Abstract
BACKGROUND:
Associations between dairy intake and body composition and cardiometabolic traits have been inconsistently observed in epidemiological studies, and the causal relationship remains ill-defined.
METHODS:
We performed Mendelian randomization analysis using an established genetic variant located upstream of the lactase gene (LCT-13910 C/T, rs4988235) associated with dairy intake as an instrumental variable (IV). The causal effects of dairy intake on body composition and cardiometabolic traits (lipids, glycemic traits, and inflammatory factors) were quantified by IV estimators among 182041 participants from 18 studies.
RESULTS:
Each 1 serving/day higher dairy intake was associated with higher lean mass [β (SE) = 0.117 kg (0.035); P = 0.001], higher hemoglobin A1c [0.009% (0.002); P = 0.001], lower LDL [−0.014 mmol/L (0.006); P = 0.013], total cholesterol (TC) [−0.012 mmol/L (0.005); P = 0.023], and non-HDL [−0.012 mmol/L (0.005); P = 0.028]. The LCT-13910 C/T CT + TT genotype was associated with 0.214 more dairy servings/day (SE = 0.047; P < 0.001), 0.284 cm higher waist circumference (SE = 0.118; P = 0.017), 0.112 kg higher lean mass (SE = 0.027; P = 3.8 × 10−5), 0.032 mmol/L lower LDL (SE = 0.009; P = 0.001), and 0.032 mmol/L lower TC (SE = 0.010; P = 0.001). Genetically higher dairy intake was associated with increased lean mass [0.523 kg per serving/day (0.170); P = 0.002] after correction for multiple testing (0.05/18). However, we find that genetically higher dairy intake was not associated with lipids and glycemic traits.
CONCLUSIONS:
The present study provides evidence to support a potential causal effect of higher dairy intake on increased lean mass among adults. Our findings suggest that the observational associations of dairy intake with lipids and glycemic traits may be the result of confounding.
Observational studies, in which reverse causation, residual confounding, and limited generalizability are often nonnegligible (1), reported an association of dairy consumption with body composition (2, 3). Meta-analyses of both observational studies (4) and randomized controlled trials (RCTs)61 (5–7) demonstrated that high dairy intake in the absence of energy restriction increased body weight. However, meta-analysis of randomized studies showed that there were no changes in cardiometabolic risk factors such as fasting glucose, insulin resistance, lipids, or C-reactive protein (CRP) (8). In contrast, another meta-analysis of controlled short-term intervention studies showed that a fermented yogurt product was associated with a 4% decrease in total cholesterol (TC) and a 5% decrease in LDL cholesterol (9). Therefore, results for cardiometabolic traits are still inconclusive. Mendelian randomization (MR) analysis (10–13), which is analogous to an RCT, when randomization to genotype takes place at conception (14), has been widely used to assess potential causal associations of lifetime variations of modifiable factors with diseases (10, 15–20).
Previous large-scale MR analyses, adopting a well-established genetic marker (LCT-13910 C/T, rs4988235) as an instrumental variable (IV) for dairy intake, demonstrated that genetically predicted high dairy intake is associated with higher body mass index (BMI) (18) but not causally related to hypertension (10), diabetes (11), and cardiovascular diseases (12, 13). However, whether dairy intake is causally associated with body composition and other important cardiometabolic traits is largely unknown.
Therefore, in the current study, we performed MR analysis among 182041 adult participants from 18 cohorts using an established dairy intake-associated genetic variant located near the lactase gene LCT62 to examine the causal association of habitual dairy intake with body composition and cardiometabolic traits such as lipids, glycemic traits, and inflammatory factors in general populations.
Materials and Methods
STUDY PARTICIPANTS
The study was conducted within the Mendelian Randomization of Dairy Consumption Working Group, represented here by 18 cohort studies including 182041 individuals in total. Detailed descriptions of each study are presented in Table 1 of the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol65/issue6. Participants from each study provided written informed consent, and local institutional review boards (see Table 2 in the online Data Supplement) granted ethical approval.
DAIRY INTAKE ASSESSMENT AND OUTCOMES
Information on intake of dairy products was collected by self-reported questionnaire in each study; detailed information on cohort-specific data collection methods is provided in Table 3 of the online Data Supplement. Total dairy products included skim/low fat milk, whole milk, ice cream, yogurt, cottage/ricotta cheese, cream cheese, other cheese, and cream. The primary outcomes are body composition (body fat percentage, waist circumference, waist to hip ratio, lean mass, and fat mass), cardiometabolic traits [lipids: HDL cholesterol, LDL cholesterol, TC, total triglyceride (TG), non-HDL cholesterol, and apolipoprotein B (apoB)], glycemic traits [fasting glucose, hemoglobin A1c (HbA1c), fasting insulin, insulin resistance, and insulin sensitivity], and inflammatory factors [regular CRP and high-sensitivity CRP (hsCRP)] at baseline or during follow-up. Detailed information on the outcome measure for each study is reported in Table 4 of the online Data Supplement.
SINGLE-NUCLEOTIDE POLYMORPHISM SELECTION AND GENOTYPING METHODS
In the present study, we chose the widely confirmed and extensively studied variant LCT-13910 C/T, rs4988235 as the IV for dairy intake (11, 12, 21). The variant rs4988235, located upstream from the LCT gene, is associated with lactase persistence and thereby with the ability to digest lactose, the primary source of carbohydrates in milk (22). The TT and TC genotypes are associated with lactase persistence, and CC is associated with nonpersistence. Therefore, lactase persistence is a dominantly inherited genetic trait. Most studies used direct genotype information on rs4988235 from previously genotyped array data. Whenever rs4988235 was not genotyped directly, we used either (a) the HapMap II reference panel-imputed genetic information for rs4988235 or (b) genotype information of proxy that are in high linkage disequilibrium with rs4988235(n = 5; r2 > 0.9). Genotyping platforms, genotype frequencies, Hardy–Weinberg equilibrium P values, and call rates (median of 98.8%) for LCT-13910 C/T are listed in Table 5 of the online Data Supplement.
STATISTICAL ANALYSIS
Our study tested the (a) observational associations of dairy intake with body composition, lipids, glycemic traits, and inflammatory factors; (b) genetic associations of the LCT-13910 C/T, rs4988235 with dairy intake and cardiometabolic traits under a dominant model (CC vs CT + TT); and (c) causal effect of dairy intake on outcomes by using the IV estimator.
A standard analysis protocol was applied to each individual study to produce comparable results. Linear regression was used to test the observational associations of dairy intake with cardiometabolic traits after adjustment for age, sex, ethnicity, region, years of follow-up, and other baseline covariates (smoking status, physical activity, total energy intake, and alcohol intake), as available. Linear regression was used to test the genetic associations of LCT-13910 C/T with dairy intake and cardiometabolic traits, respectively, after adjustment for age, sex, ethnicity, region, and total energy.
META-ANALYSIS AND BETWEEN-STUDY HETEROGENEITY
Meta-analyses were conducted using individual participant data in each study and then pooled β coefficients across studies using random-effects or fixed-effects meta-analysis. We assessed between-study heterogeneity via Cochrane’s Q and I2 statistics (23–25). We used random-effects meta-analysis if I2 > 0.25; otherwise, fixed-effects models were used (26).
SE AND INFERENCE FOR THE IV ESTIMATOR
After meta-analysis, we used the IV estimators to quantify the strength of the causal association of dairy intake with cardiometabolic traits (Fig. 1) (27). The IV estimator, which is identical to that derived by the widely used 2-stage least-squares method (28), was calculated as the of the regression coefficients MCM6 variant 4988235- outcome and MCM6 variant 4988235-dairy:
| (1) |
| (2) |
Furthermore, to explore potential sources of heterogeneity, we conducted subgroup analysis using age of participants (<50 years and ≥50 years), follow-up years (<5 years and ≥5 years), region or country (Europe and non-Europe), study design (cohort and cross-sectional), and CC genotype frequency (≤10% and >10%) as putative categorical moderators. The Bonferroni correction was conducted for multiple comparisons (P = 0.05/18 = 0.003). Statistical analyses were conducted using Stata 14.0 software. All P values reported were 2-sided.
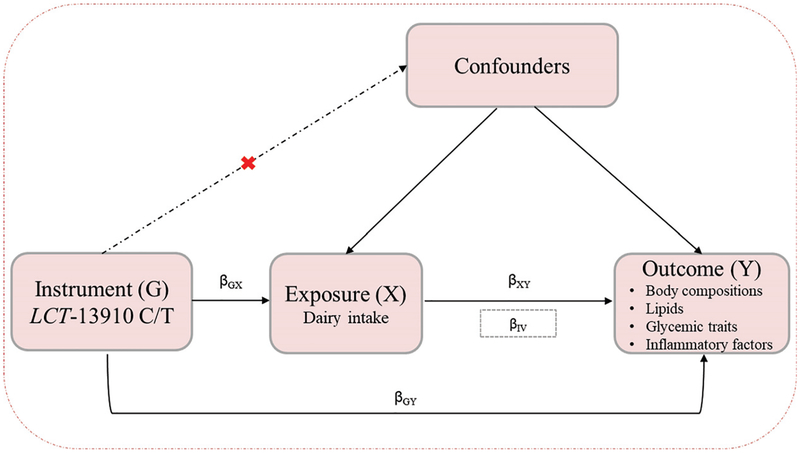
Fig. 1. A schematic description of a Mendelian randomization analysis.
MR can be used to test the hypothesis that exposure (dairy intake) causes outcomes (cardiometabolic traits). Three assumptions of MR: (a) genetic variants must be associated with dairy intake; (b) genetic variants must not be associated with confounders; and (c) genetic variants must influence cardiometabolic traits only through dairy intake, not through other pathways. The IV estimator was used to quantify the strength of the causal association of dairy intake with cardiometabolic traits using LCT-13910 C/T as an IV.
Results
BASELINE CHARACTERISTICS OF PARTICIPATING STUDIES
Baseline characteristics of the 182041 participants from 18 studies are shown in Table 1 here and Tables 6–8 of the online Data Supplement. A description of each study and additional characteristics of participants are presented in Tables 1 and 6 of the online Data Supplement. A total of 17 studies provided data for LCT-13910 C/T, and 1 study (ARIC-AA) provided results for the proxy single-nucleotide polymorphism rs1446585 (defined on the basis of r2 ≥ 0.90 with rs4988235 in individuals). The χ2 tests showed that the CCHS, CGPS, and FamHS studies did not achieve Hardy–Weinberg equilibrium (see Table 5 in the online Data Supplement).
Table 1.
Baseline characteristics of participating studies.a
| Study nameb | Study design | Number of participants, n | Follow-up, years | Age, years | Total dairy intake, serving/day | Country | Male, n (%) | BMI, kg/m2 | rs4988235, n (%) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | |||||||||
| ARIC-AAc | Cohort | 2178 | 2.8 | 53.5 (5.7) | 1.18 (1.16) | US | 793 (36.4) | 29.74 (6.01) | 1694 (77.78) | 450 (20.66) | 34 (1.56) |
| ARIC-EAd | Cohort | 8170 | 2.9 | 54.2 (5.6) | 1.82 (1.37) | US | 3882 (47.5) | 26.95 (4.78) | 1020 (12.48) | 3392 (41.52) | 3758 (46.00) |
| CCHS | Cohort | 8721 | 0 | 58.0 (15.1) | NAe | Denmark | 3911 (44.8) | 25.6 (4.32) | 548 (6.28) | 3041 (34.87) | 5132 (58.85) |
| CGPS | Cohort | 74243 | 0 | 57.0 (13.3) | 1.69 (1.21) | Denmark | 33134 (44.6) | 26.20 (4.30) | 4348 (5.86) | 26571 (35.79) | 43324 (58.35) |
| CHS | Cohort | 1863 | 4.9 | 70.9 (4.2) | 1.38 (0.70) | US | 704 (37.8) | 26.43 (4.25) | 68 (3.65) | 804 (43.16) | 991 (53.19) |
| DCH | Cohort | 8026 | NA | 2.50 (1.46) | Denmark | 3919 (48.8) | 26.84 (4.44) | 460 (5.73) | 2674 (33.32) | 4892 (60.95) | |
| DILGOM | Cohort | 1227 | 7 | 52.6 (13.1) | 5.83 (3.05) | Finland | 528 (43.0) | 26.41 (4.65) | 196 (16.0) | 332 (84) | |
| FamHS | Cohort | 2131 | 7.9 | 50.5 (13.0) | 2.05 (1.47) | US | 961 (45.1) | 27.57 (5.30) | 250 (11.73) | 893 (41.91) | 988 (46.36) |
| GESUS | Cohort | 20459 | 0 | 55.8 (13.6) | 2.37 (1.53) | Denmark | 9334 (45.6) | 26.72 (4.66) | 1212 (5.92) | 7379 (36.07) | 11868 (58.01) |
| H2000 | Cohort | 3445 | 10.9 | 49.0 (11.8) | 6.06 (2.98) | Finland | 1551 (45.0) | 26.64 (4.45) | 608 (17.65) | 1667 (48.39) | 1170 (33.96) |
| HPFS | Cohort | 6914 | 24 | 54.8 | 2.04 | US | NA | 25.32 | 841 (15.13) | 2509 (45.14) | 2208 (39.73) |
| MESA | Cohort | 4455 | 10 | 60.4 (9.5) | 1.87 (1.72) | US | 2110 (47.3) | 28.44 (5.66) | 2404 (53.96) | 1275 (28.62) | 776 (17.42) |
| NHS | Cohort | 11287 | 26 | 52.7 (6.5) | 2.05 (1.34) | US | NA | NA | 1227 (16.23) | 3600 (47.61) | 2735 (36.17) |
| PREDIMED-Valencia | Cohort | 940 | 1 −2f | 67.0 | 1.86 (1.14) | Spain | 338 (36.0) | 30.11 (4.22) | 357 (38) | 430 (45.7) | 153 (16.3) |
| RAINE | Cohort | 527 | 2.3 | 19.9 (0.3) | 1.760 (0.956) | Australia | 270 (51.2) | 24.29 (4.842) | 255 (48.39) | 202 (38.33) | 70 (13.28) |
| THISEAS | Case-control | 2565 | NA | 59.1 (0.3) | 0.83 (0.03) | Greece | 59 | 28.15 (0.10) | 78 | 20 | 1.4 |
| WGHS | Cohort | 23294 | NA | 54.7 (7.1) | 1.98 (1.36) | US | NA | 25.91 (4.96) | 2839 (12.19) | 9819 (42.15) | 10636 (45.66) |
| YFS | Cohort | 1596 | 4 | 37.7 (5.0) | 4.20 (2.49) | Finland | 714 (44.7) | 25.79 (4.57) | 243 (15.2) | 798 (50.0) | 555 (34.8) |
Mean (SD) for continuous variables, and n (%) for categorical variables.
Study abbreviation definitions and study descriptions are provided in Table 1 of the online Data Supplement.
ARIC-AA, African Ancestry.
ARIC-EA, European Ancestry.
NA, not applicable.
2 years for anthropometrics and 1 year for lipids and glycemic traits.
OBSERVATIONAL ASSOCIATIONS OF DAIRY INTAKE WITH CARDIOMETABOLIC TRAITS
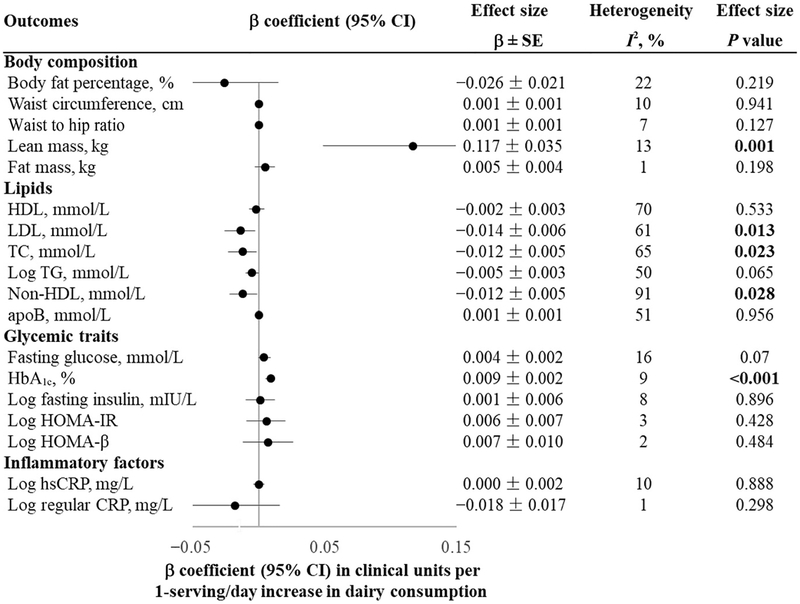
Our meta-analysis showed that high dairy intake was significantly associated with higher lean mass (β = 0.117 kg per serving/day; SE = 0.035; P = 0.001), higher HbA1c (0.009% per serving/day; SE = 0.002; P < 0.001), lower LDL (β = −0.014 mmol/L per serving/day; SE = 0.006; P = 0.013), lower TC (β = −0.012 mmol/L per serving/day; SE = 0.005; P = 0.023), and lower non-HDL (β = −0.012 mmol/L per serving/day; SE = 0.005; P = 0.028) (Fig. 2).
Fig. 2. Association between dairy intake and cardiometabolic traits among 182041 participants from 18 studies.
Linear regression was used to test the association of dairy intake (serving/day) with cardiometabolic traits after adjustment of sex, ethnicity, region, years of follow-up, and other baseline covariates if available(age, smoking status, physical activity, total energy intake, and alcohol intake)in each study. We pooled β coefficients across studies using random-effects (I2≥25%) or fixed-effects (I2<25%) meta-analyses based on the heterogeneity between studies.
GENETIC ASSOCIATION OF THE LCT-13910 C/T WITH DAIRY INTAKE AND CARDIOMETABOLIC TRAITS
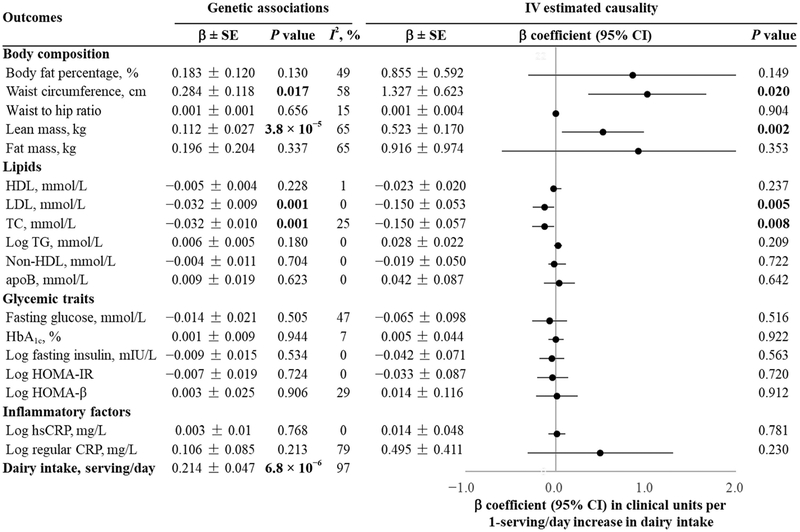
In a dominant model, we found that the LCT-13910 C/T CT + TT genotype was significantly associated with 0.214 more dairy servings/day (β = 0.214 serving/day; SE = 0.047; P = 6.8 × 106). We pooled the genetic association with cardiometabolic traits from 18 studies using fixed- or random-effects meta-analysis and found that the LCT-13910 C/T CT + TT genotype was significantly associated with 0.284 cm higher waist circumference (β = 0.284; SE = 0.118; P = 0.017), 0.112 kg higher lean mass (β = 0.112; SE = 0.027; P = 3.8 × 10−5), 0.032 mmol/L lower LDL (β = −0.032 mmol/L per serving/day; SE = 0.009; P = 0.001), and 0.032 mmol/L lower TC (β = −0.032 mmol/L per serving/day; SE = 0.010; P = 0.001) (Fig. 3).
Fig. 3. Genetic association and estimated causality between dairy intake and cardiometabolic traits.
The LCT-13910 C/T located in upstream of the lactase (LCT) gene was selected as an instrumental variable. The MR estimate was computed from the ratio of the coefficient of the association between the LCT-13910 C/T and cardiometabolic traits to that of the association between the LCT-13910 C/T and dairy intake. This IV estimate reflects the potential causal effect of dairy intake on BMI. We pooled β coefficients across studies using random-effects (I2 ≥ 25%) or fixed-effects (I2 < 25%) meta-analyses based on the heterogeneity between studies.
IV ESTIMATED CAUSALITY BETWEEN DAIRY INTAKE AND CARDIOMETABOLIC TRAITS
Fig. 3 presents the genetic association with cardiometabolic traits and the IV estimated causal effects of dairy intake on cardiometabolic traits. Genetically determined higher dairy intake was associated with increased waist circumference (β = 1.327 cm per serving/day; SE = 0.623; P = 0.020), increased lean mass (β = 0.523 kg per serving/day; SE = 0.170; P = 0.002), decreased LDL (β = −0.150 mmol/L per serving/day; SE = 0.053; P = 0.005), and decreased TC (β = −0.150 mmol/L per serving/day; SE = 0.057; P = 0.008). After correction for multiple testing, MR association of dairy intake with lean mass remained significant at P = 0.002 (0.05/18) (Fig. 3).
We further conducted stratified analyses of estimated causality by age, follow-up years, study design, ethnic group, and CC genotype frequency (Table 2). We observed significant MR associations of genetically determined higher dairy intake only on LDL and TC in studies with a patient mean age of ≥50 years and studies with follow-up time <5 years.
Table 2.
Stratified analysis on causal estimates of dairy consumption (serving/day) with cardiometabolic traits.a
| Outcomes | Age, years | Follow-up, years | Region or country | Study design | CC genotype frequency | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| ≥50 | <50 | ≥5 | <5 | Europe | non-Europe | Cross-sectional | Cohort | ≤10% | >10% | |
| Body composition | ||||||||||
| Body fat percentage, % | 0.75 (−0.54, 2.04) | 0.58 (−2.10,3.26) | 1.60 (−2.19, 5.39) | 0.55 (−0.41, 1.50) | 1.27 (−1.43, 3.97) | 0.73 (−0.54, 1.99) | 0.81 (−0.70, 2.32) | 0.95 (−1.50,3.39) | 0.77 (−0.76, 2.30) | 1.00 (−0.89, 2.90) |
| Waist circumference, cm | 1.50 (−0.16, 3.16) | 1.21 (−2.05, 4.46) | 1.26 (−1.65, 4.17) | 1.53 (0.13,2.93) | 2.23 (−0.45, 4.91) | 0.90 (−0.55, 2.34) | 1.45 (−0.35, 3.26) | 1.37(0.25,2.49) | 1.62 (−0.11, 3.35) | 1.07 (−0.05, 2.19) |
| Waist to hip ratio | 0.00 (−0.01, 0.01) | 0.02 (−0.03, 0.08) | 0.00 (−0.04, 0.04) | 0.00 (−0.01, 0.01) | 0.01 (−0.01,0.03) | 0.00 (−0.01, 0.01) | 0.01 (−0.01, 0.03) | 0.00 (−0.01,0.01) | 0.01 (−0.01, 0.03) | 0.00 (−0.01, 0.00) |
| Lean mass, kg | 0.66 (−2.10, 3.42) | 0.01 (−1.40, 1.41) | −0.81 (−3.28, 1.65) | 1.33 (−1.27, 3.93) | 4.33 (0.32, 8.34) | −0.18 (−1.33, 0.97) | 0.78 (−3.24, 4.81) | 0.10 (−1.30, 1.50) | 0.74 (−3.06, 4.54) | 0.10 (−1.27, 1.46) |
| Fat mass, kg | 0.40 (−2.89, 3.68) | 0.88 (−1.86,3.63) | −0.12 (−8.38, 8.13) | 0.85 (−0.89, 2.59) | 3.20 (−1.15, 7.54) | 0.54 (−1.37, 2.45) | 0.95 (−1.98, 3.88) | 0.85 (−2.52, 4.21) | 1.04 (−1.89, 3.96) | 0.67 (−2.08, 3.41) |
| Lipids | ||||||||||
| FIDL, mmol/L | −0.02 (−0.07, 0.02) | −0.04 (−0.16, 0.07) | 0.01 (−0.07, 0.10) | −0.05 (−0.10, −0.01) | 0.01 (−0.09, 0.10) | −0.05 (−0.10, 0.00) | 0.01 (−0.05, 0.06) | −0.05 (−0.11,0.00) | 0.00 (−0.05, 0.05) | −0.05 (−0.09, 0.00) |
| LDL, mmol/L | −0.16 (−0.31, −0.02) | 0.03 (−0.15, 0.21) | −0.06 (−0.23, 0.10) | −0.17 (−0.32, −0.03) | −0.18 (−0.39, 0.03) | −0.08 (−0.21, 0.06) | −0.13 (−0.28, 0.02) | −0.14 (−0.31, 0.04) | −0.12 (−0.26, 0.02) | −0.13 (−0.30, 0.04) |
| TC, mmol/L | −0.18 (−0.32, −0.03) | 0.08 (−0.14, 0.30) | 0.01 (−0.17, 0.20) | −0.18 (−0.34, −0.02) | −0.11 (−0.30, 0.09) | −0.12 (−0.29, 0.04) | −0.09 (−0.23, 0.06) | −0.23 (−0.47, 0.00) | −0.08 (−0.22, 0.06) | −0.22 (−0.44, 0.00) |
| Log TG, mmol/L | 0.03 (−0.02, 0.08) | 0.05 (−0.09, 0.18) | 0.01 (−0.09, 0.11) | 0.03 (−0.01, 0.08) | 0.06 (−0.02, 0.15) | 0.00 (−0.05, 0.05) | 0.05 (−0.03, 0.13) | 0.02 (−0.03, 0.06) | 0.06 (−0.01, 0.12) | 0.00 (−0.05, 0.05) |
| Non-HDL, mmol/L | −0.04 (−0.15, 0.08) | 0.09 (−0.18, 0.36) | 0.08 (−0.12, 0.29) | −0.06 (−0.17, 0.06) | −0.05 (−0.25, 0.15) | −0.01 (−0.11, 0.10) | −0.02 (−0.18, 0.15) | −0.02 (−0.14, 0.09) | −0.01 (−0.17, 0.15) | −0.02 (−0.13, 0.09) |
| apoB, mmol/L | −0.01 (−0.61, 0.60) | 0.13 (−0.26, 0.52) | 0.02 (−0.17, 0.21) | 0.09 (−1.20, 1.39) | 0.30 (−5.88, 6.49) | 0.06 (−0.21, 0.33) | 0.05 (−0.14, 0.25) | −1.80 (−7.18,3.57) | 0.05 (−0.14, 0.24) | −5.37 (−12.82, 2.09) |
| Glycemic traits | ||||||||||
| Fasting glucose, mmol/L | −0.12 (−0.41, 0.17) | 0.02 (−0.14, 0.18) | −0.37 (−1.10, 0.37) | 0.01 (−0.18, 0.20) | 0.01 (−0.42, 0.43) | −0.08 (−0.29, 0.12) | −0.12 (−0.40, 0.15) | −0.02 (−0.40, 0.37) | −0.09 (−0.33, 0.16) | −0.09 (−0.47, 0.30) |
| HbA1c, % | −0.01 (−0.09, 0.08) | 0.16 (−0.27, 0.59) | −0.12 (−0.41, 0.17) | 0.02 (−0.08, 0.12) | −0.07 (−0.19, 0.06) | 0.09 (−0.03, 0.21) | −0.10 (−0.33, 0.14) | 0.02 (−0.06, 0.10) | −0.05 (−0.16, 0.06) | 0.07 (−0.03, 0.18) |
| Log fasting insulin, mIU/L | −0.10 (−0.27, 0.08) | 0.08 (−0.16, 0.31) | −0.20 (−0.51, 0.11) | 0.01 (−0.15, 0.17) | −0.04 (−0.24, 0.16) | −0.05 (−0.28, 0.19) | −0.05 (−0.21, 0.11) | NA | −0.06 (−0.22, 0.10) | 0.07 (−0.43, 0.57) |
| Log HOMA-IRb | −0.07 (−0.30, 0.16) | 0.02 (−0.16, 0.20) | −0.03 (−0.21, 0.15) | NA | 0.01 (−0.26, 0.28) | −0.07 (−0.30, 0.16) | −0.04 (−0.23, 0.16) | NA | −0.03 (−0.22, 0.16) | −0.05 (−0.66, 0.55) |
| Log HOMA-β | −0.05 (−0.44, 0.34) | 0.07 (−0.18, 0.33) | 0.01 (−0.22, 0.25) | NA | 0.06 (−0.18, 0.31) | −0.05 (−0.44, 0.35) | 0.02 (−0.25, 0.28) | NA | −0.03 (−0.30, 0.24) | 0.26 (−0.23, 0.75) |
| Inflammatory factors | ||||||||||
| Log hsCRP, mg/L | 0.03 (−0.08, 0.13) | −0.02 (−0.36, 0.32) | −0.08 (−0.31, 0.14) | 0.08 (−0.08, 0.25) | 0.13 (−0.08, 0.35) | −0.03 (−0.12, 0.07) | −0.06 (−0.30, 0.17) | 0.08 (−0.09, 0.24) | 0.06 (−0.15, 0.27) | −0.02 (−0.11,0.08) |
| Log regularCRP, mg/L | 0.03 (−0.24, 0.30) | 1.45 (−1.66, 4.56) | 0.11 (−0.63, 0.84) | 0.74 (−0.53, 2.00) | 0.57 (−0.41, 1.55) | NA | 0.93 (−0.68, 2.54) | 0.01 (−0.23, 0.25) | 0.88 (−0.64, 2.39) | 0.01 (−0.22, 0.24) |
>β coefficients (95% CI) represent the changes in cardiometabolic traits per 1 serving/day increase in genetically predicted dairy intake; linear regression was used to test the association of MCM6 variant rs4988235 with dairy intake or cardiometabolic traits after adjustment of age, sex, ethnicity, region, total energy, and principal component for population stratification, as appropriate; We pooled β coefficients across studies using random-effect (I2≥25%) or fixed-effect (I2<25%) meta-analyses based on the heterogeneity between studies. We used the IV estimators to quantify the strength of the causal association of dairy intake and cardiometabolic traits in each study. The IV estimator, which is identical to that derived by the widely used 2-stage least-squares method, was calculated as the β of the regression coefficients MCM6 rs4988235-outcomes and MCM6 rs4988235-dairy.
HOMA: homeostatic model assessment, IR: insulin resistance, NA: not applicable.
Discussion
In thus far the largest MR analysis study, including 182041 adults from 18 cohorts, our results support a causal relationship between higher dairy intake and increased lean mass. In addition, our findings imply that the observational associations of dairy intake with lipids and glycemic traits could be the result of confounding.
In our well-powered study, we individually analyzed 182041 individuals and provided strong evidence that high dairy intake was causally associated with higher lean mass. Results from our observational analyses and our MR analyses were highly consistent, both suggesting higher lean mass in those with high intake of dairy products. In line with our findings, a previous meta-analysis of RCTs showed that dairy consumption increased lean mass (5). Several mechanisms might be responsible for the impact of dairy intake on the regulation of lean mass. First, increased protein intake from dairy products may promote maintenance of lean mass (7). Second, the hormone estrone found in dairy products may promote increases in body weight (18, 29, 30). Third, higher intake of dairy foods is associated with higher plasma insulinlike growth factor 1, which may contribute to weight gain (18, 31). However, future research is needed to further illustrate the precise mechanisms of dairy products on body composition in the context of energy restriction.
By using the LCT-13910 C/T as an instrument for dairy intake, our MR results indicated that higher dairy intake is marginally associated with decreased circulating concentrations of TC and LDL. In contrast, observational evidence from Mediterranean, Danish, and American populations suggested that milk intake was not associated with lipids (12, 13). However, our meta-analysis of observational studies showed that high dairy intake was significantly associated with lower LDL and TC. Such observations are supported by previous meta-analysis of controlled short-term intervention studies using a probiotic milk product, in which the fermented yogurt product was associated with a 4% decrease in TC and a 5% decrease in LDL cholesterol (9). Thus, it is possible that intake of probiotic milk products, fermented yogurt especially, drives the beneficial effect of intake of dairy products on lipid levels. It is worth noting that using the LCT-13910 C/T as an IV of dairy intake in general, rather than milk intake specifically, complicates the interpretation of our results. Previous studies indicate that the association between this genetic variant and dairy intake is specific to milk (1), possibly because of probiotics in some nonmilk dairy products (such as yogurt and fermented milk) that may facilitate the digestion of lactose and/or differences in lactose concentration. In the current study, the use of total dairy products including skim/low fat milk, whole milk, ice cream, yogurt, cottage/ricotta cheese, cream cheese, other cheese, and cream may largely attenuate our findings. Future study on the causal relationship between dairy-specific product and lipids is needed.
We did not find a causal association between dairy intake and glycemic traits such as fasting glucose, insulin, insulin sensitivity, and insulin resistance. Likewise, previous MR analyses demonstrated genetically high milk intake also did not influence plasma concentrations of glucose (12, 13). Our findings were also supported by a 3-week randomized crossover study indicating that both whole milk and skim milk did not affect fasting glucose or insulin in healthy adults (32). Our MR results may potentially explain the nonsignificant causal effect of high milk intake on risk of type 2 diabetes (11). However, previous MR analysis observed a significant sex difference in genetic association with fasting glucose (13), in which the T allele was significantly associated with lower fasting glucose in women but not in men. The association in women in which the T allele was associated with higher milk intake is inversely associated with fasting glucose (13). Further RCTs or MR investigations are needed to explore whether there is a true sex difference in genetic association of LCT-13910 C/T, rs4988235 with fasting glucose and whether milk intake may modulate such genetic association.
Several strengths of the current study merit consideration. First, in the present consortium-based effort involving 18 studies, we used a standardized analysis plan, which is less likely to be affected by publication bias than meta-analyses based on published reports. The large sample size allowed us to assess the consistency of associations across several studies and to gain sufficient power for conclusive estimation of causal effect. Second, the lactase-persistent variant is a well-established genetic marker for dairy intake, with solid biological basis and, therefore, a valid IV for dairy intake (10–12, 18). The instrument for carrying out this MR study has largely prevented potentially distorting influences. Our findings are of great benefit for future decision-making upon the development of novel behavioral interventions.
Furthermore, our MR results for lipids showed a suggestive causal effect of dairy consumption on improving lipids. This finding was supported by the results of a previous multicenter, randomized double-blind study among hypercholesterolemic patients demonstrating that consumption of dairy product favorably changed the lipid profile by reducing TC and LDL cholesterol (33).
Despite the convincing concept of MR analysis, several limitations have to be considered while interpreting our results. First, the MR study added to established study designs such as RCT without the ability to fully replace them. Second, we could not exclude the possibility of pleiotropic effects of the LCT genotype (a gene affects ≥2 apparently unrelated phenotypic traits). However, to our knowledge, no pleiotropic effect has been reported previously. This genetic variant is specific to milk, or at least has a stronger association with milk (1); therefore, the use of total dairy products may largely attenuate our findings. Furthermore, the associations of LCT genotype with lactase persistence and milk intake vary across populations. Although theLCT-13910 C/T is highly associated with lactase persistence and dairy intake in northern European populations, its association with dairy intake is not universal (34). Other single-nucleotide polymorphisms, including MCM6 rs3754686 at intron 15, occur more frequently in some global regions (35) and, therefore, represent plausible alternatives in diverse cohorts. Hence, bias from population stratification is deemed likely. Finally, differences in definition of total dairy products between studies might lead to the heterogeneity observed in some analyses and dilute the association.
In summary, the present study suggests a causal effect of higher dairy intake on increased lean mass. Our findings also suggest that the observational associations of dairy intake with lipids and glycemic traits could be the result of confounding. Our results emphasize that high intake of dairy may promote the maintenance of lean mass.
Supplementary Material
List of authors from the Mendelian Randomization of Dairy Consumption Working Group
Tao Huang,1,2,3*† Dianjianyi Sun,4,5† Yoriko Heianza,4,5† Helle K.M. Bergholdt,6† Meng Gao,1 Zhe Fang,1 Ming Ding,7 Alexis C. Frazier-Wood,8 Kari E. North,9 Eirini Marouli,10,11 Mariaelisa Graff,9 Caren E. Smith,12 Anette Varbo,13,14 Rozenn N. Lemaitre,15,16 Dolores Corella,17,18 Carol A. Wang,19 Anne Tjønneland,20 Kim Overvad,21,22 Thorkild I.A. Sørensen,23,24 Mary F. Feitosa,25 Mary K. Wojczynski,25 Mika Kähönen,26 Vera Mikkilä,27 TraciM.Bartz,16,28 BruceM.Psaty,29,30 DavidS.Siscovick,31 Rebecca D. Danning,32 George Dedoussis,33 Oluf Pedersen,23,34 Torben Hansen,23 Aki S. Havulinna,35 Satu Männistö,35 JeromeI.Rotter,36 LauraSares-Jäske,37 Mathew A. Allison,38 Stephen S. Rich,39 Jose V. Sorlí,17,18 Oscar Coltell,18,40 Craig E. Pennell,41,42 Peter Eastwood,43 Paul M. Ridker,32,44 Jorma Viikari,45,46 Olli Raitakari,47,48 Terho Lehtimäki,49,50 Mika Helminen,51,52 Yujie Wang,9 Panagiotis Deloukas,10,11,53 Paul Knekt,37 Noora Kanerva,35,54 Tuomas O. Kilpeläinen,23 Michael A. Province,25 the CHARGE consortium, Dariush Mozaffarian,55 Daniel I. Chasman,32,56,57 Børge G. Nordestgaard,13,14,58 Christina Ellervik,14,59,60*† and Lu Qi4,5,7*†
1 Department of Epidemiology and Biostatistics, School of Public Health, Peking University, Beijing, China; 2 Department of Global Health, School of Public Health, Peking University, Beijing, China; 3 Key Laboratory of Molecular Cardiovascular Sciences, Ministry of Education, Beijing, China; 4 Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, Tulane University, New Orleans, LA; 5 Tulane University Obesity Research Center, Tulane University, New Orleans, LA; 6 Department of Clinical Biochemistry, Naestved Hospital, Naestved, Denmark; 7 Department of Nutrition, Harvard School of Public Health, Boston, MA; 8 USDA/ARS Children’s Nutrition Research Center, Baylor College of Medicine, Houston, TX; 9 Department of Epidemiology, University of North Carolina, Chapel Hill, NC; 10 William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London, UK; 11 Centre for Genomic Health, Life Sciences, Queen Mary University of London, London, UK; 12 Jean Mayer USDA Human Nutrition Research Center on Aging at Tufts University, Boston, MA; 13 Department of Clinical Biochemistry and the Copenhagen General Population Study, Herlev and Gentofte Hospital, Copenhagen University Hospital, Copenhagen, Denmark; 14 Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark; 15 Department of Medicine, University of Washington, Seattle, WA; 16 Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA; 17 Department of Preventive Medicine and Public Health, University of Valencia, Valencia, Spain; 18 CIBER Fisiopatología de la Obesidad y Nutrición, Instituto de Salud Carlos III, Madrid, Spain; 19 School of Medicine and Public Health, Faculty of Medicine and Health, The University of Newcastle, New South Wales, Australia; 20 Danish Cancer Society Research Center, Copenhagen, Denmark; 21 Department of Public Health, Section for Epidemiology, Aarhus University, Aarhus, Denmark; 22 Aalborg University Hospital, Aalborg, Denmark; 23 Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark; 24 Department of Public Health, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark; 25 Department of Genetics, Washington University School of Medicine, Saint Louis, MO; 26 Department of Clinical Physiology, Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland; 27 Division of Nutrition, Department of Food and Environmental Sciences, Helsinki, Finland; 28 Department of Biostatistics, University of Washington, Seattle, WA; 29 Kaiser Permanente Washington Health Research Institute, Seattle, WA; 30 Cardiovascular Health Research Unit, Departments of Medicine, Epidemiology, and Health Services, University of Washington, Seattle, WA; 31 New York Academy of Medicine, New York, NY; 32 Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA; 33 Department of Nutrition and Dietetics, School of Health Science and Education, Harokopio University, Athens, Greece; 34 Faculty of Health Sciences, University of Aarhus, Aarhus, Denmark; 35 Department of Public Health Solutions, National Institute for Health and Welfare, Helsinki, Finland; 36 Institute for Translational Genomics and Population Sciences, Los Angeles BioMedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA; 37 National Institute for Health and Welfare, Helsinki, Finland; 38 Division of Preventive Medicine, Department of Family Medicine and Public Health, University of California San Diego, La Jolla, CA; 39 Center for Public Health Genomics and Department of Public Health Sciences, University of Virginia, Charlottesville, VA; 40 Department of Computer Languages and Systems, University Jaume I, Castellon, Spain; 41 School of Human Sciences, The University of Western Australia, Western Australia, Australia; 42 Western Australian Sleep Disorders Research Institute, Department of Pulmonary Physiological and Sleep Medicine, Sir Charles Gairdner Hospital, Western Australia, Australia; 43 School of Anatomy, Physiology and Human Biology, The University of Western Australia, Western Australia, Australia; 44 Division of Cardiovascular Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA; 45 Division of Medicine, Turku University Hospital, Turku, Finland; 46 Department of Medicine, University of Turku, Turku, Finland; 47 Department of Clinical Physiology and Nuclear Medicine, Turku University Hospital, Turku, Finland; 48 Research Centre of Applied and Preventive Cardiovascular Medicine, University of Turku, Turku, Finland; 49 Department of Clinical Chemistry, Fimlab Laboratories, Tampere, Finland; 50 Department of Clinical Chemistry, Finnish Cardiovascular Research Center Tampere, Faculty of Medicine and Health Technology, University of Tampere, Tampere, Finland; 51 Research, Development and Innovation Centre, Tampere University Hospital, Tampere, Finland; 52 Faculty of Social Sciences, Health Sciences, University of Tampere, Tampere, Finland; 53 Princess Al-Jawhara Al-Brahim Centre of Excellence in Research of Hereditary Disorders (PACER-HD), King Abdulaziz University, Jeddah, Saudi Arabia; 54 Department of Public Health, University of Helsinki, Helsinki, Finland; 55 Friedman School of Nutrition Science and Policy, Tufts University, Boston, MA; 56 Division of Genetics, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA; 57 Broad Institute of MIT and Harvard, Cambridge, MA; 58 The Copenhagen City Heart Study, Frederiksberg Hospital, Copenhagen University Hospital, Copenhagen, Denmark; 59 Department of Production, Research, and Innovation, Regionshuset, Soroe, Denmark; 60 Department of Laboratory Medicine, Boston Children’s Hospital and Harvard Medical School, Boston, MA.
* Address correspondence to: L.Q. at Department of Epidemiology, School of Public Health and Tropical Medicine, Tulane University, 1440 Canal St., Ste. 1724, New Orleans, LA 70112. lqi1@tulane.edu. C.E. at christina.ellervik@childrens.harvard.edu. T.H. at Department of Epidemiology and Biostatistics, School of Public Health, Peking University, Beijing, China. huangtao@bjmu.edu.cn.
† T. Huang, D. Sun, Y. Heianza, H.K.M. Bergholdt, C. Ellervik, and L. Qi contributed equally to this work.
Footnotes
Nonstandard abbreviations: RCT, randomized controlled trial; CRP, C-reactive protein; TC, total cholesterol; MR, Mendelian randomization; IV, instrumental variable; BMI, body mass index; TG, triglyceride; apoB, apolipoprotein B; HbA1c, hemoglobin A1c; hs, high sensitivity.
Human Genes: LCT, lactase; MCM6, minichromosome maintenance complex component 6.
T. Huang, Y. Heianza, D. Sun, and L. Qi designed the study, drafted the study protocol, planned analyses, and wrote the first draft of the paper. T. Huang, Y. Heianza, and D. Sun conducted the combined statistical analysis. All authors had reviewed and approved the drafts of the paper (Supplemental Table 9).
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership: None declared.
Consultant or Advisory Role: P. Eastwood, Nyxoah SA; D. Mozaffarian, GOED, Nutrition Impact, Pollock Communications, Bunge, Indigo Agriculture, Amarin, Acasti Pharma, Cleveland Clinic Foundation, America’s Test Kitchen, Danone, Elysium Health (with stock options), Omada Health, DayTwo.
Stock Ownership: None declared.
Honoraria: D. Mozaffarian, UpToDate.
Research Funding: C.A. Wang, NHMRC (to study group); M.K. Wojczynski, NHLBI and NIDDK (to study group); B.M. Psaty, NHLBI HL105756; J.I. Rotter, NIH; C.E. Pennell, NHMRC (to study group); M.A Province, NIH; L. Qi, NIH DK091718, NIH DK115679, NIH HL034594, NIH DK100383, NIH TW010790.
Expert Testimony: None declared.
Patents: D. Mozaffarian, US8889739, US9987243.
Other Remuneration: B.M. Psaty, service on the steering committee of the Yale Open Access Project funded by Johnson & Johnson.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
References
- 1.Hartwig FP, Davey Smith G. Lactase persistence and body mass index: the contribution of Mendelian randomization. Clin Chem 2018;64:4–6. [DOI] [PubMed] [Google Scholar]
- 2.Wang H, Troy LM, Rogers GT, Fox CS, McKeown NM, Meigs JB, et al. Longitudinal association between dairy consumption and changes of body weight and waist circumference: the Framingham Heart Study. Int J Obes (Lond) 2014;38:299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mozaffarian D, Hao T, Rimm EB, Willett WC, Hu FB. Changes in diet and lifestyle and long-term weight gain in women and men. N Engl J Med 2011;364:2392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louie JC, Flood VM, Hector DJ, Rangan AM, Gill TP. Dairy consumption and overweight and obesity: a systematic review of prospective cohort studies. Obes Rev 2011;12:e582–92. [DOI] [PubMed] [Google Scholar]
- 5.Abargouei AS, Janghorbani M, Salehi-Marzijarani M, Esmaillzadeh A. Effect of dairy consumption on weight and body composition in adults: a systematic review and meta-analysis of randomized controlled clinical trials. Int J Obes (Lond) 2012;36:1485–93. [DOI] [PubMed] [Google Scholar]
- 6.Chen M, Pan A, Malik VS, Hu FB. Effects of dairy intake on body weight and fat: a meta-analysis of randomized controlled trials. Am J Clin Nutr 2012;96:735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Booth AO, Huggins CE, Wattanapenpaiboon N, Nowson CA. Effect of increasing dietary calcium through supplements and dairy food on body weight and body composition: a meta-analysis of randomised controlled trials. Br J Nutr 2015;114:1013–25. [DOI] [PubMed] [Google Scholar]
- 8.Benatar JR, Sidhu K, Stewart RA. Effects of high and low fat dairy food on cardio-metabolic risk factors: a meta-analysis of randomized studies. PLoS One 2013;8:e76480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agerholm-Larsen L, Bell ML, Grunwald GK, Astrup A. The effect of a probiotic milk product on plasma cholesterol: a meta-analysis of short-term intervention studies. Eur J Clin Nutr 2000;54:856–60. [DOI] [PubMed] [Google Scholar]
- 10.Ding M, Huang T, Bergholdt HKM, Nordestgaard BG, Ellervik C, Qi L, et al. Dairy consumption, systolic blood pressure, and risk of hypertension: Mendelian randomization study. BMJ 2017;356:j1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergholdt HK, Nordestgaard BG, Ellervik C. Milk intake is not associated with low risk of diabetes or overweight-obesity: a Mendelian randomization study in 97,811 Danish individuals. Am J Clin Nutr 2015; 102:487–96. [DOI] [PubMed] [Google Scholar]
- 12.Bergholdt HK, Nordestgaard BG, Varbo A, Ellervik C. Milk intake is not associated with ischaemic heart disease in observational or Mendelian randomization analyses in 98,529 Danish adults. Int J Epidemiol 2015;44:587–603. [DOI] [PubMed] [Google Scholar]
- 13.Smith CE, Coltell O, Sorli JV, Estruch R, Martinez Gonzalez MA, Salas-Salvado J, et al. Associations of the MCM6-rs3754686 proxy for milk intake in Mediterranean and American populations with cardiovascular biomarkers, disease and mortality: Mendelian randomization. Sci Rep 2016;6:33188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133–63. [DOI] [PubMed] [Google Scholar]
- 15.Nelson CP, Hamby SE, Saleheen D, Hopewell JC, Zeng L, Assimes TL, et al. Genetically determined height and coronary artery disease. N Engl J Med 2015;372:1608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geng T, Smith CE, Li C, Huang T. Childhood BMI and adult type 2 diabetes, coronary artery diseases, chronic kidney disease, and cardiometabolic traits: a Mendelian randomization analysis. Diabetes Care 2018;41:1089–96. [DOI] [PubMed] [Google Scholar]
- 17.Ding R, Huang T, Han J. Diet/lifestyle and risk of diabetes and glycemic traits: a Mendelian randomization study. Lipids Health Dis 2018;17:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang T, Ding M, Bergholdt HKM, Wang TG, Heianza Y, Sun DJ, et al. Dairy consumption and body mass index among adults: Mendelian randomization analysis of 184802 individuals from 25 studies. Clin Chem 2018; 64:183–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang TG, Huang T, Li YP, Zheng Y, Manson JE, Hu FB, et al. Low birthweight and risk of type 2 diabetes: a Mendelian randomisationstudy.Diabetologia 2016;59:1920–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang T, Ren JJ, Huang JY, Li D. Association of homocysteine with type 2 diabetes: a meta-analysis implementing Mendelian randomization approach. BMC Genomics 2013;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Travis RC, Appleby PN, Siddiq A, Allen NE, Kaaks R, Canzian F, et al. Genetic variation in the lactase gene, dairy product intake and risk for prostate cancer in the European prospective investigation into cancer and nutrition. Int J Cancer 2013;132:1901–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I. Identification of a variant associated with adult-type hypolactasia. Nat Genet 2002; 30:233–7. [DOI] [PubMed] [Google Scholar]
- 23.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003;327: 557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. BMJ 2007;335:914–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. [DOI] [PubMed] [Google Scholar]
- 26.Borenstein M, Hedges LV, Higgins JP, Rothstein HR. A basic introduction to fixed-effect and random-effects models for meta-analysis. Res Synth Methods 2010;1: 97–111. [DOI] [PubMed] [Google Scholar]
- 27.Wald A. The fitting of straight lines if both variables are subject to error. Ann Math Stat 1940;11:284–300. [Google Scholar]
- 28.Palmer TM, Sterne JA, Harbord RM, Lawlor DA, Sheehan NA, Meng S, et al. Instrumental variable estimation of causal risk ratios and causal odds ratios in Mendelian randomization analyses. Am J Epidemiol 2011;173: 1392–403. [DOI] [PubMed] [Google Scholar]
- 29.Berkey CS, Rockett HR, Willett WC, Colditz GA. Milk, dairy fat, dietary calcium, and weight gain: a longitudinal study of adolescents. Archiv Pediatr Adolesc Med 2005;159:543–50. [DOI] [PubMed] [Google Scholar]
- 30.Remesar X, Tang V, Ferrer E, Torregrosa C, Virgili J, Masanes RM, et al. Estrone in food: a factor influencing the development of obesity? Eur J Nutr 1999;38:247–53. [DOI] [PubMed] [Google Scholar]
- 31.Ma J, Giovannucci E, Pollak M, Chan JM, Gaziano JM, Willett W, et al. Milk intake, circulating levels of insulin-like growth factor-I, and risk of colorectal cancer in men. J Nat Cancer Inst 2001;93:1330–6. [DOI] [PubMed] [Google Scholar]
- 32.Engel S, Elhauge M, Tholstrup T. Effect of whole milk compared with skimmed milk on fasting blood lipids in healthy adults: a 3-week randomized crossover study. Eur J Clin Nutr 2018;72:249–54. [DOI] [PubMed] [Google Scholar]
- 33.Mannarino E, Pirro M, Cortese C, Lupattelli G, Siepi D, Mezzetti A, et al. Effects of a phytosterol-enriched dairy product on lipids, sterols and 8-isoprostane in hyper-cholesterolemic patients: a multicenter Italian study. Nutr Metab Cardiovasc Dis 2009;19:84–90. [DOI] [PubMed] [Google Scholar]
- 34.Mulcare CA, Weale ME, Jones AL, Connell B, Zeitlyn D, Tarekegn A, et al. The T allele of a single-nucleotide polymorphism 13.9 kb upstream of the lactase gene (LCT) (C-13.9kbT) does not predict or cause the lactase-persistence phenotype in Africans. Am J Human Genet 2004;74:1102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enattah NS, Trudeau A, Pimenoff V, Maiuri L, Auricchio S, Greco L, et al. Evidence of still-ongoing convergence evolution of the lactase persistence T-13910 alleles in humans. Am J Human Genet 2007;81:615–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.