Abstract
Intrinsically disordered proteins often play an important role in protein aggregation. However, it is challenging to determine the structures and interactions that drive the early stages of aggregation because they are transient and obscured in a heterogeneous mixture of disordered states. Even computational methods are limited because the lack of ordered structure makes it difficult to ensure that the relevant conformations are sampled. We address these challenges by integrating atomistic simulations with high-resolution single-molecule measurements reported previously, using the measurements to help discern which parts of the disordered ensemble of structures in the simulations are most probable while using the simulations to identify residues and interactions that are important for oligomer stability. This approach was applied to α-synuclein, an intrinsically disordered protein that aggregates in the context of Parkinson’s disease. We simulated single-molecule pulling experiments on dimers, the minimal oligomer, and compared them to force spectroscopy measurements. Force-extension curves were simulated starting from a set of 66 structures with substantial structured content selected from the ensemble of dimer structures generated at zero force via Monte Carlo simulations. The pattern of contour length changes as the structures unfolded through intermediate states was compared to the results from optical trapping measurements on the same dimer to discern likely structures occurring in the measurements. Simulated pulling curves were generally consistent with experimental data but with a larger number of transient intermediates. We identified an ensemble of β-rich dimer structures consistent with the experimental data from which dimer interfaces could be deduced. These results suggest specific druggable targets in the structural motifs of α-synuclein that may help prevent the earliest steps of oligomerization.
Significance
Oligomers formed by disordered proteins in the initial stages of protein aggregation play important roles in many neurodegenerative diseases. However, they are difficult to characterize because they form only transiently within a heterogeneous structural population. Combining the sensitivity of high-resolution single-molecule measurements with the atomistic insight provided by computational simulations provides a powerful window on the mechanisms driving aggregation. Here, we apply this approach to α-synuclein, comparing simulations of dimers (the minimal oligomer) to force spectroscopy measurements. By identifying an ensemble of β-rich conformations consistent with experimental data, we deduce interfaces between monomer domains that can be targeted to inhibit the early, most toxic stages of aggregation.
Introduction
Most proteins adopt ordered three-dimensional structures characterized by a funnel-shaped energy landscape with a well-defined conformational minimum (1, 2, 3). However, some proteins are instead structurally disordered, featuring a flat but rough energy landscape lacking a well-defined minimum (4, 5) in which the lowest-energy states consist of an ensemble of different conformations (6). Such intrinsically disordered proteins (IDPs) play a wide range of biological roles. Structural disorder is often essential to their function, allowing IDPs to interact with a range of target proteins because of structural variations in disordered regions. IDPs may also fold when interacting with their targets (7, 8), indicative of the complex effects of the environment on IDP structure. In addition to their functional roles, however, IDPs are of great interest because of their strong association with aggregation and disease. Aggregates of misfolded IDPs are featured in many important diseases, including Alzheimer’s, Parkinson’s, Huntington’s, and type II diabetes (9), with small oligomeric aggregates generally thought to be the toxic agents (10, 11, 12). IDPs have thus become important drug targets, with the goal of inhibiting IDP aggregation to prevent the associated diseases (9, 13).
Unfortunately, structure-based drug-design strategies are hampered by the fact that IDPs are notoriously difficult to characterize structurally. Although conventional probes of static structure like crystallography and cryoelectron microscopy have been used to solve structures of fibrils made of IDPs or IDP fragments (14), they are difficult to apply to the transient oligomeric species that are likely most relevant to disease (8, 15, 16). Techniques like NMR that are capable of capturing dynamic information can provide some insight into the conformations adopted by these proteins (17). However, they are limited by averaging over the ensemble of fluctuating structures explored, and typically provide little if any information about individual conformations and their relative abundances (15), making interpretation difficult. Computational simulations provide an alternative approach that can reveal atomic-resolution structural information about IDPs, but they, too, face significant technical hurdles: obtaining adequate sampling for atomistic simulations is challenging because of the flat, rough landscape (18, 19), typically requiring the application of tempering techniques (20) and/or the use of very long simulations (up to the millisecond timescale) (21).
An approach that has been increasingly used to study IDPs is to apply single-molecule methods, which allow heterogeneous mixtures and fluctuating ensembles to be characterized more directly and with high sensitivity for rare species (5, 22). Fluorescence and force probes have been used to study the intermolecular interactions driving oligomerization, the rate of aggregation, and associated structural changes in a variety of IDPs (12, 23, 24, 25, 26, 27, 28, 29, 30, 31). It is challenging to obtain detailed structural information from such methods, however, because they typically monitor just the distance between the points on the molecule where the probes are placed. Combining single-molecule experiments with atomistic computations helps to overcome this limitation and enhance the information that can be obtained. Such a strategy has been applied previously to help interpret fluorescence (32, 33) and atomic force microscopy (AFM) experiments (30, 34, 35), but not yet to measurements using optical tweezers, which are a more sensitive probe of rare and marginally stable structures like those expected in IDPs (36, 37).
Here, we combine atomistic simulations and optical tweezers measurements of α-synuclein, which aggregates in Lewy bodies in the context of Parkinson’s disease and certain dementias (38, 39), focusing on dimer formation as the first step in aggregation. α-Synuclein is generally disordered in vitro with little secondary structure (40, 41), despite forming some long-range contacts (42, 43). It can take on a variety of more ordered structures under different conditions, from helical when interacting with lipids (44, 45) to β-rich at low pH (46), in larger oligomers (12), and in fibrils (47, 48). Metastable species with a defined secondary structure are thus energetically accessible and may be transiently observed (Fig. 1). α-Synuclein has been studied extensively with fluorescence probes, both by fluorescence correlation spectroscopy (25, 49) and Förster resonance energy transfer (12, 25, 49, 50, 51). Förster resonance energy transfer, in which the energy transfer between two nearby fluorophores is used to measure the distance between them, has also been used to constrain computational simulations (32). Force spectroscopy, in which a force probe is used to unfold the protein via mechanical tension and measure the distance between the end points of the molecule, has been used to study the conformational dynamics of α-synuclein and its small oligomers (24, 28, 29, 52, 53, 54, 55, 56, 57, 58, 59, 60), but simulations have not yet been combined with force spectroscopy data.
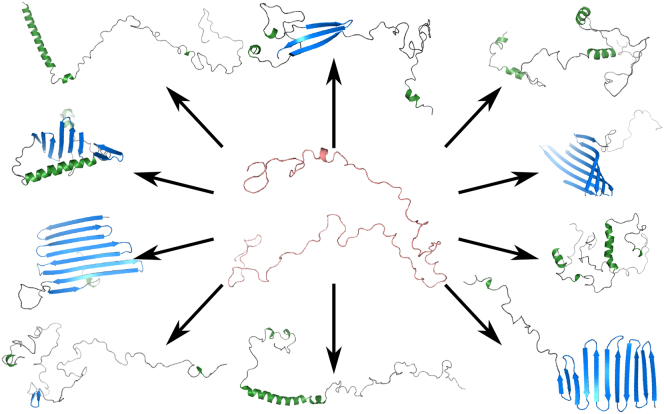
Figure 1.
Intrinsically disordered α-synuclein monomers may adopt rare, metastable species with a defined secondary structure. To see this figure in color, go online.
Previous work using optical tweezers (29, 37) found that α-synuclein dimers could be captured in metastable conformations displaying rich structural features in a nonnegligible fraction (30%) of measurements. Such well-structured conformations, although transient, are of interest because they may provide a target for drug candidates aiming to disrupt aggregation by inhibiting dimer formation or stabilizing dimers as off-pathway intermediates. Here, we characterize these transient structures by using Monte Carlo (MC) simulations to generate the structural ensemble populated by α-synuclein dimers and then testing structures showing significant ordering with simulations of force spectroscopy experiments. By comparing the simulated pulling curves to experimental data, we identify structures consistent with experiment that are therefore likely formed by dimers, and we deduce the interfaces between domains that stabilize the dimers and may thus provide drug targets for inhibiting aggregation.
Methods
Force spectroscopy experiments
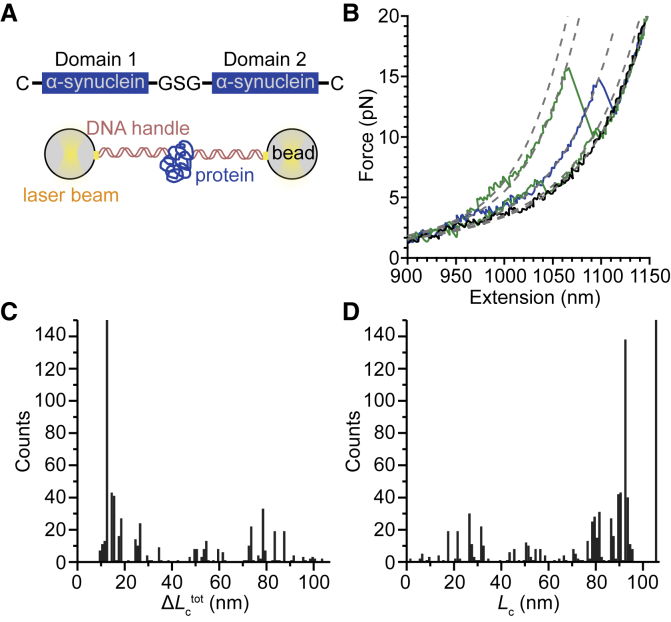
Dimers of α-synuclein were studied under tension not because force is implicated in the mechanism of protein aggregation, but because it allows the population of rarely and transiently occupied structures to be probed with high precision and sensitivity. Single-molecule force spectroscopy (SMFS) measurements of α-synuclein dimers made with high-resolution optical tweezers were reported previously (29, 37). Briefly, dimers were expressed as a single polypeptide, with the domains connected head to tail by a GSG peptide linker, then connected to beads held in optical traps via DNA handles (Fig. 2 A). Ramping the force up while recording the end-to-end extension generated a force-extension curve (FEC) in which the unfolding of structural elements appeared as sawtooth-like “rips” in the FEC (Fig. 2 B). The contour length of unfolded protein, Lc, was determined for each rupture event by fitting the rip using an extensible worm-like chain (WLC) model (61) as follows:
| (1) |
Here, F is the applied force, x is the extension, Lp is the persistence length, K is the enthalpic elasticity, and kBT is the thermal energy. Two WLCs in series were used, one for the DNA handles (Lc ∼900 nm, Lp ∼40 nm, K ∼1200 pN) and the other for the unfolded protein (Lp = 0.85 nm; K = 2000 pN) (29).
Figure 2.
Force spectroscopy of α-synuclein dimers. (A) A covalently linked α-synuclein dimer (upper image) is held under tension via DNA handles bound to beads trapped optically (lower image). (B) Ramping the force up to unfold any structures formed, most times a roughly featureless curve is seen (black), reflecting largely disordered structures. Some curves reveal rips indicating discrete unfolding of metastable structures (light colors). Different branches of the curves are fit to WLC models (dashed lines) to obtain the contour length of unfolded protein, Lc. (C) The histogram of ΔLctot from all curves with discrete rupture events shows most structures are small, but some are large. (D) The histogram of Lc for every state observed in the curves shows multiple peaks. To see this figure in color, go online.
Roughly 30% of the 1769 FECs measured displayed rupture events, indicating the presence of a mechanically stable conformation; some included more than one rip, indicating that the unfolding proceeded via at least one intermediate state. In these cases, each branch of the FEC was fit to a WLC to determine the length of unfolded protein in that conformation. The contour lengths were analyzed in two ways. First, we determined the total change in contour length measured from the most-folded state to the completely unfolded state, ΔLctot (Fig. 2 C). This total contour length change reflected the size of the structure that was unfolded. Next, we cataloged the contour length of unfolded protein in each branch of the FEC (folded, unfolded, and intermediates if present), representing the amount of the protein that remained folded at each point in the FEC (Fig. 2 D). The size of the incremental change in contour length during a given unfolding event, ΔLc, was found by subtracting the Lc values for adjacent branches of an FEC. Note that all FECs ended in the unfolded state, whose contour length (Lc = 105 nm) was known from the design of the dimer.
Simulations of the conformational ensemble
To overcome the sampling problems that can hinder molecular dynamics (MD) methods (15) for simulating proteins like α-synuclein that feature large fluctuations and conformational diversity (Fig. 1), we used MC simulations instead. An ensemble of α-synuclein dimer structures was first generated using the PROFASI software package (62), following procedures described previously (63). PROFASI employs an atomistic potential in which bond lengths and angles are fixed, but torsion angles remain as degrees of freedom. The interaction potential,
| (2) |
contains an electrostatic local backbone potential (Eloc), an excluded volume potential with 1/r12 repulsion (Eev), a potential for hydrogen bonds between the backbone and the backbone or charged side chains (Ehb), and an effective hydrophobic attraction between nonpolar side chains (Ehp). This simplified atomistic potential allowed more efficient sampling of the conformational space of the dimer and has been applied previously to study oligomer formation by IDPs (64, 65); details of the energy terms can be found in Eq. S1–S4. Because it was calibrated against structured proteins (66, 67), this potential may tend to oversample secondary structures when applied to IDPs, as with standard force fields like CHARMM22 (68) and AMBER 14 (69), but such a bias is not a major concern in the current work because the goal is to sample the possible range of conformations containing significant amounts of secondary structure (because these are the conformations being compared to experiment), rather than to obtain a properly balanced distribution of both structured and disordered conformations. An implicit solvent force field to account for water-protein interactions was used to avoid the high computational cost of explicit solvent simulations in the large simulation box needed for fully exploring the configuration space of disordered proteins. Additional information regarding the PROFASI potential is available in the literature (70, 71). The same protein sequence used in measurements was used in the simulations.
Each simulation used to construct the conformational ensemble included 5.5 million MC sweeps involving elementary steps that allowed for changes over selected degrees of freedom of the molecule. Here, the elementary steps included conformational updates involving rotations in side-chain degrees of freedom, pivots to rotate the backbone bonds, and biased Gaussian steps for semilocal internal rotation around backbone bonds (move frequencies were 22.5, 55, and 22.5%, respectively). 16 independent simulations were performed at each of three different temperatures, 310, 320, and 330 K, starting from the fully unfolded state of the protein. Note that the temperature scale used in PROFASI simulations does not represent true physical temperatures and is instead calibrated based on the Trp-cage phase transition (62). The temperature range 310–330 K was chosen to allow representative access to all of the major peaks in the energy distribution and thereby obtain sufficiently varied sampling of α-synuclein dimer structures (63). Convergence of the ensemble was verified from the evolution of the energy distribution of the ensemble, with the change in energy distribution between successive steps becoming close to zero by the end of the simulation (Fig. S1).
The secondary structure content of the structures in each simulation was analyzed using the cpptraj component (72) of AmberTools15 (69). The structures found in MC simulations tended to be richer in β-strand content compared to α-helical content, as previously reported (63). Structures containing β-sheets, α-helices, and combinations of the two were selected from the energy distributions (Fig. 3 A) for use in pulling simulations as follows. First, 200 structures were selected randomly from each peak at each temperature, then all disordered or mostly disordered structures were discarded, leaving structures with substantial structured content (∼20–70% structure). The resulting 66 structures were probed with pulling simulations.
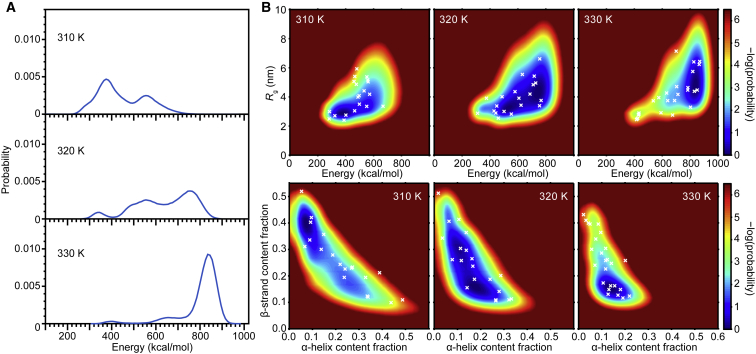
Figure 3.
α-Synuclein dimer ensembles from MC simulations at different temperatures. (A) The energy distribution shows two or three peaks for each temperature. (B) Shown are the distributions of radius of gyration (Rg) versus energy (top) and fraction of β-strand versus α-helix (bottom) at each temperature. White crosses mark the conformations used in pulling simulations. To see this figure in color, go online.
Unfolding simulations
The mechanical unfolding of each of these structures was simulated using the PROFASI MC code for constant-velocity pulling (73, 74). A harmonic force was applied to the protein ends along the vector between terminal Cα atoms, modifying the potential in Eq. 2 to (73) as follows:
| (3) |
where the total energy is increased by the addition of a harmonic term that depends on the spring constant of the force probe (k) and the distance between the ends of the molecule (x) and force probe (x0 + vt) as a function of the MC step “time,” t. Simulations were conducted with k = 37.0 pN/nm and a velocity of v = 0.05 fm/step, values used in previous PROFASI pulling simulations (62, 74), and at a temperature of 318 K that corresponds to the phase-transition temperature for the dimers found in the conformational-ensemble simulations. The force constant was larger than that used experimentally to reduce the time needed for computations.
MC sweeps were continued until the protein was fully extended. Elementary steps included rotations in side-chain degrees of freedom and biased Gaussian steps for the semilocal rotation of backbone bonds, each with a frequency of 50%. We ran 12 independent simulations for each of the 66 structures to capture the random nature and variations in the unfolding, resulting in 792 pulling trajectories. The force, extension, and structure were collected at each MC step in a trajectory. The force and extension were plotted as a running average over 5000 steps to generate an FEC similar to experiments (Fig. 4, A–F). Simulated FECs were analyzed in the same way as experimental FECs, fitting each rupture event in the FEC to Eq. 1 to determine Lc for each state in the pulling curve. We note that MC simulations of pulling experiments provide only a crude representation of the microscopic dynamics but should capture the essential barriers and wells during the unfolding process when small steps are used (74, 75). We therefore focus the comparisons to experiment on the structures observed in the simulations rather than dynamical information reflected in the unfolding forces.
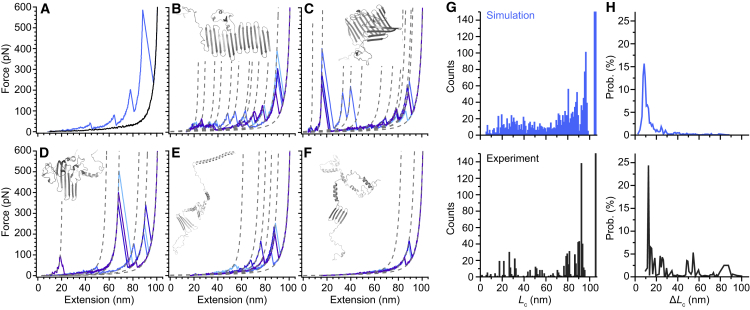
Figure 4.
Simulated pulling curves for α-synuclein dimers. (A) Some FECs show discrete rupture events (light blue), reflecting the unfolding of metastable structures; whereas others do not (black), reflecting largely disordered states. (B–F) Replicates of pulling curves simulated for a selection of structures studied are shown. Curves were fit with WLC models (dashed lines) to identify Lc for each state. Replicate trajectories visited the same set of intermediates, as found from Lc values, indicating reproducibility. (G) The histograms of Lc for all rupture events found in every FEC show generally similar values in simulations (upper panel) compared to experiments (lower panel). (H) The histograms of the probability of a given ΔLc value show larger transitions are more likely to occur in experiments (lower panel) than simulations (upper panel). To see this figure in color, go online.
Results
SMFS measurements of α-synuclein dimers displayed a wide variety of behavior, as would be expected for a disordered protein. Most FECs showed evidence only of marginally stable, rapidly fluctuating structures consistent with NMR observations of a compact-coil state (42), but ∼30% displayed rips reflecting the unfolding of mechanically stable structures that varied significantly between pulls (Fig. 2 B), reflecting metastable structures captured during the pulling (29). We first used MC simulations to generate a diverse ensemble of structured conformations of the α-synuclein dimer. Examining the distribution of energies for the dimer conformations in these simulations at 310–330 K (Fig. 3 A), we found two or three peaks. To ensure that a diverse subset of the structures in these distributions was used in pulling simulations for comparison with the experiments, we selected 200 conformations at random from each peak in the energy distribution and discarded those that were mostly disordered, thereby obtaining 66 structures containing substantial (up to 72%) metastable secondary structure from the different peaks (because only structures with a substantial amount of secondary structure would be likely to resist force). These structures were distributed broadly with respect to energy, radius of gyration, and secondary-structure content (Fig. 3 B). As a general rule, highly ordered β-rich structures were present at lower energies, whereas helical content and more disordered structures became more likely at higher energies.
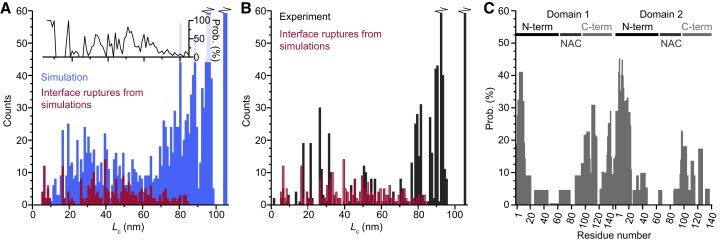
Pulling simulations were performed on each of the 66 structures selected, using 12 replicas per structure to capture stochastic fluctuations in the unfolding. Simulated FECs showed qualitatively similar features to the experimental FECs: some curves contained well-defined rips corresponding to the unfolding of mechanically stable domains before the fully unfolded state was reached at a contour length of 104–105 nm (Fig. 4 A, light blue), whereas others did not (Fig. 4 A, black). Sets of FECs for selected structures are displayed in Fig. 4, B–F; the replicate trajectories included in each panel illustrate the variability between individual pulls but show that rips tended to recur at the same contour lengths found from WLC fits (dotted lines), confirming that the same characteristic structural transitions were being probed. The distribution of values for the contour length of unfolded protein at each well-defined state in the FEC was also broadly similar for simulations (Fig. 4 G, upper panel) and experiments (Fig. 4 G, lower panel). However, a number of quantitative differences from the experimental FECs were observed. Unfolding forces were higher in the simulations than experiments, presumably reflecting a much higher effective loading rate, although the approximate dynamics in the MC simulations makes quantitative comparisons unreliable. Simulated FECs also tended to include many more intermediate states than experimental FECs: as many as 10 rupture events were observed in a single simulated FEC compared to a maximum of three in experimental FECs (Fig. S2). As a result, fewer transitions with large ΔLc were seen in simulations (Fig. 4 H, upper panel) than in experiments (Fig. 4 H, lower panel).
No discrete rupture events were observed in FECs from 345 of the 792 simulated pulling trajectories. These FECs included most of the pulling simulations starting from conformations with a lower content of stable secondary structure (<20% β-strand content, 305 trajectories). Occasionally, a disordered structure rearranged during pulling to form antiparallel β-strands, producing a low-force rupture (Fig. S3). Interestingly, conformations consisting primarily of helices (e.g., a structure with 47% α-helical content and 4% β-strand content) also lacked discrete rupture events, instead producing FECs featuring continuous (noncooperative) unfolding transitions (Fig. S4). Discrete ruptures were thus observed in 447 pulling trajectories, all resulting from structures with primarily β-sheet content.
We next refined the subset of simulated FECs containing discrete ruptures to identify which ones were consistent with the experimental FECs before examining the structural information about the dimers encoded in the simulations. As a first step, we calculated the total ΔLc unfolded across all ruptures in a given FEC, as a measure of the size of the structure that unfolded, comparing each simulated FEC to the set of values obtained experimentally (Fig. 2 D). If the simulated FEC had a ΔLctot within ±2 nm of a value observed experimentally, then it was judged to be consistent. For each of the simulated FECs passing this test, we then compared the set of Lc values for the intermediates in the FEC to the set of values observed in every experimental FEC to determine which simulated FECs were consistent with which experimental FECs. This analysis is illustrated in Fig. 5; again, Lc values were taken as matching if they agreed within ±2 nm. As a final filter, we eliminated trajectories in which a disordered structure rearranged during pulling to produce low-force ruptures as reflecting low-probability events (when involving two or fewer of the replicate trajectories for a given structure). After these steps, we were left with 429 simulated FECs containing unfolding transitions consistent with the experimental data.
Figure 5.
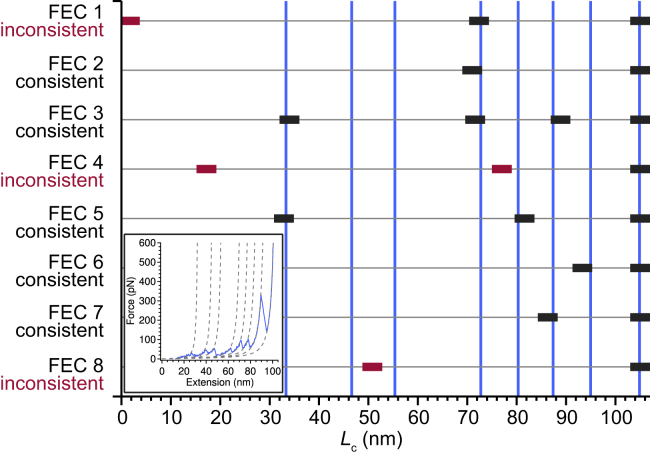
Illustration of the comparison of Lc values obtained from a simulated pulling curve to experimental results. A simulated FEC (inset) identifying eight Lc values (inset: dashed lines; main figure: vertical lines) was compared to Lc values obtained from a set of experimental FECs (solid bars) to determine if the simulation was consistent with experimental Lc values within experimental uncertainty (±2 nm). Values that match are shown in black, whereas those that do not are shown in dark red. To see this figure in color, go online.
The example in Fig. 5 illustrates many of the common features observed in the simulations. The last structural elements to unfold, rupturing at Lc ∼91–98 nm, were most commonly three-strand antiparallel β-sheets formed from ∼20 to 40 consecutive residues (Fig. S5), matching the commonly observed transitions at Lc ∼90–95 nm in the experimental FECs (Fig. 4 G) and agreeing with previous simulations of α-synuclein monomers (74). These three-strand sheets were not restricted to a limited part of the protein, instead forming across the N-terminal and NAC regions of each monomer domain, commonly in residues 20–100 (Fig. S6). Earlier unfolding transitions (Lc ∼80–90 nm) often involved the peeling of two or more strands from the edge of a larger sheet (typically with five or seven strands) or the rupture of a large sheet into two smaller ones (Fig. S5). Fewer commonalities could be discerned in the earliest unfolding transitions because of the large amount of structural variability. Not all regions of the protein sequence were equally likely to form stable secondary structures that led to a rupture (Fig. S6), with the C-terminus of each monomer domain and the linker between domains almost always remaining unstructured.
In many ways, the most interesting feature of the simulations is that they can identify the interfaces between the monomer units stabilizing the dimer structures, because these are likely to help detect the interactions that drive the initial step in aggregation and could be targeted to disrupt oligomerization. A concrete example is found in Fig. 6, in which the rip occurring at Lc = 35 nm breaks the interface between a sheet in each of the two monomer domains. Unfolding transitions involving the rupture of interfaces were identified for 23 structures, with discrete interface ruptures occurring in 266 trajectories. Most interfaces ruptured in a single event, but in eight structures, there were interfaces present at two distinct locations (Fig. S7) that ruptured in separate transitions. Cataloging the Lc values at which interface ruptures occurred (Fig. 7 A), we found that they tended to occur before much of the protein had unfolded, at relatively low Lc. Looking at the fraction of rips in the simulations that involved interface rupture (Fig. 7 A, inset), we found that the rips at the shortest Lc were almost exclusively interface ruptures, and a high proportion of rips (31%) in the second quartile of Lc also involved interfaces, but this fraction was smaller in the third quartile (4%) and no interface ruptures occurred in the fourth quartile. Most of the interface ruptures were consistent with Lc values of transitions observed experimentally (Fig. 7 B), with the possible exception of those at Lc ∼40 and 60 nm, at which few experimental transitions were observed.
Figure 6.
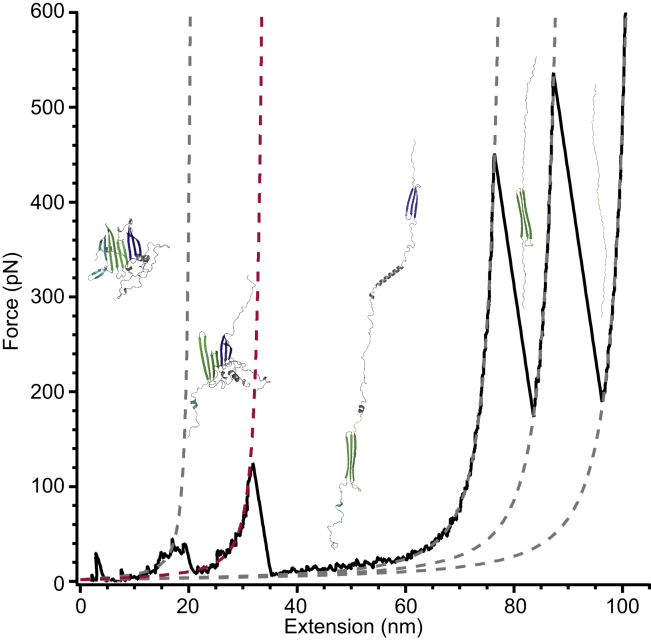
Simulated FEC illustrating the structural changes occurring at each rupture event. The protein structure changes from highly ordered at the start (left) to fully unfolded state at the end (right), unfolding via discrete rupture events at each rip in the FEC. WLC fits (dashed lines) quantified the unfolded protein Lc in each state (21, 35, 80, 91, 105 nm). The rupture of the interface between domain 1 (light green) and domain 2 (dark blue) is shown with a red dashed line at ∼30 nm. To see this figure in color, go online.
Figure 7.
Analysis of interface ruptures. (A) Comparing the Lc values at interface ruptures from simulations (dark red) to those for all discrete unfolding events in simulations (light blue) shows that interface ruptures occur at a wide range of lengths. The inset shows the probability that an unfolding event at a given Lc involved the rupture of an interface. (B) A comparison of the interface rupture events found in simulations (dark red) to all unfolding events found experimentally (black) finds significant overlap. (C) The probability that a given residue is involved in an interdomain interface is highest in the two terminal regions of each domain and lowest in the NAC region. The inset shows a schematic the of protein domains. To see this figure in color, go online.
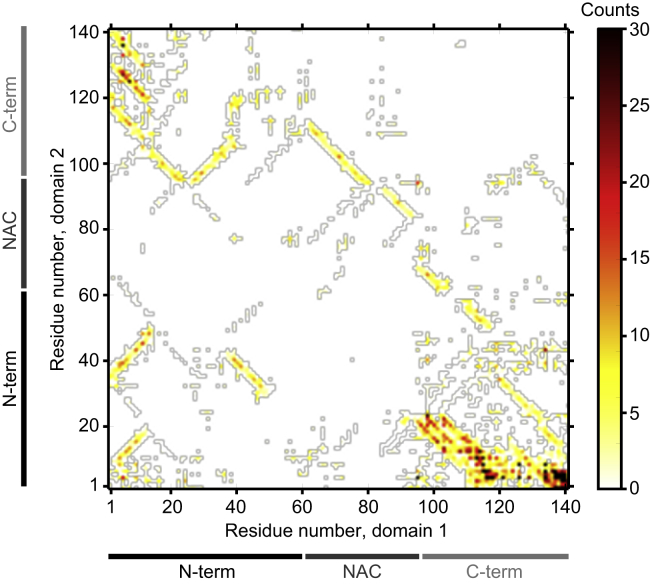
Turning to the regions of the protein involved in forming the interfaces that ruptured in pulling curves (Fig. 7 C), we found that there was a distinct position dependence. In both domains, residues near the N-terminus (∼1–20), the C-terminus (∼125–140), and the junction between the NAC region and acidic tail (∼90–120) were the most likely to form interfaces. Residue contact maps show that certain interface contacts were more prevalent than others (Fig. 8; full contact map shown in Fig. S8). Interface contacts often involved antiparallel β-sheets formed between the C-terminus of domain 1 and the N-terminus of domain 2, as expected from the close physical proximity due to the linker between the domains. The N-terminus of domain 1 (especially residues 1–12) could interact with the second domain in both the N-terminus (antiparallel β-sheets) and the C-terminal tail (parallel β-sheets), as illustrated in the gallery of interface structures (Fig. S7). The contact map also shows specific regions of the α-synuclein dimer that were devoid of interface interactions: the C-terminus of domain 2 did not contribute many interface contacts, and a negligible number were observed in the NAC region. Notably, most of the interfaces that we observed involved an edge-to-edge interaction of β-strands from the two domains; in contrast, only three of the interfaces involved stacking of β-sheets from each domain (Fig. S7).
Figure 8.
Contact map showing interactions between residues at interfaces. Contacts are concentrated in the termini of each domain. Most contacts involved antiparallel β-strands (lines of adjacent contacts with negative slope in the contact map), although a few involved parallel β-strands (lines with positive slope). To see this figure in color, go online.
Discussion
This work identifies possible structural models for the dimer (minimal oligomer) of an IDP by comparing the “fingerprint” for mechanical unfolding of structures found by computational simulation to the analogous fingerprint observed experimentally by SMFS, thereby deducing structures that are consistent with the observations. In contrast to previous attempts to match the observed fingerprints to structural models obtained experimentally (29), which found that over 30% of the curves could not be matched, by combining the experiments with computation, we were able to account for all of the fingerprints in 99% of experimental pulling curves. This result underlines the power of using simulations to extend the interpretation of single-molecule data, especially in the case of IDPs, for which insufficient structural information may be available from experimental methods.
The ability to distinguish between structures in this approach depends in part on the complexity of the mechanical fingerprint: the more complex the fingerprint is because of the presence of partially unfolded intermediates, the easier it is to distinguish. Our simulations of α-synuclein found fairly complex fingerprints reflecting multiple intermediates, qualitatively similar to the behavior seen in previous simulations of IDPs like aβ (35) and α-synuclein (74), suggesting that the approach may be generally viable for studying IDP oligomerization. However, discrimination between structural hypotheses will be most effective when the resolving power for detecting intermediates is similar in the experiment, highlighting one of the most important practical challenges of the approach: the discrepancy between the number of intermediates detected in experiments compared to simulations. Many more intermediates were typically observed per pulling curve in the simulations than in the experiments (Fig. S2), although the measurements were made with optical tweezers, which are one of the most sensitive methods for detecting intermediates (36) and did see much more complex behavior than studies using other methods like AFM (24, 28, 54, 57, 59, 76). A similar discrepancy was found between simulations and measurements of aβ dimers, for which simulations again found many more intermediates than did the experiments (34). This difference between experiment and simulation may reflect insufficient resolving power in the experiments (especially for intermediates that are particularly short-lived and hence might be overlooked), an underestimation in the simulation of the interactions driving cooperativity (such that the unfolding is less cooperative in simulations than in experiments), or an oversampling of highly ordered structures in the simulations as compared to experiments (possibly reflecting an overestimation of hydrogen-bond interactions), and it might be improved through approaches allowing more efficient sampling of IDP conformational space (77).
The dimer structures that we found to be mechanically stable were all β-rich. The notion that β-sheet structures form during the early stages of α-synuclein oligomerization is consistent with several strands of evidence, including circular dichroism spectroscopy studies (52), fluorescence correlation measurements of small oligomers (12), and MD simulations of dimers (78). Many of the β-structures we found involved a meander motif in which consecutive residues formed antiparallel strands linked via loops. Such antiparallel strand arrangements have been proposed in dimers of aβ and Sup35 (30) based on comparison of AFM measurements and MC simulations analogous to our work, in which pulling was used to examine the interfaces in segments of amyloid-β dimers and Sup35 dimers. However, previous studies of α-synuclein dimers were interpreted instead in terms of symmetric, parallel strand arrangements (57, 59), of which we found few examples consistent with optical trapping experiments.
The lack of any helical structures among those simulated FECs that match the experimental observations is very interesting because all of the helical structures unfolded via noncooperative (continuous) transitions or at sufficiently low force so as to be undetectable. Such behavior is clearly not intrinsic to helical structures: despite the fact that helical structures generally unfold at lower forces than β-rich structures and thus may be more difficult to resolve (79), many helical proteins do have well-defined rips denoting cooperative unfolding when pulled mechanically in both experimental (80, 81, 82) and computational (83, 84, 85) studies. However, noncooperative unfolding of helices has been observed recently in a combined experimental and computational study of single α-helix domains (85). To ensure that the noncooperative behavior was not an artifact of our simulation protocols, we tested that the same result held for the helical α-synuclein dimer structures under different simulation conditions, varying the probe stiffness (k) from 37 to 0.37 pN/nm and pulling speed (v) from 0.03 to 0.05 fm/step, finding no change. As a positive control, we also simulated the mechanical unfolding of a different helical protein, acyl-coenzyme A binding protein (ABP), for which both experimental and ratcheted MD studies show cooperative unfolding (86). Following the same procedure as applied to α-synuclein, we did indeed observe discrete rupture events in our simulations for ABP (Fig. S9), indicating that the lack of cooperative unfolding in helical α-synuclein is not simply an artifact but instead reflects unusually weak interactions within the transient helices formed in α-synuclein. The fact that the forces for unfolding ABP were noticeably lower than for the β-rich α-synuclein conformers is consistent with experimental observations that helical proteins unfold at lower forces (87), although it is also possible that the sensitivity of the potential to helical structures is lower than to β-structures.
Considering finally the interfaces between domains, previous work has reported somewhat conflicting results for small oligomers of α-synuclein. Measurements of α-synuclein dimers using AFM that identified ordered states at low pH (24, 60), in the presence of cations (56), and in the presence of other molecules like spermidine (57) were interpreted in terms of an interface consisting of parallel strands arranged side-on to form a β-sheet. The interface was suggested to be symmetric, involving the same residues in each monomer domain, with regions in the C-terminal end of the NAC region and N-terminal end of the acidic region acting as a mechanical clamp (57). In contrast, MD simulations of α-synuclein dimers found that interfaces were formed preferentially not by the NAC region itself but rather by the part of the N-terminal domain adjacent to the NAC region (78), although these simulations agreed with our findings that interfaces were stabilized by edge-to-edge interactions. Our results are more consistent with the latter picture, finding few interface interactions in the NAC region and most in the N-terminal or the C-terminal regions (Fig. 8), suggesting that electrostatic interactions between charged residues in the latter are more important than interactions between hydrophobic residues of the NAC region. Notably, we do not find that the same sites on the two domains always interact, as shown by the lack of contacts along the diagonal in the contact map (Fig. 8). Instead, we find that a wide variety of possible interfaces (Fig. S6) is needed to account for the diversity observed in the experimental FECs, suggesting that a symmetric model may be too simplistic for explaining such rich behavior. The interfaces identified in our work also differ from those observed in fibrils by NMR: one model found sheets interacting face-on rather than edge-on (47), whereas another found edge-on interactions form intermolecular sheets but with a parallel arrangement of strands (48).
Such differences with previous work are not surprising given the differences between the experimental conditions. For example, the structures of oligomers often change as the oligomers mature into fibrils (9, 12). The influence of the C- to N-terminal linker in our work should also be taken into account: the linker constrains the dimer conformations that are explored, likely biasing the distribution of structures in favor of interactions between the C-terminus of domain 1 and N-terminus of domain 2 and against C-to-C and N-to-N interactions. Evidence of such a bias can be seen in the contact map (Fig. 8), which shows a higher likelihood of contacts between the C-terminus of domain 1 and N-terminus of domain 2 than the converse, and in the interface map (Fig. 7 C), which shows a higher probability of involvement of residues in the domain 1 C-terminus and domain 2 N-terminus than the reverse. However, this bias does not prevent the formation of long-range interactions, even those that it disfavors such as N-to-N interactions or interactions between the domain 1 N-terminus and domain 2 C-terminus, and all of the conformations observed are clearly possible outcomes for dimers composed of free monomers. Moreover, previous studies comparing tandem α-synuclein dimers to unlinked monomers found that whereas the linkage accelerated the nucleation of aggregates, the outcomes in terms of fibril morphology and formation of membrane-permeabilizing pores were similar (88, 89), suggesting that the bias introduced by the linker has relatively minor effects overall. The structures found through our analysis thus provide valuable information about regions of the protein that are important for dimerization, even if they do not sample the full conformational space of dimers.
Conclusion
Determining the structure of IDPs is inherently difficult, particularly as they interact to form transient oligomers that may contribute to disease pathology. To address this challenge, we have integrated atomistic MC simulations with high-resolution SMFS measurements of an IDP dimer, comparing their mechanical fingerprints for unfolding α-synuclein dimers to identify structures that are consistent with the experiments. This comparison found a diverse set of β-rich structures with meander motifs that interacted primarily edge-on to stabilize the dimer. By identifying a set of interfaces, we have established a database of key protein contacts to target for disruption in future drug-design work, with the goal of inhibiting the earliest oligomerization step in the α-synuclein aggregation pathway. The same strategy of combining high-resolution single-molecule experiments and computational simulations may also prove fruitful for identifying potential druggable targets for inhibiting the oligomerization of other disease-related IDPs.
Author Contributions
C.D.M.C., J.A.T., and M.T.W. designed research. C.D.M.C., M.A.H., and J.P. performed research and analyzed data. C.D.M.C., J.P., J.A.T., and M.T.W. wrote the manuscript.
Acknowledgments
We thank Compute Canada for access to its computational resources.
This work was supported by the Alberta Prion Research Institute, Alberta Innovates Health Solutions, Alberta Innovates Technology Futures, and the National Research Council Canada.
Editor: Wendy Shaw.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.08.013.
Contributor Information
Jack A. Tuszynski, Email: jackt@ualberta.ca.
Michael T. Woodside, Email: michael.woodside@ualberta.ca.
Supporting Material
References
- 1.Wolynes P.G., Onuchic J.N., Thirumalai D. Navigating the folding routes. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 2.Stefani M., Dobson C.M. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. (Berl.) 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 3.Lu Q., Lu H.P., Wang J. Exploring the mechanism of flexible biomolecular recognition with single molecule dynamics. Phys. Rev. Lett. 2007;98:128105. doi: 10.1103/PhysRevLett.98.128105. [DOI] [PubMed] [Google Scholar]
- 4.Keskin O., Gursoy A., Nussinov R. Principles of protein-protein interactions: what are the preferred ways for proteins to interact? Chem. Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- 5.Brucale M., Schuler B., Samorì B. Single-molecule studies of intrinsically disordered proteins. Chem. Rev. 2014;114:3281–3317. doi: 10.1021/cr400297g. [DOI] [PubMed] [Google Scholar]
- 6.Vendruscolo M. Determination of conformationally heterogeneous states of proteins. Curr. Opin. Struct. Biol. 2007;17:15–20. doi: 10.1016/j.sbi.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 8.Wright P.E., Dyson H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knowles T.P., Vendruscolo M., Dobson C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 10.Lashuel H.A., Lansbury P.T., Jr. Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q. Rev. Biophys. 2006;39:167–201. doi: 10.1017/S0033583506004422. [DOI] [PubMed] [Google Scholar]
- 11.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 12.Cremades N., Cohen S.I., Klenerman D. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell. 2012;149:1048–1059. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S.W., Drakulic S., Cremades N. Structural characterization of toxic oligomers that are kinetically trapped during α-synuclein fibril formation. Proc. Natl. Acad. Sci. USA. 2015;112:E1994–E2003. doi: 10.1073/pnas.1421204112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenberg D.S., Sawaya M.R. Structural studies of amyloid proteins at the molecular level. Annu. Rev. Biochem. 2017;86:69–95. doi: 10.1146/annurev-biochem-061516-045104. [DOI] [PubMed] [Google Scholar]
- 15.Bhowmick A., Brookes D.H., Head-Gordon T. Finding our way in the dark proteome. J. Am. Chem. Soc. 2016;138:9730–9742. doi: 10.1021/jacs.6b06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uversky V.N., Dunker A.K. Understanding protein non-folding. Biochim. Biophys. Acta. Proteins Proteomics. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mittag T., Forman-Kay J.D. Atomic-level characterization of disordered protein ensembles. Curr. Opin. Struct. Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Zuckerman D.M. Equilibrium sampling in biomolecular simulations. Annu. Rev. Biophys. 2011;40:41–62. doi: 10.1146/annurev-biophys-042910-155255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orozco M., Noy A., Pérez A. Recent advances in the study of nucleic acid flexibility by molecular dynamics. Curr. Opin. Struct. Biol. 2008;18:185–193. doi: 10.1016/j.sbi.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Pan A.C., Weinreich T.M., Shaw D.E. Demonstrating an order-of-magnitude sampling enhancement in molecular dynamics simulations of complex protein systems. J. Chem. Theory Comput. 2016;12:1360–1367. doi: 10.1021/acs.jctc.5b00913. [DOI] [PubMed] [Google Scholar]
- 21.Piana S., Lindorff-Larsen K., Shaw D.E. Atomic-level description of ubiquitin folding. Proc. Natl. Acad. Sci. USA. 2013;110:5915–5920. doi: 10.1073/pnas.1218321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann A., Neupane K., Woodside M.T. Single-molecule assays for investigating protein misfolding and aggregation. Phys. Chem. Chem. Phys. 2013;15:7934–7948. doi: 10.1039/c3cp44564j. [DOI] [PubMed] [Google Scholar]
- 23.Orte A., Birkett N.R., Klenerman D. Direct characterization of amyloidogenic oligomers by single-molecule fluorescence. Proc. Natl. Acad. Sci. USA. 2008;105:14424–14429. doi: 10.1073/pnas.0803086105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu J., Malkova S., Lyubchenko Y.L. α-Synuclein misfolding: single molecule AFM force spectroscopy study. J. Mol. Biol. 2008;384:992–1001. doi: 10.1016/j.jmb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Nath S., Meuvis J., Engelborghs Y. Early aggregation steps in α-synuclein as measured by FCS and FRET: evidence for a contagious conformational change. Biophys. J. 2010;98:1302–1311. doi: 10.1016/j.bpj.2009.12.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim B.H., Palermo N.Y., Lyubchenko Y.L. Single-molecule atomic force microscopy force spectroscopy study of Aβ-40 interactions. Biochemistry. 2011;50:5154–5162. doi: 10.1021/bi200147a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neupane K., Yu H., Woodside M.T. Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 2011;39:7677–7687. doi: 10.1093/nar/gkr305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hervás R., Oroz J., Carrión-Vázquez M. Common features at the start of the neurodegeneration cascade. PLoS Biol. 2012;10:e1001335. doi: 10.1371/journal.pbio.1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neupane K., Solanki A., Woodside M.T. Diverse metastable structures formed by small oligomers of α-synuclein probed by force spectroscopy. PLoS One. 2014;9:e86495. doi: 10.1371/journal.pone.0086495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Lyubchenko Y.L. The structure of misfolded amyloidogenic dimers: computational analysis of force spectroscopy data. Biophys. J. 2014;107:2903–2910. doi: 10.1016/j.bpj.2014.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lv Z., Krasnoslobodtsev A.V., Lyubchenko Y.L. Direct detection of α-synuclein dimerization dynamics: single-molecule fluorescence analysis. Biophys. J. 2015;108:2038–2047. doi: 10.1016/j.bpj.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nath A., Sammalkorpi M., Rhoades E. The conformational ensembles of α-synuclein and tau: combining single-molecule FRET and simulations. Biophys. J. 2012;103:1940–1949. doi: 10.1016/j.bpj.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metskas L.A., Rhoades E. Conformation and dynamics of the troponin I C-terminal domain: combining single-molecule and computational approaches for a disordered protein region. J. Am. Chem. Soc. 2015;137:11962–11969. doi: 10.1021/jacs.5b04471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lovas S., Zhang Y., Lyubchenko Y.L. Molecular mechanism of misfolding and aggregation of Aβ(13-23) J. Phys. Chem. B. 2013;117:6175–6186. doi: 10.1021/jp402938p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Hashemi M., Lyubchenko Y.L. Self-assembly of the full-length amyloid Aβ42 protein in dimers. Nanoscale. 2016;8:18928–18937. doi: 10.1039/c6nr06850b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie D.B., Woodside M.T. Probing the structural dynamics of proteins and nucleic acids with optical tweezers. Curr. Opin. Struct. Biol. 2015;34:43–51. doi: 10.1016/j.sbi.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solanki A., Neupane K., Woodside M.T. Single-molecule force spectroscopy of rapidly fluctuating, marginally stable structures in the intrinsically disordered protein α-synuclein. Phys. Rev. Lett. 2014;112:158103. doi: 10.1103/PhysRevLett.112.158103. [DOI] [PubMed] [Google Scholar]
- 38.Spillantini M.G., Schmidt M.L., Goedert M. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 39.Breydo L., Wu J.W., Uversky V.N. α-Synuclein misfolding and Parkinson’s disease. Biochim. Biophys. Acta. 2012;1822:261–285. doi: 10.1016/j.bbadis.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Weinreb P.H., Zhen W., Lansbury P.T., Jr. NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 41.Uversky V.N. α-Synuclein misfolding and neurodegenerative diseases. Curr. Protein Pept. Sci. 2008;9:507–540. doi: 10.2174/138920308785915218. [DOI] [PubMed] [Google Scholar]
- 42.Dedmon M.M., Lindorff-Larsen K., Dobson C.M. Mapping long-range interactions in α-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J. Am. Chem. Soc. 2005;127:476–477. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 43.Bertoncini C.W., Jung Y.S., Zweckstetter M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured α-synuclein. Proc. Natl. Acad. Sci. USA. 2005;102:1430–1435. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eliezer D., Kutluay E., Browne G. Conformational properties of α-synuclein in its free and lipid-associated states. J. Mol. Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 45.Wang W., Perovic I., Hoang Q.Q. A soluble α-synuclein construct forms a dynamic tetramer. Proc. Natl. Acad. Sci. USA. 2011;108:17797–17802. doi: 10.1073/pnas.1113260108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uversky V.N., Li J., Fink A.L. Evidence for a partially folded intermediate in α-synuclein fibril formation. J. Biol. Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- 47.Vilar M., Chou H.T., Riek R. The fold of α-synuclein fibrils. Proc. Natl. Acad. Sci. USA. 2008;105:8637–8642. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuttle M.D., Comellas G., Rienstra C.M. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trexler A.J., Rhoades E. Single molecule characterization of α-synuclein in aggregation-prone states. Biophys. J. 2010;99:3048–3055. doi: 10.1016/j.bpj.2010.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferreon A.C., Moran C.R., Deniz A.A. Alteration of the α-synuclein folding landscape by a mutation related to Parkinson’s disease. Angew. Chem. Int.Engl. 2010;49:3469–3472. doi: 10.1002/anie.201000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferreon A.C.M., Gambin Y., Deniz A.A. Interplay of α-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. USA. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McAllister C., Karymov M.A., Lyubchenko Y.L. Protein interactions and misfolding analyzed by AFM force spectroscopy. J. Mol. Biol. 2005;354:1028–1042. doi: 10.1016/j.jmb.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 53.Ray C., Akhremitchev B.B. Conformational heterogeneity of surface-grafted amyloidogenic fragments of α-synuclein dimers detected by atomic force microscopy. J. Am. Chem. Soc. 2005;127:14739–14744. doi: 10.1021/ja052932e. [DOI] [PubMed] [Google Scholar]
- 54.Sandal M., Valle F., Samorì B. Conformational equilibria in monomeric α-synuclein at the single-molecule level. PLoS Biol. 2008;6:e6. doi: 10.1371/journal.pbio.0060006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu J., Lyubchenko Y.L. Early stages for Parkinson’s development: α-synuclein misfolding and aggregation. J. Neuroimmune Pharmacol. 2009;4:10–16. doi: 10.1007/s11481-008-9115-5. [DOI] [PubMed] [Google Scholar]
- 56.Yu J., Warnke J., Lyubchenko Y.L. Nanoprobing of α-synuclein misfolding and aggregation with atomic force microscopy. Nanomedicine (Lond.) 2011;7:146–152. doi: 10.1016/j.nano.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Krasnoslobodtsev A.V., Peng J., Lyubchenko Y.L. Effect of spermidine on misfolding and interactions of α-synuclein. PLoS One. 2012;7:e38099. doi: 10.1371/journal.pone.0038099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pivato M., De Franceschi G., Bubacco L. Covalent α-synuclein dimers: chemico-physical and aggregation properties. PLoS One. 2012;7:e50027. doi: 10.1371/journal.pone.0050027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krasnoslobodtsev A.V., Volkov I.L., Lyubchenko Y.L. α-Synuclein misfolding assessed with single molecule AFM force spectroscopy: effect of pathogenic mutations. Biochemistry. 2013;52:7377–7386. doi: 10.1021/bi401037z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lv Z., Krasnoslobodtsev A.V., Lyubchenko Y.L. Effect of acidic pH on the stability of α-synuclein dimers. Biopolymers. 2016;105:715–724. doi: 10.1002/bip.22874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang M.D., Yin H., Block S.M. Stretching DNA with optical tweezers. Biophys. J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Irbäck A., Mohanty S. PROFASI: a Monte Carlo simulation package for protein folding and aggregation. J. Comput. Chem. 2006;27:1548–1555. doi: 10.1002/jcc.20452. [DOI] [PubMed] [Google Scholar]
- 63.Healey M.A., Woodside M.T., Tuszynski J.A. Phase transitions and structure analysis in wild-type, A30P, E46K, and A53T mutants of α-synuclein. Eur. Biophys. J. 2016;45:355–364. doi: 10.1007/s00249-015-1103-0. [DOI] [PubMed] [Google Scholar]
- 64.Li D.W., Mohanty S., Huo S. Formation and growth of oligomers: a Monte Carlo study of an amyloid tau fragment. PLoS Comput. Biol. 2008;4:e1000238. doi: 10.1371/journal.pcbi.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitternacht S., Staneva I., Irbäck A. Monte Carlo study of the formation and conformational properties of dimers of Aβ42 variants. J. Mol. Biol. 2011;410:357–367. doi: 10.1016/j.jmb.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 66.Irbäck A., Mitternacht S., Mohanty S. An effective all-atom potential for proteins. PMC Biophys. 2009;2:2. doi: 10.1186/1757-5036-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petrlova J., Bhattacherjee A., Irbäck A. Conformational and aggregation properties of the 1-93 fragment of apolipoprotein A-I. Protein Sci. 2014;23:1559–1571. doi: 10.1002/pro.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 69.Case D.A., Babin V., Cerutti D.S. University of California; San Francisco: 2014. AMBER 14. [Google Scholar]
- 70.Irbäck A., Samuelsson B., Wallin S. Thermodynamics of α- and β-structure formation in proteins. Biophys. J. 2003;85:1466–1473. doi: 10.1016/S0006-3495(03)74579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Irbäck A., Mohanty S. Folding thermodynamics of peptides. Biophys. J. 2005;88:1560–1569. doi: 10.1529/biophysj.104.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 73.Mitternacht S., Luccioli S., Irbäck A. Changing the mechanical unfolding pathway of FnIII10 by tuning the pulling strength. Biophys. J. 2009;96:429–441. doi: 10.1016/j.bpj.2008.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jónsson S.Æ., Mitternacht S., Irbäck A. Mechanical resistance in unstructured proteins. Biophys. J. 2013;104:2725–2732. doi: 10.1016/j.bpj.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vitalis A., Pappu R.V. Methods for Monte Carlo simulations of biomacromolecules. In: Wheeler R.A., editor. Volume 5. Elsevier Science Bv; 2009. pp. 49–76. (Annual Reports in Computational Chemistry). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brucale M., Sandal M., Samorì B. Pathogenic mutations shift the equilibria of α-synuclein single molecules towards structured conformers. ChemBioChem. 2009;10:176–183. doi: 10.1002/cbic.200800581. [DOI] [PubMed] [Google Scholar]
- 77.Zhao L., Suarez I.P., Wang J. The key role of electrostatic interactions in the induced folding in RNA recognition by DCL1-A. Phys. Chem. Chem. Phys. 2018;20:9376–9388. doi: 10.1039/C7CP07889G. [DOI] [PubMed] [Google Scholar]
- 78.Zhang T., Tian Y., Zhang J. Molecular dynamics study to investigate the dimeric structure of the full-length α-synuclein in aqueous solution. J. Chem. Inf. Model. 2017;57:2281–2293. doi: 10.1021/acs.jcim.7b00210. [DOI] [PubMed] [Google Scholar]
- 79.Oberhauser A.F., Carrión-Vázquez M. Mechanical biochemistry of proteins one molecule at a time. J. Biol. Chem. 2008;283:6617–6621. doi: 10.1074/jbc.R700050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stigler J., Ziegler F., Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 81.Yu H., Gupta A.N., Woodside M.T. Energy landscape analysis of native folding of the prion protein yields the diffusion constant, transition path time, and rates. Proc. Natl. Acad. Sci. USA. 2012;109:14452–14457. doi: 10.1073/pnas.1206190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gao Y., Zorman S., Zhang Y. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brockwell D.J., Beddard G.S., Radford S.E. Mechanically unfolding the small, topologically simple protein L. Biophys. J. 2005;89:506–519. doi: 10.1529/biophysj.105.061465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharma D., Perisic O., Li H. Single-molecule force spectroscopy reveals a mechanically stable protein fold and the rational tuning of its mechanical stability. Proc. Natl. Acad. Sci. USA. 2007;104:9278–9283. doi: 10.1073/pnas.0700351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wolny M., Batchelor M., Peckham M. Stable single α-helices are constant force springs in proteins. J. Biol. Chem. 2014;289:27825–27835. doi: 10.1074/jbc.M114.585679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Heidarsson P.O., Valpapuram I., Cecconi C. A highly compliant protein native state with a spontaneous-like mechanical unfolding pathway. J. Am. Chem. Soc. 2012;134:17068–17075. doi: 10.1021/ja305862m. [DOI] [PubMed] [Google Scholar]
- 87.Hoffmann T., Dougan L. Single molecule force spectroscopy using polyproteins. Chem. Soc. Rev. 2012;41:4781–4796. doi: 10.1039/c2cs35033e. [DOI] [PubMed] [Google Scholar]
- 88.Li X., Dong C., Woodside M.T. Early stages of aggregation of engineered α-synuclein monomers and oligomers in solution. Sci. Rep. 2019;9:1734. doi: 10.1038/s41598-018-37584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dong C., Hoffmann M., Woodside M.T. Structural characteristics and membrane interactions of tandem α-synuclein oligomers. Sci. Rep. 2018;8:6755. doi: 10.1038/s41598-018-25133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.