Abstract
Skeletal muscle is a highly metabolic and dynamic tissue that is formed through the complex and well-organised process of myogenesis. Although there is a good understanding about the role of the Muscle Regulatory Factors during myogenesis, little is known about the potential interplay of other metabolic proteins. The aim of this study was to determine the endogenous mRNA expression profile for a novel group of genes, recently associated with β2-adrenergic agonist (BA) induced muscle hypertrophy in pigs [1], during myogenic differentiation in C2C12 cells and their response to dibutyryl cyclic-AMP (dbcAMP). These genes included mitochondrial phosphoenolpyruvate carboxykinase (PCK2/PEPCK-M), genes involved in serine biosynthesis (Phosphoglycerate dehydrogenase, PHGDH; Phosphoserine aminotransferase-1, PSAT1; Phosphoserine phosphatase, PSPH) and those involved in an integrated stress response (Asparagine synthetase, ASNS; Sestrin-2, SESN2; and Activating transcription factor-5, ATF5).
A coordinated peak in endogenous PCK2, PHGDH, PSAT1, PSPH, ASNS, ATF5 and SESN2 mRNA expression was observed at day 2 of differentiation (P < 0.001) in C2C12 cells, which coincided with the peak in myogenin mRNA. Myotube hypertrophy was induced with dbcAMP (1 mM) treatment from day 0, thereby mimicking the in vivo BA response. Although dbcAMP treatment from day 0 induced larger myotubes and increased both myosin heavy chain-IIB (MyHC-IIB) and pyruvate carboxylase (PC) mRNA, the expression of PCK2, PHGDH, PSAT1 and ASNS mRNA were all unaffected. Treatment with dbcAMP from day 4 increased MyHC-IIB mRNA, however this was less dramatic compared to the response observed following treatment from day 0, but there was no effect on PC mRNA. There was also no effect of dbcAMP treatment from day 4 on PCK2, PHGDH, PSAT1 and ASNS mRNA.
To conclude, the coordinated day 2 peak in endogenous expression of PCK2, PHGDH, PSAT1, PSPH, ASNS, ATF5 and SESN2 mRNA may relate to a shift in biosynthetic demand required to initiate myogenic differentiation. However, dbcAMP had no effect on the expression of these genes in vitro suggesting that the effects observed in BA-treated pigs might be via other signalling pathways from the activation of the β2-adrenergic receptor, but independent of cAMP, or that there are species differences in the response.
Keywords: C2C12, dbcAMP, Hypertrophy, Myogenesis, PCK2/PEPCK-M, PHGDH
Highlights
-
•
ASNS, PCK2, PHGDH, PSAT1, PSPH show similar mRNA expression patterns in C2C12 cells.
-
•
Novel peak in C2C12 cell endogenous mRNA expression for all these genes at day 2.
-
•
Novel peak in mRNA expression at day 2 coincided with peak in Myogenin mRNA.
-
•
dbcAMP treatment from day 0 significantly increased both MyHC-IIB and PC mRNA.
-
•
ASNS, PCK2, PHGDH and PSAT1 mRNA was unaffected by dbcAMP treatment from day 0 or 4.
1. Introduction
Growth promoters, including β2-adrenergic receptor agonists (BA) and recombinant porcine growth hormone (GH), are used commercially in Australia, India and the USA to improve livestock feed efficiency [1]. BA and GH are repartitioning agents that increase weight gain by redirecting nutrients to muscle growth and reducing fat deposition, but via different mechanisms, which have yet to be fully elucidated [[2], [3], [4]]. We previously compared the effects of Ractopamine (BA) and Reporcin (GH) administered to growing gilts for up to 27 days on porcine skeletal muscle transcriptomes to identify mechanistic differences in the responses [5,6]. GH had a whole-body growth effect and increased liver weight, whereas BA induced greater increases in muscle weights. Both BA and GH induced a coordinated upregulation of a novel group of genes in muscle at day 3, but only BA induced a sustained effect (>7 days). This novel group of genes included those encoding proteins involved in serine biosynthesis (phosphoglycerate dehydrogenase, PHGDH; phosphoserine aminotransferase-1, PSAT1; phosphoserine phosphatase, PSPH), a mitochondrial gluconeogenic enzyme (phosphoenolpyruvate carboxykinase, PCK2/PEPCK-M) [5] and proteins involved in an integrated stress response (asparagine synthetase, ASNS; sestrin-2, SESN2; activating transcription factor-5, ATF5; and CCAAT/enhancer binding protein-γ, CEBPG/C/EBP-γ) [6]. This demonstrated, for the first time, that coordinated upregulation of this novel group of genes was associated with hypertrophic growth of terminally differentiated muscle, whereas previous studies had associated upregulation of PHGDH and PEPCK-M with cancer.
There has been increased interest in the genes involved in the serine biosynthesis pathway, since PHGDH was observed to be upregulated in certain breast cancers and reported to be key for cancer cell metabolism and survival [[7], [8], [9], [10]]. Serine is synthesised from 3-phosphoglycerate, an intermediate of glycolysis, through a three-step enzymatic process involving PHGDH, PSAT1 and PSPH. Serine plays an important role in growth and metabolism and can subsequently be used for the synthesis of amino acids, phospholipids, and nucleotides [11]. The upregulation of PHGDH indicates that serine biosynthesis is important during times of high metabolic activity and tumour growth, however little is known about the role of serine biosynthesis in muscle cell metabolism. Similarly there has been increased interest in the mitochondrial isoform of phosphoenolpyruvate carboxykinase (PEPCK-M) in recent years as it too is upregulated in certain cancers [12,13]. Phosphoenolpyruvate carboxykinase (PEPCK) is a gluconeogenic enzyme involved in the regeneration of phosphoenolpyruvate from oxaloacetate, but has also been identified to play a role in glyceroneogenesis, amino acid synthesis and cataplerosis [14]. There are two genes that encode for the two PEPCK isoforms: PCK1 encodes for cytosolic PEPCK (or PEPCK-C) and PCK2 for PEPCK-M. For many years, studies of PEPCK function focussed on the role of PEPCK-C and the specific role of PEPCK-M was overlooked, however PEPCK-M has been recently identified as a key regulator of cancer cell metabolism. Upregulation of PEPCK-M has been associated with the rewiring of energy metabolism to reroute glycolytic intermediates into serine biosynthesis in order to sustain cancer cell growth and survival through the activation of an endoplasmic reticulum stress response [12,15]. PCK2, but not PCK1, was identified in the same gene cluster as PHGDH, showing the same response profile to BA treatment in pigs [5]; and a similar mRNA expression profile was reported for ASNS, ATF5, CEBPG and SESN2 [6], which have all been linked to an integrated stress response [16,17]. The apparent co-ordinated upregulation of this novel group of genes suggests the possible transcriptional regulation by a common transcription factor or factors. A plausible candidate is Activating transcription factor-4 (ATF4), which forms a heterodimer with C/EBP-γ and regulates the transcription of various genes, including ASNS, ATF5 and SESN2 [16]. ATF4 has also been shown to upregulate genes involved in glycolysis, serine biosynthesis and amino acid transport, including PHGDH [18] and PCK2 [12], therefore ATF4 or ATF5 could potentially be associated or directly involved in co-ordinately regulating the transcription of this novel group of genes.
As mentioned, upregulation of these genes (PHGDH, PCK2, ASNS) has previously been observed in certain cancers, but we were the first to report the co-ordinated upregulation and transitional change associated with hypertrophic growth of terminally differentiated skeletal muscle [5,6]. The aim of the current study was to characterise the expression profiles for the novel group of genes (PCK2, PHGDH, PSAT1, PSPH, ASNS, SESN2, ATF5, CEBPG) during myogenic differentiation in vitro. Myogenesis is a highly coordinated process that involves the fusion of mononuclear muscle progenitor cells (myoblasts) to form multinucleated myotubes (in vitro) and muscle fibres (in vivo). The myogenic regulatory factors (MRFs), Myf5, MyoD, Myogenin and MRF4, are a family of basic helix-loop-helix transcription factors that are key for the transcriptional regulation of myogenesis [19] and each is expressed in a specific pattern [20]. Myf5 is the earliest MRF to be expressed, followed closely by MyoD [20], and both are essential for proliferation, specification and commitment to a muscle cell fate; whereas Myogenin and MRF4 are expressed later [20], associated with their role in the initiation of myogenic differentiation, leading to upregulation of various muscle-specific genes and contractile proteins, such as Myosin Heavy Chain (MyHC) isoforms. Therefore selected myogenesis associated genes were measured alongside the novel group of genes. The mouse myoblast C2C12 cell line is often used to model skeletal muscle differentiation in vitro as it provides a robust system that has the capacity to generate well-established myotube networks [21]. Therefore, we utilised C2C12 cells to perform comprehensive time-course experiments to study the endogenous mRNA expression profiles for this novel group of genes, as well as genes associated with myogenesis and skeletal muscle metabolism. Despite expressing β2-adrenergic receptors (β2-AR), it has been reported that C2C12 cells do not display a hypertrophic response to BA treatment [22]. Hence, C2C12 cells were treated with dibutyryl cyclic AMP (dbcAMP) from day 0 (as differentiation was initiated) or day 4 of differentiation (terminally differentiated myotubes) to induce down-stream signalling from the β2-AR and determine the effects on mRNA expression for the same group of genes.
2. Materials and methods
2.1. Cell culture materials and compounds
High glucose Dulbecco's modified eagle's medium (DMEM), dbcAMP and cell culture grade water were purchased from Sigma-Aldrich (Poole, UK) and other cell culture reagents were purchased from Fisher Scientific (Loughborough, UK). Cell culture grade water was used to resuspend dbcAMP sodium salt (CAS #: 16980-89-5; LOT #: SLBJ2610V), filter-sterilised through a 0.2 μm filter and stored in aliquots at −20 °C. dbcAMP was freshly diluted (1:100) into differentiation media (DM) on each day of treatment.
2.2. Cell culture
C2C12 cells were seeded onto 6-well plates in growth medium (DMEM supplemented with 10% foetal bovine serum and 1% penicillin/streptomycin) at a density of 1.5 × 105 cells per well and cultured as described previously [20]. At day 0 (~80% confluence), cells were switched to DM (DMEM supplemented with 2% horse serum and 1% penicillin/streptomycin), to induce myogenic differentiation and DM refreshed every 48 h.
To study endogenous mRNA expression, cells were harvested every 24 h. To investigate the effect of dbcAMP treatment, dbcAMP was diluted in DM then added from either day 0 (onset of differentiation) or day 4 (differentiated myotubes) until the end of each time-course. Preliminary work determined 1 mM as an effective dose of dbcAMP [23] and cell culture grade water was used as the vehicle control. Cells were harvested every 12 h or 24 h as appropriate.
2.3. Cell harvest
Medium was removed and cells harvested into 200 μl per well of ice cold RNase-free phosphate buffered saline at the indicated time-points. Samples were then stored at −80 °C until RNA extraction was performed. The number of replicate wells harvested per time-point and treatment varied between n = 3 and 6 (see Results for detail).
2.4. RNA extraction
Samples collected for endogenous mRNA expression were thawed into 800 μl TRIzol® reagent (Invitrogen, Paisley, UK) at room temperature and gently vortexed, then total RNA was extracted and DNase treated (Promega, Southampton, UK) as recommended in the manufacturer's guide. Total RNA was extracted from dbcAMP study samples using the High Pure RNA Isolation Kit (Roche Diagnostics, Mannheim, Germany) as described previously [20].
2.5. cDNA synthesis
NanodropTM 2000 spectrometer (Thermo Scientific, Wilmington, USA) was used to quantify RNA concentrations of each sample and then all samples were diluted to 50 ng/μl. RNA (500 ng) was reverse transcribed into cDNA using random hexamer primers, as supplied in the Transcriptor First Strand cDNA Synthesis kit (Roche Diagnostics, Mannheim, Germany), for the endogenous mRNA expression study; whereas RevertAid RT First Strand cDNA Synthesis kit (Thermo Scientific, Vilnius, Lithuania) was used for the dbcAMP study samples. In both cases, synthesis of cDNA was performed according to the manufacturer's guide.
2.6. Real-time quantitative PCR and quantification of total cDNA
All reagents for real-time quantitative PCR (QPCR) were purchased from Roche Diagnostics (Burgess Hill, UK). QPCR was performed using SYBR Green reagent to quantify mRNA transcript abundance, as previously described [20]. QPCR primers were designed using the Mus musculus genome (GRCm38) and tested in-house by analysis of the melt curve and gel electrophoresis. QPCR primers used in this study can be found in Table 1. QPCR primer sequences for myosin heavy chain (MyHC)-I [24], MyHC-IIB [25], Myf5 and myogenin [20] have been reported elsewhere. We previously reported [20] the use of OliGreen® reagent (Life technologies, Oregon, USA) to quantify total cDNA concentration and thereby normalise mRNA transcript abundance for each gene, due to problems in finding suitable reference genes that showed no changes in expression over the C2C12 differentiation time-course.
Table 1.
Forward and reverse murine primer sequences for QPCR.
| Gene | Forward primer (5’ → 3′) | Reverse primer (5’ → 3′) |
|---|---|---|
| ASNS | GAAGGAACTCTACCTGTTTGATGTT | GGGACTCTCAGTTCGAGACCGT |
| ATF5 | GAGAGGGAGGTCTCGTGTACGT | GATGGACTGGATAGGAAAGTGGAA |
| CEBPG | GAGGCGCAGGTACATGTGAA | CCTTTGCCAACACAGAATAGGTAGA |
| ENO3 | GCCTGCTCCTGAAGGTCAAC | TGCAAGTTTACAGGCCTGGAT |
| IDH2 | TTGAGGCTGAGGCTGCTCAT | CCGGCCCTTCTGGTGTT |
| PC | CCAACTTCGCTCACGTCTCA | GCGTTCTCATAGCCTACCTGCTT |
| PCK2 | GCAGAGCACATGCTGATTTTG | GGAAAGCAGCTGCCACGTA |
| PHGDH | CGTGAACTTGGTGAACGCTAAG | GTGGGAGGTGGTGACATTGAG |
| PSAT1 | CGTGCTTCAGCATCTACGTCAT | GCCCCGCCGTTGTTCT |
| PSPH | GGCATAAGGGAGCTGGTAAGC | GCCACCAGAGATGAGGAACAC |
| SESN2 | TCAGCGAGGTCAACAAGTTACTG | CAAGGCCTGGATATGCTCCTT |
2.7. Statistical analyses
For endogenous mRNA expression, one-way analysis of variance (ANOVA) was performed on normalised data using GraphPad Prism (7.03) software. Dunnett's multiple comparison test was used when appropriate (ANOVA P < 0.05) to compare time-points to day 0. For dbcAMP experiments, two-way ANOVA (time x treatment) was performed on normalised data using Genstat (19th edition), followed by Bonferroni post-hoc test when appropriate (ANOVA P < 0.05). Data are presented as means ± standard error of the mean (SEM). Significance was accepted at P < 0.05. Matrices for Pearson's r correlation coefficients were generated in Microsoft Excel.
3. Results
3.1. Endogenous mRNA expression in C2C12 cells across the time-course of myogenic differentiation
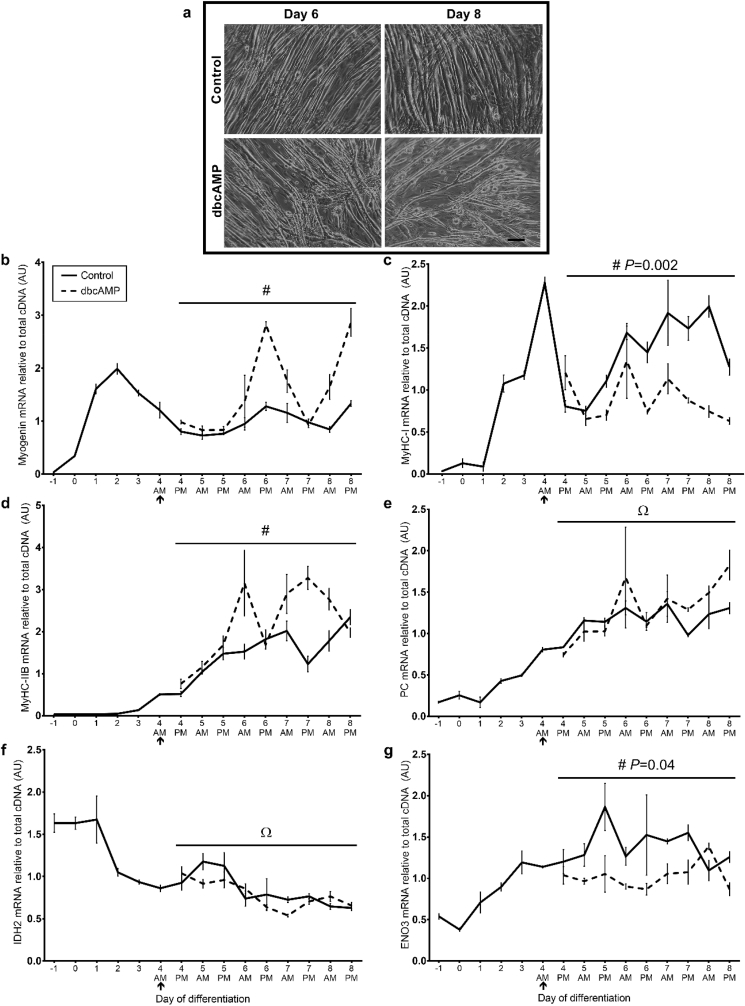
Early C2C12 myotubes were observed at day 2 of differentiation that increased in size with time resulting in a large extensive network of C2C12 myotubes by day 8 of differentiation (Fig. 1a). As we reported previously [20], Myf5 mRNA was significantly higher in proliferating C2C12 myoblasts compared to differentiating myotubes (P < 0.001; Fig. 1b), whereas myogenin mRNA expression was minimal in proliferating myoblasts, then as Myf5 mRNA decreased, myogenin mRNA increased and peaked between days 1 and 2 of differentiation followed by a gradual decline thereafter (P < 0.001; Fig. 1c). Similarly, the mRNA expression of the myosin heavy chain isoforms (MyHC-I and -IIB) showed similar patterns as those we previously reported [20]. MyHC-I mRNA was upregulated as differentiation was initiated, showing a similar profile to myogenin (P < 0.001; Fig. 1d), whereas MyHC-IIB was expressed much later, from day 3 of differentiation (P < 0.001; Fig. 1e). Relative mRNA expression of isocitrate dehydrogenase-2 (IDH2), a mitochondrial enzyme involved in the TCA cycle, enolase-3 (ENO3), a glycolytic enzyme, and pyruvate carboxylase (PC), a gluconeogenic enzyme, were determined as indicators of oxidative, glycolytic and gluconeogenic metabolism respectively. Relative IDH2 mRNA was higher in proliferating myoblasts and decreased with differentiation (P < 0.001; Fig. 1f), whereas ENO3 and PC were lower in proliferating myoblasts and increased in differentiated myotubes (both P < 0.001; Fig. 1g and h).
Fig. 1.
Endogenous mRNA expression profiles of genes associated with myogenesis and metabolism during C2C12 myogenic differentiation.
(a) Bright field photographs show C2C12 cells on days 0, 2 and 8 of differentiation. Photographs were captured at 6.3X magnification. Scale bar: 100 μm. Day 0 indicates the switch from growth to differentiation media. Relative mRNA expression of (b) Myf5, (c) myogenin, (d) myosin heavy chain (MyHC)-I, (e) MyHC-IIB, (f) isocitrate dehydrogenase-2 (IDH2), (g) enolase-3 (ENO3) and (h) pyruvate carboxylase (PC) were normalised to OliGreen® as a measure of total cDNA. Data is means (n = 4) ± SEM. Significant differences between individual time-points and day 0 of differentiation are indicated: P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***).
A strong positive correlation was observed between Myf5 and IDH2 (r = 0.955) but a negative relationship between Myf5 and both ENO3 (r = −0.739) and PC (r = −0.658), which suggests that as C2C12 cells differentiate they might switch from oxidative-dependent metabolism to more glycolytic. There were also strong positive correlations between MyHC-I and ENO3 (r = 0.728) and MyHC-IIB and PC (r = 0.893). Pearson's r correlation coefficients are displayed in Table A.1.
Relative mRNA expression levels for PCK2, ASNS, PHGDH, PSAT1 and PSPH were higher in proliferating myoblasts (between days −2 and −1 of differentiation) and decreased in differentiated myotubes from day 4 onwards (all P < 0.001; Fig. 2a-e). In contrast, SESN2 and ATF5 showed similar mRNA levels in proliferating myoblasts and differentiated myotubes, but there was a significant effect of time, due to increased expression at day 2 of differentiation (both P < 0.001; Fig. 2f and g). Interestingly, a peak in endogenous mRNA expression was observed at day 2 of differentiation for all 7 genes, which coincided with the peak in myogenin mRNA, suggesting a potential coordinated role in myogenic differentiation. In contrast, relative mRNA expression of CEBPG was low in proliferating C2C12 myoblasts, tended to be higher in differentiated myotubes and demonstrated a slightly later peak around day 3 of differentiation (P < 0.001; Fig. 2h). ATF5 is a transcription factor that has previously been reported to transcriptionally regulate similar gene targets as ATF4 in response to stress associated with nutrient deprivation [26]. Similar to ATF4, ATF5 forms a heterodimer with different C/EBPs that binds to regulate transcription of target genes. Pearson's r coefficient indicated positive correlations between ATF5 and PCK2 (r = 0.692), ASNS (r = 0.741), PHGDH (r = 0.504), PSAT1 (r = 0.664), PSPH (r = 0.715) and SESN2 (r = 0.922), whereas a negative correlation was observed between ATF5 and CEBPG (r = −0.796). This suggests that ATF5 might act as a possible transcriptional regulator of this group of genes during C2C12 cell myogenic differentiation; but CEBPG is unlikely to be a transcriptional regulator of these genes.
Fig. 2.
Endogenous mRNA expression profiles of serine biosynthesis and integrated stress response genes during C2C12 differentiation.
Relative mRNA expression of (a) phosphoenolpyruvate carboxykinase-2 (PCK2), (b) asparagine synthetase (ASNS), (c) phosphoglycerate dehydrogenase (PHGDH), (d) phosphoserine aminotransferase-1 (PSAT1), (e) phosphoserine phosphatase (PSPH), (f) sestrin-2 (SESN2), (g) activating transcription factor-5 (ATF5) and (h) CCAAT/enhancer binding protein (CEBPG) were normalised to OliGreen® as a measure of total cDNA. Data is means (n = 4) ± SEM. Significant differences between individual time-points and day 0 of differentiation are indicated: P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***).
The next step was to investigate the association between the mRNA expression of this group of genes with muscle cell hypertrophic growth in vitro. This was performed by treating C2C12 cells with dbcAMP, to mimic the downstream signalling effects of β2-AR stimulation, both at the onset of differentiation (from day 0) and once myotubes were sufficiently differentiated (from day 4).
3.2. Treatment of C2C12 cells with dbcAMP from day 0 of differentiation
Treatment with dbcAMP from day 0 resulted in larger myotubes after 2 and 5 days (Fig. 3a). After 5 days the cells were sparser but much larger myotubes were observed, as well as round bulging cells, suggesting that cells had migrated to form more mature myotubes, but had also started to contract off the plate.
Fig. 3.
Effect of dbcAMP treatment from day 0 of differentiation on C2C12 cells and mRNA expression.
(a) Bright-field images captured at days 2 and 5 of treatment with 1 mM dbcAMP or vehicle control (water) from day 0 of differentiation. Photographs were captured at 6.3X magnification. Scale bar: 100 μm. Relative mRNA expression of (b) myogenin, (c) myosin heavy chain (MyHC)-I isoform, (d) MyHC-IIB, (e) pyruvate carboxylase (PC), (f) IDH2 and (g) ENO3 were normalised to OliGreen® as a measure of total cDNA. Data is means ± SEM for n = 6 (day −1), n = 5 (day 0), n = 4 (day 2, 4 & 5 a.m.; day 3 dbcAMP a.m.) or n = 3 (day 1 a.m. & p.m.; day 2, 3, 4, 5 & 6 p.m.; day 3 control a.m.). #P < 0.001 (time × treatment interaction). Ω P < 0.01 (time effect). Φ P < 0.01 (treatment effect). ↑ indicates the start of treatment. See Table A.2 for Bonferroni post-hoc multiple comparisons.
There was no significant time × treatment interaction for myogenin mRNA (P > 0.1), but as seen previously myogenin was low in proliferating myoblasts, peaked around day 2 and then decreased with time (P < 0.001; Fig. 3b), while dbcAMP treatment significantly increased myogenin mRNA (P < 0.001). There were significant time × treatment interactions for relative mRNA expression of MyHC-I, MyHC-IIB and PC (all P < 0.001). Bonferroni post-hoc tests were performed and results can be found in Table A.2. As differentiation was initiated MyHC-I mRNA was upregulated, demonstrating a similar trend to myogenin, but with an additional large induction at day 6 (Fig. 3c). Treatment with dbcAMP from day 0 appeared to reduce this spike in MyHC-I mRNA at day 6 compared to cells treated with vehicle control. As observed previously, MyHC-IIB mRNA expression progressively increased with myogenic differentiation (Fig. 3d). Interestingly, PC mRNA expression showed a very similar pattern to MyHC-IIB mRNA (Fig. 3e) and treatment with dbcAMP resulted in a dramatic induction of both PC and MyHC-IIB mRNA from day 2 and day 3 of differentiation respectively. Strong positive correlations were observed between MyHC-IIB and PC mRNA expression in cells treated from day 0 with either vehicle control (r = 0.984) or dbcAMP (r = 0.946). Additional Pearson's r correlation coefficients can be found in Table A.3.
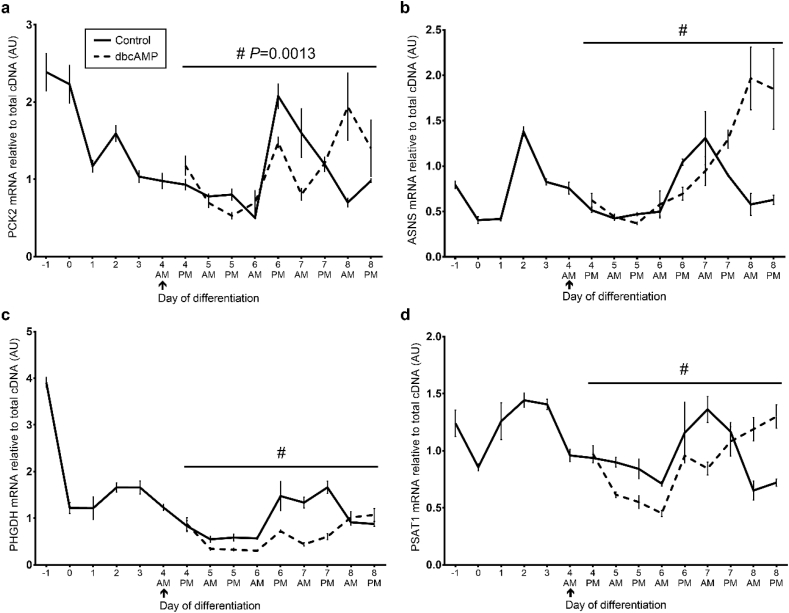
There were significant time × treatment interactions for expression of both IDH2 (P = 0.021; Fig. 3f) and ENO3 mRNA (P = 0.002; Fig. 3g). Treatment with dbcAMP from day 0 generally increased ENO3 mRNA at most time-points, whereas there was an inconsistent effect on IDH2. As in the previous experiment, expression of PCK2, PHGDH, PSAT1 and ASNS mRNA were generally higher in proliferating myoblasts than differentiated myotubes and there was again a peak at day 2 (all P < 0.01). Unexpectedly, dbcAMP treatment from day 0 reduced PCK2 and PHGDH mRNA compared to the control (P < 0.01; Fig. 4a and c) and had no effect on ASNS or PSAT1 mRNA (P > 0.1; Fig. 4b and d).
Fig. 4.
Effect of dbcAMP treatment from day 0 on serine biosynthesis and integrated stress response genes.
Relative mRNA expression of (a) PCK2, (b) ASNS (c) PHGDH and (d) PSAT1 following treatment with 1 mM dbcAMP or vehicle control (water) from day 0 of differentiation were normalised to OliGreen® as a measure of total cDNA. Data is mean ± SEM for. n = 6 (day −1), n = 5 (day 0), n = 4 (day 2, 4 & 5 a.m.; day 3 dbcAMP a.m.) or n = 3 (day 1 a.m. & p.m.; day 2, 3, 4, 5 & 6 p.m.; day 3 control a.m.). Ω P < 0.01 (time effect). Φ P < 0.01 (treatment effect). ↑ indicates the start of treatment.
Since treatment with dbcAMP from day 0 did not induce the expected coordinate upregulation of these genes, the next experiment used C2C12 cells differentiated for 4 days prior to treatment with dbcAMP.
3.3. Treatment of C2C12 cells with dbcAMP from day 4 of differentiation
Treating C2C12 myotubes with dbcAMP from day 4 of differentiation gave rise to sparser C2C12 myotubes as a result of clumping after 2 and 4 days of treatment (Fig. 5a). There appeared to be fewer myotubes per field of view, possibly due to migration of cells or, more likely, loss of mature myotubes due to them contracting off the plate. There was a significant time × treatment interaction for expression of myogenin and MyHC-I mRNA (both P < 0.01). Bonferroni post-hoc tests were performed and results can be found in Table A.4. Myogenin mRNA showed a similar pattern as observed previously, but the more extensive time-course identified two less prominent peaks around days 6 and 8, which dbcAMP treatment from day 4 further induced (Fig. 5b). This would appear to suggest multiple rounds of differentiation. MyHC-I mRNA showed a similar trend as previously reported, but with the addition of a spike in expression at day 4 a.m. followed by a decrease then a gradual increase up to day 8. Treatment with dbcAMP from day 4 of differentiation reduced MyHC-I mRNA, particularly between days 6 p.m. and 8 p.m.
Fig. 5.
Effect of dbcAMP treatment from day 4 of differentiation on C2C12 cells and mRNA expression.
(a) Bright-field images captured after 2 and 4 days of treatment with 1 mM dbcAMP or vehicle control (water) from day 4 of differentiation. Photographs were captured at 6.3X magnification. Scale bar: 100 μm. Relative mRNA expression of (b) myogenin, (c) MyHC-I, (d) MyHC-IIB, (e) PC, (f) IDH2 and (g) ENO3 were normalised to OliGreen® as a measure of total cDNA. Data is means ± SEM for. n = 6 (day 0; day 8 p.m.), n = 5 (days −1, 1, 2, 3; day 4 a.m.), n = 4 (days 7 a.m., 8 a.m.) or n = 3 (day 4 p.m., 7 p.m.; days 5, 6 a.m. and p.m. for each). #P < 0.001 (time × treatment interaction). Ω P < 0.01 (time effect). ↑ indicates the start of treatment. See Table A.4 for Bonferroni post-hoc multiple comparisons.
There was a significant time × treatment interaction for MyHC-IIB mRNA (P < 0.001; Fig. 5d), but not PC mRNA (P = 0.44; Fig. 5e). As expected, expression of both MyHC-IIB and PC (P = 0.0014) was low in proliferating myoblasts and generally increased during differentiation. Although less dramatic than treatment from day 0, there was a significant increase in MyHC-IIB mRNA on days 6 (a.m.) and 7 (p.m.) following dbcAMP treatment from day 4 (time × treatment interaction P < 0.001), whereas there was no effect of dbcAMP on PC mRNA (P = 0.157). There was a significant time × treatment interaction for ENO3 mRNA (P = 0.04; Fig. 5g), but not IDH2 (P > 0.05; Fig. 5f). As previously observed, ENO3 mRNA increased with differentiation, but this was reduced with dbcAMP; while IDH2 mRNA decreased with differentiation (P < 0.001), and there was no effect of dbcAMP (P = 0.145).
There were significant time × treatment interactions (P < 0.01) for expression of PCK2, ASNS, PHGDH and PSAT1 mRNA (Fig. 6a-d). Bonferroni post-hoc tests were performed and results can be found in Table A.4. As observed previously, mRNA expression was higher in proliferating myoblasts compared to differentiated myotubes and there was a peak at day 2, but also a later peak between days 6 and 7 of differentiation (similar to myogenin expression). Treatment with dbcAMP from day 4 appeared to reduce PHGDH and PSAT1 mRNA levels between days 5 and 7 but had little effect on PCK2 or ASNS mRNA. Pearson's r correlation coefficients can be found in Table A.5.
Fig. 6.
Effect of dbcAMP treatment from day 4 on serine biosynthesis and integrated stress response genes.
Relative mRNA expression of (a) PCK2, (b) ASNS, (c) PHGDH and (d) PSAT1 following treatment with 1 mM dbcAMP or vehicle control (water) from day 4 of differentiation were normalised to OliGreen® as a measure of total cDNA. Data is means ± SEM for n = 6 (day 0; day 8 p.m.), n = 5 (days −1, 1, 2, 3; day 4 a.m.), n = 4 (days 7 a.m., 8 a.m.) or n = 3 (day 4 p.m., 7 p.m.; days 5, 6 a.m. and p.m. for each). #P < 0.001 (time × treatment interaction). ↑ indicates the start of treatment. See Table A.4 for Bonferroni post-hoc multiple comparisons.
4. Discussion
This study involved extensive time-course experiments to determine changes in endogenous gene expression during C2C12 myogenic differentiation. It made the novel observation of a prominent day 2 peak in mRNA expression for a group of genes (PCK2, PHGDH, PSAT1, PSPH, ASNS, SESN2, ATF5) that had previously been shown to have a coordinate increase in expression associated with BA-induced muscle fibre hypertrophy in pigs [5,6]. The peak in endogenous expression at day 2 coincided with the peak in myogenin mRNA, therefore indicating a potential role for these genes in myogenic differentiation. Although it was already known that dbcAMP induced hypertrophy in C2C12 cells, this is the first time that the dramatic induction and strong positive correlation between MyHC-IIB and PC mRNA expression has been reported in response to dbcAMP treatment from day 0 of differentiation.
The transcription factor ATF4 has been described as regulating the expression of various genes, including PCK2, PHGDH, PSAT1, PSPH, ASNS, SESN2 and ATF5 [16,27]. However, in BA treated pigs no change in ATF4 gene expression was detected, therefore ATF4 was not measured in the current study. As seen previously in BA-stimulated muscle, there was an increase in the mRNA expression of the transcription factor, ATF5, coincident with an increase in PCK2, PHGDH, PSAT1, PSPH, ASNS and SESN2 at day 2 of differentiation in C2C12 cells. The coordinate upregulation of the same group of genes observed in this study and in response to BA treatment in the pig trial [5,6] indicates a potential role in myogenic differentiation in vitro, as well as hypertrophic muscle growth in vivo. Myogenic differentiation involves dramatic morphological changes, where mononucleated myoblasts fuse to form multinucleated myotubes. A transient peak between days −1 and 2 for a large group of genes related to cell adhesion, cell-cycle arrest and cell fusion has previously been reported [28], highlighting this as a key time for the switch from a proliferative to differentiated state. This transition is likely to be a highly demanding time, therefore the synchronous peak in PEPCK-M, serine biosynthesis and other metabolic genes might demonstrate the route by which biosynthetic demands are met. Muscle hypertrophy mainly occurs due to the recruitment and fusion of satellite cells, as well as the associated changes in the rate of protein synthesis, resulting in increased muscle mass [29]. Therefore, the induction of these metabolic genes observed during myogenic differentiation (at day 2) in vitro and in BA-stimulated muscle in vivo [5,6], might be associated with the biosynthetic demands related to cell fusion and hypertrophic growth. By upregulating this group of genes, we postulate that the flux of carbons through glycolysis and TCA cycle is subsequently diverted towards the synthesis of molecules that may become limiting (e.g. serine, asparagine, sphingolipids, etc.), rather than being used to produce lactic acid or being completely oxidised for the generation of ATP.
Although hypertrophic growth was observed in C2C12 cells treated with dbcAMP from day 0 and day 4 of differentiation, associated with the induction of myogenin and MyHC-IIB mRNA, dbcAMP treatment had little effect on the expression of PCK2, PHGDH, PSAT1 and ASNS mRNA in vitro. Treatment from day 0 induced an increase in ENO3 mRNA, followed by the dramatic induction of MyHC-IIB mRNA, which reflects observations in the pig trial [5], and coupled with a similar induction of PC mRNA. However, this was not observed when differentiated C2C12 myotubes were treated with dbcAMP from day 4. The difference in responses might demonstrate a species difference between pigs and mice, but it might also relate to the treatment used. The BA response in vivo involves activation of the β2-adrenergic receptor (β2-AR), which leads to downstream signalling mechanisms and increased intracellular cyclic AMP (cAMP) [30]. As mentioned previously, C2C12 cells are not responsive to BA treatment [22], therefore the addition of dbcAMP mimics the downstream response to BA by increasing intracellular cAMP, but without activating the β2-AR. Bypassing the β2-AR prevents downregulation of the receptor and therefore inhibition of the response, however alternative signalling pathways that branch from β2-AR activation, independent of cAMP, may also be bypassed. This might indicate that the upregulation in this group of genes resulting in muscle hypertrophic growth in pigs [5,6] is via another pathway, such as Ras/MAPK or PI3K/AKT [31]; however, further work is needed to prove or disprove this hypothesis.
As C2C12 cells progressed through myogenic differentiation we observed similar changes in mRNA expression for Myf5 and myogenin as reported previously [32,33]. This was associated with a decrease in IDH2 and an increase in ENO3 mRNA levels, suggesting a change in metabolic potential to support glycolytic metabolism and would agree with the observation of increased fast-glycolytic MyHC-IIB isoform mRNA as C2C12 myotubes mature. However, it has previously been reported that C2C12 cells become more oxidative as they differentiate, as indicated by an increased number and remodelling of the mitochondria [34,35]. This is supported by observations using Seahorse technology [36], which indicates some discrepancy between the direct metabolic and indirect gene expression data. Seahorse technology measures glycolysis based on the production of lactic acid, while oxidative metabolism is measured as oxygen consumption, but these measurements may not be truly representative of the flux of carbons through glycolysis and oxidative metabolism, especially given the other observed changes in gene expression (e.g. serine and asparagine biosynthesis). A good example of this are cancer cells as they demonstrate a dependence on increased glycolytic flux, but not all of the glycolytic flux results in lactate production [37]. Cancer cells alter their metabolic profile in order to sustain proliferation and growth by diverting glycolytic intermediates towards biosynthetic pathways, such as serine biosynthesis for protein synthesis and other macromolecules [10,15,38,39]. As lactate is not produced, Seahorse technology is unable to account for glycolytic flux feeding into these different pathways, and therefore most likely underestimates actual glycolytic flux. C2C12 myoblasts are highly proliferative so possibly demonstrate similar metabolic characteristics to generate biosynthetic intermediates and sustain myogenic differentiation, which would also be overlooked by Seahorse technology. Therefore we hypothesise that this glycolytic flux feeding into other biosynthetic pathways during proliferation could be maintained or even increased during myogenic differentiation, to meet the demands of C2C12 myoblast fusion and myotube maturation.
5. Conclusion
In summary, the same genes (PCK2, PHGDH, PSAT1, PSPH, ASNS, SESN2, ATF5) that were upregulated with BA-induced hypertrophy in pigs [5,6] demonstrated a coordinated upregulation in endogenous mRNA expression at day 2 of differentiation in C2C12 cells, that coincided with the peak in myogenin mRNA. This day 2 peak appears to be a key point of myogenic differentiation, where proliferative myoblasts become post-mitotic, and differentiate, potentially leading to a shift in biosynthetic demand. Therefore, this coordinated induction of metabolic gene expression may help sustain requirements during cell fusion and hypertrophy induced by BA treatment. The effects of dbcAMP treatment on mRNA expression in C2C12 cells were not the same as BA-induced effects on pig muscles. We hypothesise that these differences indicate that physical binding of a BA to the β2-AR is essential to activate alternative downstream signalling pathways (i.e. those not activated via cAMP) and induce the observed coordinate upregulation of these metabolic genes in vivo [5,6].
Funding
This work was supported by the Biotechnology and Biological Sciences Research Council [grant number BB/MM001385/1] and by Zoetis (formerly Pfizer Animal Health). MCB was supported by a PhD studentship funded by Zoetis and the University of Nottingham. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of competing interest
This research was funded by the BBSRC and Zoetis (formally Pfizer Animal Health).
Acknowledgements
MCB, JMB, TP and PTL conceived the experimental designs. MCB wrote the manuscript, prepared figures and performed data and statistical analyses. JMB and TP edited the manuscript. MCB and CL performed cell culture and RNA extractions. MCB and ZCTRD performed QPCR analysis. JMB, TP and PTL are the grant holders for this project.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2019.100694.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Brameld J.M., Parr T. Improving efficiency in meat production. Proc. Nutr. Soc. 2016;75(03):242–246. doi: 10.1017/S0029665116000161. [DOI] [PubMed] [Google Scholar]
- 2.Buttery P.J., Dawson J.M. Growth promotion in farm animals. Proc. Nutr. Soc. 1990;49(3):459–466. doi: 10.1079/pns19900054. [DOI] [PubMed] [Google Scholar]
- 3.Johnson B.J., Smith S.B., Chung K.Y. Historical overview of the effect of β-adrenergic agonists on Beef cattle production. AJAS (Asian-Australas. J. Anim. Sci.) 2014;27(5):757–766. doi: 10.5713/ajas.2012.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devesa J., Almengló C., Devesa P. Multiple effects of growth hormone in the body: is it really the hormone for growth? Clin. Med. Insights Endocrinol. Diabetes. 2016;9:47–71. doi: 10.4137/CMED.S38201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown D.M., Williams H., Ryan K.J., Wilson T.L., Daniel Z.C., Mareko M.H. Mitochondrial phosphoenolpyruvate carboxykinase (PEPCK-M) and serine biosynthetic pathway genes are co-ordinately increased during anabolic agent-induced skeletal muscle growth. Sci. Rep. 2016;6:28693. doi: 10.1038/srep28693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown D., Ryan K., Daniel Z., Mareko M., Talbot R., Moreton J. The Beta-adrenergic agonist, Ractopamine, increases skeletal muscle expression of Asparagine Synthetase as part of an integrated stress response gene program. Sci. Rep. 2018;8(1):15915. doi: 10.1038/s41598-018-34315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Possemato R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locasale J.W., Grassian A.R., Melman T., Lyssiotis C.A., Mattaini K.R., Bass A.J. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 2011;43(9):869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J., Chung F., Yang G., Pu M., Gao H., Jiang W. Phosphoglycerate dehydrogenase is dispensable for breast tumor maintenance and growth. Oncotarget. 2013;4(12):2502–2511. doi: 10.18632/oncotarget.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonov A., Agostini M., Morello M., Minieri M., Melino G., Amelio I. Bioinformatics analysis of the serine and glycine pathway in cancer cells. Oncotarget. 2014;5(22):11004–11103. doi: 10.18632/oncotarget.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalhan S.C., Hanson R.W. Resurgence of serine: an often neglected but indispensable amino acid. J. Biol. Chem. 2012;287(24):19786–19791. doi: 10.1074/jbc.R112.357194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Méndez-Lucas A., Hyroššová P., Novellasdemunt L., Viñals F., Perales J.C. Mitochondrial phosphoenolpyruvate carboxykinase (PEPCK-M) is a pro-survival, endoplasmic reticulum (ER) stress response gene involved in tumor cell adaptation to nutrient availability. J. Biol. Chem. 2014;289(32):22090–22102. doi: 10.1074/jbc.M114.566927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leithner K., Hrzenjak A., Trötzmüller M., Moustafa T., Köfeler H.C., Wohlkoenig C. PCK2 activation mediates an adaptive response to glucose depletion in lung cancer. Oncogene. 2015;34(8):1044–1050. doi: 10.1038/onc.2014.47. [DOI] [PubMed] [Google Scholar]
- 14.Yang J., Kalhan S.C., Hanson R.W. What is the metabolic role of phosphoenolpyruvate carboxykinase? J. Biol. Chem. 2009;284(40):27025–27029. doi: 10.1074/jbc.R109.040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent E.E., Sergushichev A., Griss T., Gingras M.-C., Samborska B., Ntimbane T. Mitochondrial phosphoenolpyruvate carboxykinase regulates metabolic adaptation and enables glucose-independent tumor growth. Mol. Cell. 2015;60(2):195–207. doi: 10.1016/j.molcel.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 16.Huggins C.J., Mayekar M.K., Martin N., Saylor K.L., Gonit M., Jailwala P. C/EBPγ is a critical regulator of cellular stress response networks through heterodimerization with ATF4. Mol. Cell. Biol. 2015;36(5):693–713. doi: 10.1128/MCB.00911-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A.M. The integrated stress response. EMBO Rep. 2016;17(10):1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams C.M. Role of the transcription factor ATF4 in the anabolic actions of insulin and the anti-anabolic actions of glucocorticoids. J. Biol. Chem. 2007;282(23):16744–16753. doi: 10.1074/jbc.M610510200. [DOI] [PubMed] [Google Scholar]
- 19.Brameld J.M., Buttery P.J., Dawson J.M., Harper J.M.M. Nutritional and hormonal control of skeletal-muscle cell growth and differentiation. Proc. Nutr. Soc. 1998;57(02):207–217. doi: 10.1079/pns19980033. [DOI] [PubMed] [Google Scholar]
- 20.Brown D.M., Parr T., Brameld J.M. Myosin heavy chain mRNA isoforms are expressed in two distinct cohorts during C2C12 myogenesis. J. Muscle Res. Cell Motil. 2012;32(6):383–390. doi: 10.1007/s10974-011-9267-4. [DOI] [PubMed] [Google Scholar]
- 21.Burattini S., Ferri P., Battistelli M., Curci R., Luchetti F., Falcieri E. C2C12 murine myoblasts as a model of skeletal muscle development: morpho-functional characterization. Eur. J. Histochem. 2004;48(3):223–233. [PubMed] [Google Scholar]
- 22.Wannenes F., Magni L., Bonini M., Dimauro I., Caporossi D., Moretti C. In vitro effects of Beta-2 agonists on skeletal muscle differentiation, hypertrophy, and atrophy. World Allergy Organ J. 2012;5(6):66–72. doi: 10.1097/WOX.0b013e31825eff8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown D.M. The University of Nottingham; 2015. Molecular Regulation of Skeletal Muscle Myosin Heavy Chain Isoforms.http://eprints.nottingham.ac.uk/28406/1/DAVID BROWN PhD THESIS.pdf Ph. D. Thesis. Available from. [Google Scholar]
- 24.Zhou Y., Liu D., Kaminski H.J. Myosin heavy chain expression in mouse extraocular muscle: more complex than expected. Investig. Ophthalmol. Vis. Sci. 2010;51(12):6355–6363. doi: 10.1167/iovs.10-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Da Costa N., Edgar J., Ooi P.-T., Su Y., Meissner J.D., Chang K.-C. Calcineurin differentially regulates fast myosin heavy chain genes in oxidative muscle fibre type conversion. Cell Tissue Res. 2007;329(3):515–527. doi: 10.1007/s00441-007-0441-3. [DOI] [PubMed] [Google Scholar]
- 26.Kilberg M.S., Balasubramanian M., Fu L., Shan J. The transcription factor network associated with the amino acid response in mammalian cells. Adv. Nutr. 2012;3(3):295–306. doi: 10.3945/an.112.001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quirós P.M., Prado M.A., Zamboni N., D'Amico D., Williams R.W., Finley D. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 2017;216(7):2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomczak K.K., Marinescu V.D., Ramoni M.F., Sanoudou D., Montanaro F., Han M. Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J. 2004;18(2):403–405. doi: 10.1096/fj.03-0568fje. [DOI] [PubMed] [Google Scholar]
- 29.Blaauw B., Reggiani C. The role of satellite cells in muscle hypertrophy. J. Muscle Res. Cell Motil. 2014;35(1):3–10. doi: 10.1007/s10974-014-9376-y. [DOI] [PubMed] [Google Scholar]
- 30.Rosenbaum D.M., Rasmussen S.G.F., Kobilka B.K. The structure and function of G-protein-coupled receptors. Nature. 2009;459(7245):356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belcheva M.M., Coscia C.J. Diversity of G Protein-Coupled receptor signaling pathways to ERK/MAP kinase. Neurosignals. 2002;11:34–44. doi: 10.1159/000057320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dedieu S., Mazères G., Cottin P., Brustis J.J. Involvement of myogenic regulator factors during fusion in the cell line C2C12. Int. J. Dev. Biol. 2002;46(2):235–241. [PubMed] [Google Scholar]
- 33.Ferri P., Barbieri E., Burattini S., Guescini M., D'Emilio A., Biagiotti L. Expression and subcellular localization of myogenic regulatory factors during the differentiation of skeletal muscle C2C12 myoblasts. J. Cell. Biochem. 2009;108(6):1302–1317. doi: 10.1002/jcb.22360. [DOI] [PubMed] [Google Scholar]
- 34.Wagatsuma A., Sakuma K. Mitochondria as a potential regulator of myogenesis. Sci. World J. 2013;2013:1–9. doi: 10.1155/2013/593267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sin J., Andres A.M., Taylor D.J.R., Weston T., Hiraumi Y., Stotland A. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy. 2016;12(2):369–380. doi: 10.1080/15548627.2015.1115172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong J., Kim B.-W., Choo H.-J., Park J.-J., Yi J.-S., Yu D.-M. Mitochondrial complex I deficiency enhances skeletal myogenesis but impairs insulin signaling through SIRT1 inactivation. J. Biol. Chem. 2014;289(29):20012–20025. doi: 10.1074/jbc.M114.560078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 38.Kit S. The biosynthesis of free Glycine and serine by tumors. Cancer Res. 1955;15(11):715–718. [PubMed] [Google Scholar]
- 39.Tedeschi P.M., Markert E.K., Gounder M., Lin H., Dvorzhinski D., Dolfi S.C. Contribution of serine, folate and glycine metabolism to the ATP, NADPH and purine requirements of cancer cells. Cell Death Dis. 2013;4(10):e877. doi: 10.1038/cddis.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.