJ; Pellegrino JM; Favier LS; Raimondi MP; Gattuso MA; Zacchino SA Phytomedicine. 2013, 20, 1230–1239. [DOI] [PubMed] [Google Scholar]
- (21).Zhang L; Tan K; Zhang Y; Huang R; Bian J; Zheng C; Sun H; Chen Z; Sun N; An R; Min F; Zhao W; Zhuo Y; You J; Song Y; Yu Z; Liu Z; Yang K; Gao H; Dai H; Zhang Z; Wang J; Fu C; Pei G; Liu J; Zhang S; Goodfellow M; Jiang Y; Kuai J; Zhou G; Chen Z PNAS 2007, 104, 4606–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Mahieu NG; Patti GJ Anal. Chem 2017, 89, 10397–10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Brualdi RA Introductory Combinatorics, 5 ed.; Pearson Prentice Hall, 2010, p 648. [Google Scholar]
- (24).Cha J-D; Jeong M-R; Choi K-M; Park J-H; Cha S-M; Lee K-Y Adv. Biosci. Biotechnol 2013, 4, 283. [Google Scholar]
- (25).Cha J-D; Lee J-H; Choi KM; Choi S-M; Park JH Evid. Based Complement. Alternat. Med 2014, 2014, 450572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Jamshidi-Aidji M; Morlock GE Anal. Chem 2016, 88, 10979–10986. [DOI] [PubMed] [Google Scholar]
- (27).Lee J-W; Ji Y-J; Lee S-O; Lee I-SJ Microbiol. 2007, 45, 350–357. [PubMed] [Google Scholar]
- (28).Wang B-QJ Med. Plant Res. 2010, 4, 2813–2820. [Google Scholar]
- (29).Xu S; Liu P Expert Opin. Ther. Pat 2013, 23, 149–153. [DOI] [PubMed] [Google Scholar]
- (30).Ara I; Siddiqui BS; Faizi S; Siddiqui S Planta Med. 1990, 56, 84–86. [DOI] [PubMed] [Google Scholar]
- (31).Chao K-P; Hua K-F; Hsu H-Y; Su Y-C; Chang S-T Planta Med. 2005, 71, 300–305. [DOI] [PubMed] [Google Scholar]
- (32).Dávila M; Stenier O; Hinojosa N Revista Boliviana de Química 2014, 31, 22–28. [Google Scholar]
- (33).Gao J; Han G Phytochemistry 1997, 44, 759–761. [Google Scholar]
- (34).Li A; She X; Zhang J; Wu T; Pan X Tetrahedron 2003, 59, 5737–5741. [Google Scholar]
- (35).Rodríguez B Magn. Res. Chem 2003, 41, 741–746. [Google Scholar]
- (36).Wang S-Y; Wu J-H; Shyur L-F; Kuo Y-H; Chang S-T Holzforschung 2002, 56, 487–492. [Google Scholar]
- (37).Kang HS; Chung HY; Jung JH; Kang SS; Choi JS Arch. Pharmacal Res 1997, 20, 496. [DOI] [PubMed] [Google Scholar]
- (38).Sairafianpour M; Christensen J; Staerk D; Budnik BA; Kharazmi A; Bagherzadeh K; Jaroszewski JW J. Nat. Prod. 2001, 64, 1398–1403. [DOI] [PubMed] [Google Scholar]
- (39).Boles BR; Thoendel M; Roth AJ; Horswill AR PLoS One 2010, 5, e10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).CLSI. In Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically--Tenth Edition:Approved Standard M7-A10: Wayne, PA, 2015. [Google Scholar]
- (41).Ettefagh KA; Burns JT; Junio HA; Kaatz GW; Cech NB Planta Med. 2010, 77, 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).van Vuuren S; Viljoen A Planta Med. 2011, 77, 1168–1182. [DOI] [PubMed] [Google Scholar]
- (43).Pluskal T; Castillo S; Villar-Briones A; Orešič M BMC Bioinform. 2010, 11, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Kvalheim OM; Chan H-Y; Benzie IF; Szeto Y-T; Tzang AH-C; Mok DK-W; Chau F-T Chemom. Intell. Lab. Syst 2011, 107, 98–105. [Google Scholar]
- (45).Caesar LK; Kvalheim OM; Cech NB Anal. Chim. Acta 2018, 1021, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Lee DS; Lee SH; Noh JG; Hong SD Biosci. Biotechnol. Biochem 1999, 63, 2236–2239. [DOI] [PubMed] [Google Scholar]
- (47).Efferth T; Koch E Curr. Drug Targets 2011, 12, 122–132. [DOI] [PubMed] [Google Scholar]
- (48).Gilbert B; Alves LF Curr. Med. Chem 2003, 10, 13–20. [DOI] [PubMed] [Google Scholar]
- (49).Ulrich-Merzenich G; Panek D; Zeitler H; Vetter H; Wagner H Indian J. Exp. Biol 2010, 48, 208–219. [PubMed] [Google Scholar]
- (50).Wagner H; Ulrich-Merzenich G Phytomedicine 2009, 16, 97–110. [DOI] [PubMed] [Google Scholar]
- (51).Eliopolous GM.; Moellering R In Antibiotics in Laboratory Medicine, Lorian V, Ed.: Baltimore MD, 1996, pp 330–396. [Google Scholar]
- (52).Cottarel G; Wierzbowski J Trends Biotechnol. 2007, 25, 547–555. [DOI] [PubMed] [Google Scholar]
- (53).Goñi P; López P; Sánchez C; Gómez-Lus R; Becerril R; Nerín C Food Chem. 2009, 116, 982–989. [Google Scholar]
- (54).EUCAST. Clin. Microbiol. Infect 2000, 6, 503–508. [DOI] [PubMed] [Google Scholar]
- (55).Odds FC J. Antimicrob. Chemother 2003, 52, 1. [DOI] [PubMed] [Google Scholar]
- (56).Rajalahti T; Arneberg R; Kroksveen AC; Berle M; Myhr K-M; Kvalheim OM Anal. Chem 2009, 81, 2581–2590. [DOI] [PubMed] [Google Scholar]
- (57).Chang HM; Cheng KP; Choang TF; Chow HF; Chui KY; Hon PM; Tan FWL; Yang Y; Zhong ZP J. Org. Chem 1990, 55, 3537–3543. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.